Abstract
To determine whether the diversity of pyrene-degrading bacteria in an aged polycyclic aromatic hydrocarbon-contaminated soil is affected by the addition of inorganic nutrients or by slurrying the soil, various incubation conditions (all including phosphate buffer) were examined by mineralization studies and stable-isotope probing (SIP). The addition of nitrogen to either continuously mixed slurry or static field-wet soil incubations increased the rate and extent of mineralization of [14C]pyrene, with the most rapid mineralization observed in slurried, nitrogen-amended soil. Microcosms of slurry and static field-wet soil amended with nitrogen were also examined by SIP with [U-13C]pyrene. Recovered 13C-enriched deoxyribonucleic acid (DNA) was analyzed by denaturing-gradient gel electrophoresis (DGGE) and 16S ribosomal ribonucleic acid (rRNA) gene clone libraries. DGGE profiles of 13C-enriched DNA fractions from both incubation conditions were similar, suggesting that pyrene-degrading bacterial community diversity may be independent of treatment method. The vast majority (67 of 71) of the partial sequences recovered from clone libraries were greater than or equal to 97% similar to one another, 98% similar to sequences of pyrene-degrading bacteria previously detected by SIP with pyrene in different soil, and only 89% similar to the closest cultivated genus. All of the sequences recovered from the field-wet incubation and most of the sequences recovered from the slurry incubation were in this clade. Of the four sequences from slurry incubations not within this clade, three possessed greater than 99% similarity to the 16S rRNA gene sequences of phylogenetically dissimilar Caulobacter spp.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polycyclic aromatic hydrocarbons (PAH) are regulated by the US Environmental Protection Agency (EPA) as hazardous substances, and many of the higher-molecular-weight PAH (four to six rings) are known or suspected carcinogens [2]. These compounds are ubiquitous environmental pollutants that generally exist as complex mixtures and result from the incomplete combustion of both natural and anthropogenic organic materials. Failure to remediate PAH contamination in soils can result in PAH accumulation in plants, groundwater contamination, and overall increased human and environmental health risk [2, 32].
A variety of methods are currently employed for the remediation of PAH-contaminated soil. Natural attenuation is one such method, whereby naturally occurring physical, chemical, and biological processes are monitored but otherwise allowed to govern contaminant removal without human intervention [49]. Biostimulation encourages the growth of indigenous microorganisms acclimated for the removal of specific contaminants through the addition of nutrients or other amendments to increase rates of contaminant degradation [7, 19, 41, 50]. For example, ex situ biostimulation in slurry (S)-phase bioreactors has proven effective in PAH-contaminated soil remediation [18, 26, 29, 40, 44].
Previous studies have indicated that the treatment method and/or nutrient status can be responsible for changes in microbial community dynamics or the rate or extent of substrate removal. For example, Viñas et al. [50] found that the change in bacterial community structure in creosote-contaminated soil was attributed to both factors. However, PAH-degrading populations were more abundant in microcosms without nutrient addition than in those to which nutrients had been added, and the removal of most PAHs after 200 days in microcosms with nutrients was not significantly different from those without nutrients [50]. In addition to the treatment method and nutrient status, soil type, moisture content, and PAH exposure history can also affect microbial community dynamics [7, 20, 50].
Many fungal and bacterial species have been studied for their PAH-degrading ability [9]. Most microorganisms that have been isolated and characterized as using pyrene as the sole carbon and/or energy source via traditional techniques are among the Actinobacteria [12, 17, 22, 24, 27, 38, 51], but some pyrene-degrading γ-Proteobacteria have also been isolated [15, 22]. However, the overall significance of these organisms in the systems from which they were isolated has not been evaluated. The application of stable-isotope probing (SIP) to bioremediation studies has greatly facilitated efforts to identify microorganisms capable of degrading particular substrates without isolation, and SIP has been applied in several recent cases to identify PAH-degrading bacteria in field-contaminated soils. SIP has been used to identify naphthalene degraders in coal tar-contaminated sediment in situ [21] and to identify organisms capable of degrading naphthalene, phenanthrene, or pyrene in a bioreactor treating soil from a former manufactured-gas plant [44, 45].
In the only previous SIP study targeting pyrene-degrading microorganisms, we identified pyrene degraders in an S-phase bioreactor treating soil from a former manufactured-gas plant site in which the soil was amended with nitrogen and phosphorus [45]. The primary pyrene degraders identified in that study were uncultivated β- and γ-Proteobacteria. In the present study, we investigated the effects of inorganic nutrient addition and physical treatment on pyrene mineralization and the pyrene-degrading community in PAH-contaminated soil from a former wood-preserving facility. Bacteria responsible for the biotransformation and biodegradation of pyrene in the soil amended with inorganic nutrients were identified using SIP and found to be in close phylogenetic association with other uncultivated pyrene-degrading bacteria identified by SIP in our earlier work on the S-phase bioreactor [45].
Methods
Chemicals
Pyrene (highest purity available) and [4, 5, 9, 10-14C]pyrene (>98% purity) were obtained from Sigma Chemical (St. Louis, MO). [4, 5, 9, 10-14C]Pyrene was diluted in methanol and stored at −20°C until used. [U-13C]Pyrene was synthesized as described elsewhere [45].
Soil Samples
A sample of creosote-contaminated soil was obtained from the Reilly Tar and Chemical Superfund site in St. Louis Park, MN, which is the site of a former coal tar-distilling and wood-preserving facility. Properties of this soil are summarized in Table 1. Soil class, texture, pH, and percent organic matter were determined by the University of Wisconsin—Madison Soil and Plant Analysis Laboratory (Madison, WI) in accordance with standard procedures [6, 43, 52]. PAHs were analyzed in triplicate by Eno River Laboratories (Durham, NC) using EPA method 8270. The soil sample was stored in the dark at 4°C until needed. Subsamples were sieved (2mm) and likewise stored for experimental use. Soil from this site is known to contain a pyrene-degrading microbial community [16].
Field Capacity Approximation
The field or water-holding capacity (WHC) of the contaminated soil was measured by a modified container capacity procedure [8]. Each container consisted of a 15-cm-diameter water-saturated filter paper (Whatman no. 6, 0.2μm pore size) shaped to fit a glass funnel held upright by a 250-mL glass beaker. Three 5-g aliquots of soil were dried at 105°C for 24 h. Each dried soil aliquot was added to a separate container, saturated with water, covered loosely with aluminum foil, and left to drain for 2 h at room temperature. WHC was determined from the difference in mass between water-saturated filter paper containing drained soil and water-saturated filter paper without soil.
Addition of Pyrene to Soil
Pyrene was added to air-dried soil as described by others [30, 39]. Briefly, for the mineralization experiment a target amount of 20,000 dpm, [4, 5, 9, 10-14C]pyrene in methanol per gram soil was dispensed into a clean 250-mL glass beaker and placed in a fume hood to completely volatilize the solvent. Subsequently, an acetone solution containing 17 mg unlabeled pyrene per milliliter and 20% of the air-dried soil were added to that same beaker. The contents were mixed manually for 1min using a clean glass rod. The beaker was covered loosely with aluminum foil and placed in a fume hood for 24 h for solvent evaporation. The remaining 80% of the air-dried soil was subsequently mixed into the beaker in 20% increments for 30 s per increment. The final concentration of added pyrene in the soil was 1 mg/g. An additional quantity of soil was prepared in the same manner with only unlabeled pyrene to prepare control microcosms.
Pyrene Mineralization and SIP Microcosms
Mineralization was tested under four conditions, and the results were used to determine the endpoint for the SIP experiment. Soil was either slurried in 3 mL of buffer (10 mM phosphate, pH7.5) or field-wet (FW). FW samples were wetted with 75 mM phosphate buffer (pH7.5) to a moisture content of 70% WHC. In addition to samples amended with buffer only, additional sets of S and FW samples were amended with buffer and nitrogen (500 μg N as NH4Cl/g soil). Each of the four experimental conditions was prepared in triplicate, with 1 g of soil containing radiolabeled pyrene (20,000 dpm, prepared as described above) in 40-mL glass EPA vials. Inside each vial was a 12 mm (diameter) × 75-mm glass culture tube containing a 6.5-cm2 filter paper saturated with 2N KOH as a CO2 trap [3], and the vials were sealed with Teflon-coated silicon disk-lined screw caps. Duplicate parallel microcosms for each condition were prepared with 1 g of soil containing only unlabeled pyrene. Controls consisted of duplicate S and FW incubations without nitrogen amendment or pyrene addition and killed controls (one S and one FW) containing soil spiked with radiolabeled pyrene. Killed controls were achieved with the addition of phosphoric acid to pH < 2. S microcosms were agitated at 250 rpm, whereas FW microcosms remained static. All microcosms were incubated in the dark at room temperature for 49 days. Trapped 14CO2 was measured at selected intervals by placing each trap in 10 mL Ultima Gold XR liquid scintillation cocktail (PerkinElmer Life and Analytical Sciences, Boston, MA) and counting in a Packard TriCarb 1900TR Liquid Scintillation Counter (Packard Instruments, Meriden, CT). New traps were added to the incubation vials after each sampling event.
SIP was performed only with S and FW incubations in which nitrogen was added. Microcosms for SIP were prepared and incubated as described above except that no radiolabeled pyrene or CO2 trap was used. Experimental microcosms were prepared in duplicate, and only [U-13C]pyrene was added to experimental vessels (1 mg/g soil). Duplicate parallel microcosms and killed controls for each condition were also prepared with unlabeled pyrene. Based on the results of the mineralization experiments, all SIP microcosms were terminated at day 28.
DNA Extraction and 13C-enriched DNA Isolation
Total community deoxyribonucleic acid (DNA) was extracted on day 49 from microcosms containing unlabeled pyrene in the mineralization experiment and on day 28 from all microcosms in the SIP experiment, using an Ultraclean Soil DNA Kit (MoBio Laboratories, Carlsbad, CA) per the manufacturer’s instructions, and stored in Tris–EDTA (TE) buffer (10 mM Tris HCl, 1mM EDTA; pH8.0). DNA was quantified using a NanoDrop ND-3300 fluorospectrometer (NanoDrop Technologies, Wilmington, DE) with a Quant-iT PicoGreen dsDNA Quantitation Kit (Molecular Probes, Eugene, OR). Unlabeled and 13C-enriched DNA were separated by cesium chloride density-gradient ultracentrifugation, fractionated, and purified as previously described [44]. Purified DNA was resuspended in 75 μL of TE buffer per fraction. Before ultracentrifugation, unlabeled genomic DNA from E. coli K-12 was added to each centrifuge tube as an indicator of separation efficiency [44]. Polymerase chain reaction (PCR) screening of DNA extracted from the soil with a primer set specific to the 16S ribosomal ribonucleic acid (rRNA) gene of E. coli [42] confirmed that E. coli was not present in the soil. 13C-enriched DNA was screened by PCR for archaeal 16S rRNA genes and fungal 18S rRNA genes using domain-specific primers 344fGC [37] and 806r [47] and 817f and 1196r [5], respectively.
Bacterial Community Profiles
PCR for denaturing-gradient gel electrophoresis (DGGE) was performed with primers P63f and P518r [14]. The resulting products were loaded onto a 6% polyacrylamide gel with a 30 to 60% urea-formamide denaturing gradient. The gel was run on a Bio-Rad DCode system at 60°C and 60V for 16h and post-stained with ethidium bromide.
Analysis of 13C-enriched DNA
Clone libraries of 16S rRNA genes were constructed from fractions containing 13C-labeled DNA using primers 8f [13] and 1492r [25]. The PCR program consisted of 25 cycles of 1 min at 94°C, 1 min at 50°C, and 3 min at 72°C. PCR products were cloned into an Invitrogen TA cloning kit (Carlsbad, CA) per the manufacturer’s instructions. Inserts were partially sequenced with primer 8f by SeqWright DNA Technology Services (Houston, TX). A neighbor-joining phylogenetic tree was constructed from the resulting 71 partial 16S rRNA gene sequences (34 S and 37 FW) using ClustalX [48] and bootstrapped 1,000 times without considering gaps [31]. Chimeras were resolved using the CHIMERA_CHECK tool within Ribosomal Database Project II Release 8.1 [11]. All sequences were compared to public sequence databases using BLASTN [4] to identify related sequences and confirm chimeras.
Accession Numbers
The partial 16S rRNA gene sequences recovered from this work were submitted to GenBank with accession numbers DQ906936–DQ907006.
Results
Rate and Extent of Pyrene Mineralization
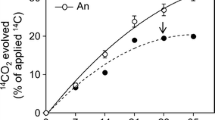
Mineralization of [4, 5, 9, 10-14C]pyrene was monitored for each incubation condition (S, S with nitrogen [S + N], static FW [FW], and static FW with nitrogen [FW + N]) over a 49-day incubation period (Fig. 1). In the first 3 days, no significant amounts of pyrene were mineralized in any of the incubation conditions. After 7 days, the S incubation amended with nitrogen showed significant mineralization, but the S incubations without nitrogen and both FW conditions (with and without nitrogen) did not. Excluding the lag in mineralization, both incubations with nitrogen (S and FW) had higher mineralization rates than incubations without nitrogen. By day 28, the mineralization rate in the S + N condition began to decline. The mineralization experiment was allowed to proceed beyond 28 days to determine whether the extent of mineralization in the other microcosms would approach that observed in the S + N condition, although this was not the case after 21 days of additional incubation.
A significant increase (two-tailed t test, p < 0.001) in the extent of mineralization after 49 days was seen in each nitrogen-amended microcosm compared to those without nitrogen amendment. Pyrene was mineralized to a significantly greater extent (p < 0.001) in S microcosms compared to FW microcosms, whether nitrogen was added or not, suggesting that the extent of mineralization over this period was influenced by the nature of the incubation and the availability of inorganic nutrients.
Inorganic Nutrient Effects on Bacterial Community
DNA extracts from parallel microcosms amended with unlabeled pyrene and nitrogen yielded more total-community DNA per gram of dry soil than microcosms without nitrogen amendment. S incubations with nitrogen contained 7.0 ± 0.22 μg of extracted DNA compared to 2.0 ± 0.31 μg DNA for S incubations without amended nitrogen. Similarly, under FW conditions, an average of 7.0 ± 0.22 and 2.0 ± 0.26 μg of DNA were extracted from nitrogen-amended and unamended soils, respectively. PCR-amplified bacterial rRNA genes from the four microcosm conditions were compared with the respective controls by DGGE (Fig. 2). The profiles for the FW and S communities for the incubations in which pyrene was added in the absence of nitrogen (lanes 4–5 and 9–10, respectively) were similar to those for the incubations in which pyrene was not added (lanes 1 and 6, respectively). Profiles from the incubations in which pyrene and nitrogen were added (lanes 2–3 and 7–8) had bands of greater intensity than in the profiles from the other conditions, particularly bands lower on the gel (the bands low on the gel are nearly absent in lanes 9 and 10 in Fig. 2, but all bands in those lanes are relatively faint).
Negative image of DGGE profile of microbial communities from unlabelled pyrene enrichments after 49 days of incubation. Lane designations are as follows: 1, field-wet without nitrogen or pyrene; 2–3, field-wet with nitrogen and pyrene; 4–5, field-wet with pyrene and no nitrogen; 6, slurry without nitrogen or pyrene; 7–8, slurry with nitrogen and pyrene; 9–10, slurry with pyrene and no nitrogen
Stable-isotope Probing with Pyrene
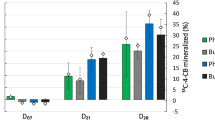
Because the DNA yield was greater and certain 16S rRNA gene sequences were enriched by the addition of nitrogen (as judged by greater band intensity in the DGGE gel shown in Fig. 2), we decided to perform SIP with [U-13C]pyrene for the S and FW incubations only under the condition in which nitrogen was added. Based on results from the mineralization experiment, SIP incubations were terminated after 28 days. After ultracentrifuging the extracted DNA, fractions collected from the ultracentrifuge tubes were evaluated by PCR using bacterial primers and a primer set specific to E coli. E. coli DNA was detected by PCR amplification only in fraction 6 from each condition, and that fraction was therefore considered to contain unlabeled DNA in all cases. Based on PCR analyses to identify the lowest fraction in each centrifuge tube containing amplifiable amounts of DNA, fractions 3 or 4 were designated the 13C-enriched (“heavy”) fractions for the FW and S conditions, respectively (Fig. 3). No archaeal 16S or fungal 18S rRNA gene sequences were recovered by PCR of the “heavy” fractions, although archaeal sequences were present in the original soil used as an inoculum for the SIP experiment (data not shown).
Negative image of DGGE gel delineating 16 S rRNA genes in selected fractions after ultracentrifugation of DNA isolated from SIP incubations. Lane designations are as follows: lanes 1–2, duplicates of field-wet fraction 3 (“heavy” fraction); lanes 3–4, duplicates of slurry fraction 3; lanes 5–6, duplicates of field-wet fraction 4; lanes 7–8, duplicates of slurry fraction 4 (“heavy” fraction); lane 9, E. coli K-12 DNA; lanes 10–11, field-wet fraction 6 (“light” fraction); lanes 12–13, slurry fraction 6 (“light” fraction)
Phylogenetic Analysis of 16S rRNA Gene Clone Libraries
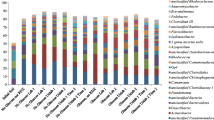
A total of 73 partial 16S rRNA gene sequences were obtained from 13C-enriched DNA fractions, two of which were determined to be chimeras and excluded from further analyses. All of the sequences from the FW microcosms (37 of 37) and the majority of sequences from the S microcosms (30 of 34) grouped with the γ-Proteobacteria (Fig. 4). These sequences were 98% similar to sequences associated with previously described members of “Pyrene Group 2” (PG2), a clade of uncultivated bacteria previously implicated in pyrene degradation by SIP of PAH-contaminated soil from a manufactured-gas plant [45]. The remaining four sequences recovered from the S incubations grouped with the α-Proteobacteria and were less than 87% similar to any of the other sequences recovered in this study. No attempt was made to evaluate the cloned sequences by DGGE, so that the clone library results cannot be compared directly to the DGGE profiles in Fig. 3.
Neighbor-joining phylogenetic tree of partial 16 S rRNA gene sequences of pyrene-degrading organisms identified by SIP in this study (in bold), close phylogenetic relatives, and selected reference sequences. Parentheses include the number of sequences represented and GenBank accession number. Closed and open circles at nodes indicate greater than or equal to 50 and greater than or equal to 95% bootstrap support, respectively
Discussion
We evaluated the effects of the treatment method (either as S or static) and the addition of nitrogen on pyrene mineralization by microbial communities in a PAH-contaminated soil. In other studies, the addition of inorganic nutrients to enhance biodegradation has met with mixed results [7, 19, 34, 41, 50]. In this study, adding nitrogen to the incubations increased the rate of pyrene mineralization for both types of incubations, and in the S microcosms, nitrogen amendment decreased the lag period before significant pyrene mineralization occurred (Fig. 1). Microcosms with nitrogen addition yielded more total-community DNA than microcosms without nitrogen amendment, suggesting a higher level of microbial growth on the added pyrene. In addition, microcosms amended with nitrogen displayed a greater extent of mineralization over 49 days than those without nitrogen amendment, which is consistent with the effects of nutrient amendment in previous studies on the treatment of soil from the same site in compost reactors [34].
S incubation led to greater pyrene mineralization than the corresponding static incubation whether nitrogen was added or not (Fig. 1). The greater rates and extents of pyrene mineralization in S microcosms might be attributable to their constant agitation [53]. Agitation increases the likelihood of contact between soil particles, bacteria, and pyrene in the aqueous phase and also increases oxygen transfer. Although pyrene has limited solubility in water (0.13 mg/L), the potential for soil-associated pyrene to be in contact with the aqueous phase increases the bacterial community’s ability to metabolize the substrate, whereas aeration promotes biomass growth and metabolic activity. Agitation may also have delayed sorption of the added pyrene by disrupting any weak association with the soil organic matter or the contaminant matrix. In addition, it has been proposed that moisture content and aeration are more important than nutrient addition with regard to substrate removal [50].
The highest concentration of DNA per gram of soil (and presumably the largest microbial community) was recovered from the S incubation amended with nitrogen (S + N), which also displayed the highest rate and greatest extent of pyrene mineralization. This correlation between microbial concentration and rate and extent of mineralization differs from the findings of previous studies [7, 19, 34] and indicates that inorganic nutrient or treatment status alone may be predictive of extent of substrate removal when bioavailability does not govern degradation kinetics.
DGGE profiles were similar among the conditions; however, relative to controls (no added pyrene or nitrogen) and to the incubations amended with pyrene but no nitrogen, bands low on the gel were more prominent in the incubations to which nitrogen was added (Fig. 2). There was little difference between physical treatments (continuously mixed S or static FW) on the apparent community diversity with or without nitrogen addition. SIP experiments with both S and static incubations amended with nitrogen resulted in only slightly different communities of pyrene-degrading bacteria. The majority of recovered sequences in clone libraries derived from “heavy” DNA of both incubation conditions were closely related to sequences of uncultured γ-Proteobacteria, including those described in earlier work as PG2 [45]. Organisms in this as-yet uncultivated clade were previously found to be a significant component of the “heavy” DNA after SIP with 13C-pyrene of a bioreactor community treating PAH-contaminated soil from a different source than the soil used in the present study [45]. Also similar to the previous study in which PG2 was first described, members of this group appear to be a significant part of the community in the contaminated soil used as inoculum in the present study. In a separate analysis to be reported elsewhere, nearly 16% of the sequences in a 16 S rRNA gene clone library constructed from the contaminated soil (without pyrene amendment) grouped phylogenetically within PG2 (Swanson, unpublished data).
Although the γ-Proteobacterial PG2 were the most prominent sequences recovered in 13C-enriched DNA under both incubation conditions (67 of the 71 sequences in the combined clone libraries), α-Proteobacterial sequences were also present in the S treatment (4 of 34 sequences). Three were very similar to sequences of Caulobacter species, two of which cluster with Caulobacter leidyi. This genus has been described as both paraphyletic [1] and polyphyletic [46], and its members are known to thrive in nonstatic, high-moisture, oxygen-rich environments [33], which is consistent with the S environment. Various analyses, including 16 S rRNA gene sequencing, suggest that C. leidyi is likely a member of the genus Sphingomonas [1, 36]. Therefore, organisms represented by the clone designated slurry 2-03 may also be sphingomonads. Sphingomonas spp. have been implicated in pyrene transformation in other studies [23, 28, 45].
The presence of several sequences in the clone library from “heavy” DNA associated with S incubation during the SIP experiment that were not in the clone library from the static incubation might have been reflected in differences in the DGGE profiles of the “heavy” DNA from the two incubation conditions (Fig. 3). However, we did not perform DGGE analysis on clonal sequences, so that specific sequences cannot be matched with specific bands on the DGGE gel. Similarly, it is not possible to determine whether slight sequence variations among the PG2 sequences might have led to the two most prominent bands in the “heavy” DNA from either incubation condition.
Given the relatively long incubation time in the SIP experiment (28 days), we cannot be certain that the few singleton sequences recovered in the S microcosms resulted from growth on pyrene itself or whether growth might have occurred on one or more products of pyrene transformation. However, the fact that the vast majority of clones recovered from both static and S incubations were from a single clade suggests that organisms in this group are able to grow on pyrene itself. The 28-day incubation was required to achieve a high extent of pyrene mineralization, which is expected to parallel pyrene assimilation during active growth. Although the 13C-labeled pyrene added to the soil was likely more bioavailable than pyrene in a weathered contaminated soil, it was added in an amount greatly in excess of its aqueous solubility (approximately 0.13 mg/L), which would have made it available to the pyrene-degrading bacteria much more slowly than a water-soluble growth substrate.
It is interesting that in this and the only previous study in which SIP was performed with pyrene [45], no Actinobacteria were identified as significant pyrene degraders. Mycobacterium spp. and other Gram-positive organisms have been isolated from a number of field-contaminated soils when enriched on pyrene as a sole carbon source [10, 24, 27, 35], but none were identified by SIP in this study; we did not attempt to isolate any pyrene degraders from the soil investigated in this study. The contrasting findings of SIP studies and isolation methods highlight the differences in these two approaches to studying environmentally relevant microorganisms. Both SIP and culture-based methods can introduce selection bias when identifying microorganisms able to grow on a particular carbon source. Both approaches can lead to identification of organisms capable of degrading a particular compound but cannot by themselves indicate how those organisms respond to the compound under in situ conditions. However, SIP eliminates the requirement for growth outside the natural microenvironment, thereby reducing biases associated with growth in artificial media; its major bias is in exposing an indigenous microbial community to quantities of growth substrates in excess of what would be experienced under natural conditions. Furthermore, the bias introduced by high concentrations of a water-soluble substrate are expected to be attenuated with poorly soluble substrates such as pyrene and other PAHs. Nevertheless, any added hydrophobic substance such as pyrene is likely to be more bioavailable than the native, aged contaminant. Further research with cultivation-independent techniques must be conducted before the limits of either approach are fully appreciated.
References
Abraham W, Strompl C, Meyer H, Lindholst S, Moore ERB, Christ R, Vancanneyt M, Tindall BJ, Bennasar A, Smit J, Tesar M (1999) Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis Maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Intl J Syst Bacteriol 49:1053–1073
Agency for Toxic Substances and Disease Registry (1995) Public health statement for polycyclic aromatic hydrocarbons. US Department of Health and Human Services, Public Health Service, Atlanta, GA
Aitken MD, Stringfellow WT, Nagel RD, Kazunga C, Chen SH (1998) Characteristics of phenanthrene-degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can J Microbiol 44:743–752
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Borneman J, Hartin RJ (2000) PCR primers that amplify fungal rRNA genes from environmental samples. Appl Environ Microbiol 66:4356–4360
Bouyoucos GJ (1962) Hydrometer method improved for making particle size analysis of soils. Agron J 54:464–465
Carmichael LM, Pfaender FK (1997) The effect of inorganic and organic supplements on the microbial degradation of phenanthrene and pyrene in soils. Biodegradation 8:1–13
Cassel DK, Nielsen DR (1986) Field capacity and available water capacity. In: Klute A (ed) Methods of soil analysis. Part 1. Physical and mineralogical methods. Agronomy monograph no. 9. 2nd edn. American Society of Agronomy, Wisconsin, pp 901–926
Cerniglia CE (1992) Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351–368
Cheung P, Kinkle BK (2001) Mycobacterium diversity and pyrene mineralization in petroleum-contaminated soils. Appl Environ Microbiol 67:2222–2229
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The ribosomal database project (RDP-II): Previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Dean-Ross D, Cerniglia CE (1996) Degradation of pyrene by Mycobacterium flavescens. Appl Microbiol Biotechnol 46:307–312
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
El Fantroussi S, Verschuere L, Verstraete W, Top EM (1999) Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16 S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol 65:982–988
Eriksson M, Dalhammar G, Mohn WW (2002) Bacterial growth and biofilm production on pyrene. FEMS Microbiol Ecol 40:21–27
Guthrie EA, Pfaender FK (1998) Reduced pyrene bioavailability in microbially active soils. Environ Sci Technol 32:501–508
Heitkamp MA, Franklin W, Cerniglia CE (1998) Microbial metabolism of polycyclic aromatic hydrocarbons: Isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol 54:2549–2555
Huesemann MH, Hausmann TS, Fortman TJ (2004) Does bioavailability limit biodegradation? A comparison of hydrocarbon biodegradation and desorption rates in aged soils. Biodeg 15:261–274
Hwang S, Cutright TJ (2002) Biodegradability of aged pyrene and phenanthrene in a natural soil. Chemosphere 47:891–899
Hwang S, Cutright TJ (2003) Preliminary exploration of the relationships between soil characteristics and PAH desorption and biodegradation. Environ Intl 29:887–894
Jeon CO, Park W, Padmanabhan P, DeRito C, Snape JR, Madsen EL (2003) Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc Natl Acad Sci USA 100:13591–13596
Kanaly RA, Harayama S (2000) Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J Bacteriol 182:2059–2067
Kazunga C, Aitken MD (2000) Products from the incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon-degrading bacteria. Appl Environ Microbiol 66:1917–1922
Kim Y, Freeman JP, Moody JD, Engesser K, Cerniglia CE (2005) Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Appl Microbiol Biotechnol 67:275–285
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid sequencing techniques in bacterial systematics. Wiley, New York, pp 115–175
MacLeod CT, Daugulis AJ (2003) Biodegradation of polycyclic aromatic hydrocarbons in a two-phase partitioning bioreactor in the presence of a bioavailable solvent. Appl Microbiol Biotechnol 62:291–296
Miller CD, Hall K, Liang YN, Nieman K, Sorensen D, Issa B, Anderson AJ, Sims RC (2004) Isolation and characterization of polycyclic aromatic hydrocarbon-degrading Mycobacterium isolates from soil. Microb Ecol 48:230–238
Mueller JG, Devereux R, Santavy DL, Lantz SE, Willis SG, Pritchard PH (1997) Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie van Leeuwenhoek 71:329–343
Mueller JG, Lank SE, Blattmann BO, Chapman PJ (1991) Bench-scale evaluation of alternative biological treatment processes for the remediation of pentachlorophenol- and creosote-contaminated materials: Slurry-phase bioremediation. Environ Sci Technol 25:1055–1061
Northcott GL, Jones KC (2000) Developing a standard spiking procedure for the introduction of hydrophobic organic compounds into field-wet soil. Environ Toxicol Chem 19:2409–2417
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Appl Biosci 12:357–358
Parrish ZD, White JC, Isleyen M, Gent MPN, Iannucci-Berger W, Eitzer BD, Kelsey JW, Mattina MI (2006) Accumulation of weathered polycyclic aromatic hydrocarbons (PAHs) by plant and earthworm species. Chemosphere 64:609–618
Poindexter JS (1981) The Caulobacters: ubiquitous unusual bacteria. Microbiol Rev 45:123–179
Potter CL, Glaser JA, Chang LW, Meier JR, Dosani MA, Herrmann RF (1999) Degradation of polynuclear aromatic hydrocarbons under bench-scale compost conditions. Environ Sci Technol 33:1717–1725
Ramirez N, Cutright T, Ju L (2001) Pyrene biodegradation in aqueous solutions and soil slurries by Mycobacterium PYR-1 and enriched consortium. Chemosphere 44:1079–1086
Rappe MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394
Raskin L, Stromley JM, Rittman BE, Stahl DA (1994) Group-specific 16 S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60:1232–1240
Rehmann K, Noll HP, Steinberg CEW, Kettrup AA (1998) Pyrene degradation by Mycobacterium sp. strain KR2. Chemosphere 36:2977–2992
Reid BJ, Northcott GL, Jones KC, Semple KT (1998) Evaluation of spiking procedures for the introduction of poorly water soluble contaminants into soil. Environ Sci Technol 32:3224–3227
Risk Reduction Engineering Laboratory, US EPA (1993) Pilot-scale demonstration of a slurry-phase biological reactor for creosote-contaminated soil: applications analysis report. EPA/540/A5-91/009
Roling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, Head IM (2002) Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68:5537–5548
Sabat G, Rose P, Hickey WJ, Harkin JM (2000) Selective and sensitive method for PCR amplification of Escherichia coli 16 S rRNA genes in soil. Appl Environ Microbiol 66:844–849
Schulte EE, Hopkins BG (1996) Estimation of soil organic matter by weight loss-on-ignition. In: Magdoff FR, Tabatabai MA, Hanlon EA Jr (eds) Soil organic matter: analysis and interpretation. Soil Science Society of America, Wisconsin, pp 21–31
Singleton DR, Powell SN, Sangaiah R, Gold A, Ball LM, Aitken MD (2005) Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil. Appl Environ Microbiol 71:1202–1209
Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD (2006) Identification and quantification of uncultivated proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ Microbiol 8:1736–1745
Sly LI, Cox TL, Beckenham TB (1999) The phylogenetic relationships of Caulobacter, Asticcacaulis and Brevundimonas species and their taxonomic implications. Intl J Syst Bacteriol 49:483–488
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
US EPA Office of Solid Waste and Emergency Response (1999) Use of monitored natural attenuation at Superfund, RCRA corrective action, and underground storage tank sites. OSWER Directive 9200.4-17P
Viñas M, Sabaté J, Espuny MJ, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018
Walter U, Beyer M, Klein J, Rehm H (1991) Degradation of pyrene by Rhodococcus sp. UW1. Appl Microbiol Biotechnol 34:671–676
Watson ME, Brown JR (1998) pH and lime requirement. In: Brown JR (ed) Recommended chemical test procedures for north central region, NCR Publication no. 221 (revised), Missouri Agr 0045p Sta SB 1001. NCR, Missouri, pp 13–16
White JC, Alexander M, Pignatello JJ (1999) Enhancing the bioavailability of organic compounds sequestered in soil and aquifer solids. Environ Toxicol Chem 18:182–187
Acknowledgments
This study was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program (5 P42 ES05948) and by the National Science Foundation (BES 0221836). MDJ was supported by the Research Education Support Program, a project of the National Science Foundation Alliance for Graduate Education and the Professoriate (HRD0450099).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jones, M.D., Singleton, D.R., Carstensen, D.P. et al. Effect of Incubation Conditions on the Enrichment of Pyrene-degrading Bacteria Identified by Stable-isotope Probing in an Aged, PAH-contaminated Soil. Microb Ecol 56, 341–349 (2008). https://doi.org/10.1007/s00248-007-9352-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9352-9