Abstract
Knowledge of the distribution of Paenibacillus larvae spores, the causative agent of American foulbrood (AFB), among individual adult honey bees is crucial for determining the appropriate number of adult bees to include in apiary composite samples when screening for diseased colonies. To study spore distribution at the individual bee level, 500 honey bees were collected from different parts of eight clinically diseased colonies and individually analyzed for P. larvae. From the brood chamber and from the super, bees were randomly collected and individually put in Eppendorf vials. The samples were frozen as soon as possible after collection. Concurrently with sampling, each colony was visually inspected for clinical symptoms of AFB. The number of clinically diseased cells in the colony was visually estimated. All samples were cultured in the laboratory for P. larvae. The results demonstrate that the spores are not randomly distributed among the bees; some bees have much higher spore loads than others. It is also clear that as the proportion of contaminated bees increase, the number of spores from each positive bee also increases. The data also demonstrated a relationship between the number of clinically diseased cells and the proportion of positive bees in individual colonies. This relationship was used to develop a mathematical formula for estimating the minimum number of bees in a sample to detect clinical disease. The formula takes into account the size of the apiary and the degree of certainty with which one aims to discover clinical symptoms. Calculations using the formula suggest that adult bee samples at the colony level will detect light AFB infections with a high probability. However, the skewed spore distribution of the adult bees makes composite sampling at the apiary level more problematic, if the aim of the sampling is to locate lightly infected individual colonies within apiaries. The results suggest that false-negative culturing results from composite samples of adult bees from individual colonies with clinical symptoms of AFB are highly improbable. However, if single colonies have light infections in large apiaries, the dilution effect from uncontaminated bees from healthy colonies on the positive bees from diseased colonies may yield false-negative results at the apiary level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
American foulbrood (AFB) is caused by the bacterium Paenibacillus larvae [6]. The pathogen infects and kills larvae of the honey bee (Apis mellifera). AFB is widely distributed throughout the world and is a problem for many beekeepers, causing great economic losses [4, 10]. To decrease the impact of AFB, it is important to have efficient screening methods for diseased colonies so they can be removed from the apiary before the disease is transmitted to adjacent colonies. Although honey has been widely used to monitor for AFB infections [9], Nordström et al. [14] showed that culturing of colony-level composite samples of adult bees is more sensitive than culturing of honey samples from the same colony. Hornitzky and Karlovskis [12] introduced the method of culturing adult honey bees for AFB and demonstrated that spores could also be detected from colonies without clinical symptoms. These false positives represent colonies that are infected by the pathogen but where clinical symptoms are not manifested. From an epidemiological perspective, such subclinical infections should not be neglected because they may be responsible for considerable horizontal disease transmission within beekeeping operations, also where clinical disease symptoms have not appeared, as beekeepers move material between colonies. Adult bees from the brood chamber has been reported to contain more spores per bee compared with samples from the supers [8], but the differences are small and may not be of practical importance when sampling for diseased colonies [13].
Whereas honey, on occasion, may produce false-negative results (no bacterial growth in spite of clinical disease symptoms), samples of adult bees have not produced false negatives when sampling of adult bees, and field inspection for clinical symptoms have been done at the same time [13]. However, the issue of false negatives is a problem that depends on the desired detection level and how large a sample one is prepared to collect and process.
To understand how a pathogen is transmitted and how it is distributed in the population is crucial for understanding the epidemiology of the disease. Furthermore, an understanding of the epidemiology is crucial for disease control and disease management. Some studies on the transmission and distribution of AFB spores at the colony and apiary level have been published [5, 8, 11, 13], but no studies explicitly investigate the spore load and spore distribution among individual honey bees. The fate of P. larvae spores in individual honey bees after they have been ingested has been studied [16], but the spore distribution in the colony or among bees remains elusive. In a previous study, the distribution of P. larvae spores based on colony-level and apiary-level sampling was described [13]. To understand how the spores of P. larvae are distributed among adult honey bees, this article investigates individual bee-level sampling. The results are compared with data from the colony level for proper dimensioning of adult bee sampling for AFB detection.
Materials and Methods
Individual honey bees of unidentified mixed race from eight randomly selected, naturally infected colonies were collected from June to August of 2002 and individually put in Eppendorf® vials to prevent cross-contamination. The colonies belonged to a commercial beekeeper outside Uppsala in central Sweden and had not been treated for any other diseases before sampling. The brood chamber in the sampled colonies were allowed two deep ten-frame Langstroth boxes. Approximately 30 bees (range 30–32) were randomly collected from the supers (n = 8) and about 30 bees (range 28–33) from the brood chambers (n = 8). A total of 500 individual bees were sampled. The colonies were visually inspected at the time of sampling and all were clinically diseased with different intensities of AFB. The number of clinically diseased cells that could be detected upon careful visual inspection ranged from 1 to more than 400. Both ropey larval remains and dried-in scales were counted as clinical symptoms and added together to give the total number of clinically diseased cells.
Each bee was cultured individually following the protocol in the study of Lindström and Fries [13] but adjusted to fit for samples of one bee only. To culture a sample, a bee was put in a fine mesh bag inside a plastic bag, 1 mL of sterile water was added, and the bee was crushed. The liquid, filtered by the fine mesh bag to keep larger body parts such as legs out, was poured into a centrifugal tube and centrifuged (27,000×g, 10 min). The supernatant was discarded and the pellet resuspended in 200μl of sterile water. To reduce contamination, the pellet was put in a water bath (Grant™ GLS 400) for 10 min at 91°C. Dilution series (1/1,000, 1/10,000, and 1/100,000) was plated on MYPGP agar [3] with 3 μg/mL nalidixic acid. Plates were incubated for 7 days in 36°C with 5% CO2. Bacterial colonies were counted manually.
Spore numbers where log e transformed. To test for randomness, the Poisson goodness-of-fit test was employed. The Mann–Whitney U test was used to test for differences in spore load between supers and brood chambers and the two-proportions test to test for differences in proportion of positive bees. Regression analyses were used to test for relationships between other parameters. SAS Institute statistical software (version 9.1.3) was used for statistical analyses. To calculate the number of bees needed to detect infected apiaries on different detection levels and apiary sizes, the formula:
was used, where DC is the degree of certainty and p is the proportion of infected bees in a sample [1].
Results
The results show that the spores are not randomly distributed among the bees (p < 0.001) when all bees from all colonies were tested in a Poisson goodness-of-fit test. The test instead suggests that the spores are aggregated among the bees (Fig. 1). Furthermore, it is worth noting that the variation in spore load per bee ranges over several orders of magnitude (Fig. 1).
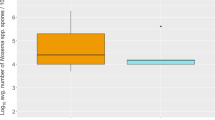
There was no significant difference between the spore loads of individual bees in supers and brood chambers (Mann–Whitney U test, p = 0.88, n = 16; Fig. 2). Nor did the proportions of positive or contaminated bees differ significantly between supers and brood chambers (two-proportions test, p = 0.131, n = 16). However, there is a highly significant relationship between the proportion of positive bees of a given colony and the spore load of individual bees from that colony described by the equation \(y = 0.01e^{{6.96x}} {\left( {r^{2} = 0.89} \right)}\) with higher spore loads per bee as the proportion of positive bees increase as shown in Fig. 3.
The relationship between the approximate number of clinically diseased brood cells in the colony and the spore load of the individual bees was also highly significant described by the equation \(y = 1.05{\text{Ln}}{\left( x \right)} + 0.77\;{\left( {r^{2} = 0.63} \right)}\). The results demonstrate that the more clinical symptoms that are detected, the heavier is the spore load found on individual bees (Fig. 4). The relationship between the number of clinically diseased cells and the proportion of positive bees (Fig. 5) can be described by a function:
where p is the proportion of positive bees in a sample and x is the number of clinically diseased cells in the colony where the sample was taken. If Eq. 2 is combined with Eq. 1, we get:
which allows us to sum the proportions of positive bees for each colony in the apiary based on the clinical symptoms and calculate the proportion of positive bees in the composite sample. In the denominator, n i denotes the number of clinically diseased colonies sampled, and n h denotes the number of healthy colonies sampled. This formula assumes that the colonies that do not show any clinical symptoms are spore-free and can therefore be used to calculate the worst-case scenario. In Fig. 6, the formula has been used to calculate sample sizes of adult bees for different detection levels and apiary sizes. It should be pointed out that this formula is based on data from a limited number of colonies and do not represent the great variation in virulence of the bacterium nor the variation in resistance on behalf of the bees. It is therefore likely that if this experiment is repeated in other populations, results will vary accordingly.
The lowest proportion of positive bees in a sample from a clinically diseased colony according to Eq. 2 is 12%, corresponding to one clinically diseased cell (Fig. 5). If Eq. 1 is solved for DC, the probability for a positive result can be calculated. The empirical data suggest that if 100 bees are sampled from a colony with one clinically diseased cell the probability of a positive culturing result is result is greater than 99.99% (Fig. 7).
Discussion
The spores of P. larvae are not randomly distributed among adult bees. This distribution is important to know for dimensioning sample sizes when colonies are monitored for AFB, using cultivation for the causative agent from adult bees. The presented results strongly suggest that samples of adult bees from individual colonies are highly effective in detecting clinically diseased colonies (Fig. 5). However, the skewed distribution of spores among bees in colonies with few visibly infected cells make composite samples including many colonies more problematic. In samples of 100 bees from individual colonies, the chance of a false negative is negligible (Fig. 6). With inclusion of noninfected colonies in an apiary composite sample, the probability of a false negative result will increase.
There were no significant difference in spore load between the brood chamber and the super. This is congruent with what has been reported by Lindström and Fries [13] but contrary to that of Goodwin et al. [8]. However, Goodwin et al. [8] reported that when clustering bees are shaken from the brood combs, the remaining young bees have a significantly lower spore load than the bees from the super. The reported difference corresponds to a few positive bees (~130 colonies/plate). This supports the data from this study that the spores are unevenly distributed. During their lifetime, honey bees have different tasks in the colony [17], which would expose them to spores to varying degrees.
The figures also show that a number of bees will be positive for P. larvae without any clinical symptoms manifested, in the colony so-called false-positive samples. Because the spores are not uniformly distributed, single honey bee individuals that contain high spore levels may be sampled and present a skewed image of the situation in the colony.
If we use the model from Fig. 5, we can calculate the proportions of positive bees for different levels of clinical disease. The calculations show that already with six clinically diseased cells, 50% of the bees are positive and with 36 and 112 clinically diseased cells, 99 and 100% of the bees in the colony, respectively, will be positive for P. larvae. This means that as the level of clinical disease increase in a colony included in an apiary composite bee sample, the probability of false negative results decrease drastically. Provided an equal number of bees from each colony is included in the apiary composite bee sample to be investigated, we can make the following calculation: In an apiary composite sample from ten colonies with one colony containing one clinically diseased cell and all other colonies being free of spores (unlikely because of subclinical infections), 12% of the adult bees from the clinically diseased colony are positive (Fig. 5). The probability that the composite sample will give a false-negative result is approximately 30% if 100 bees are cultured as a subsample of the composite sample. In Fig. 6, it can be seen that if we want to be 95% sure to detect one colony diseased with one cell in a composite sample from ten colonies, we have to take a subsample that consists of 245 bees. Thus, if the lowest level of infection should be detected with a high probability, the results demonstrate that individual colony samples should be preferred. However, the data also suggest that composite apiary samples could be useful for more general screening purposes.
Using composite samples of adult bees from individual colonies, Lindström and Fries [13] report no false negatives from colony-level samples. However, Goodwin et al. [8] report 2.5% false-negative samples from individual colonies. Partly, the results of Goodwin et al. [8] may be attributed to culturing the bacteria for 3 days only and inspections for symptoms at a time different from the sampling of bees. It has been shown that samples have to be incubated for at least a week to let the majority of spores germinate [15].
The results show no significant difference in spore load or proportion of spore carriers between bees from the brood chamber and the supers. This is congruent with earlier data both at the colony and apiary level of sampling [13]. Although bees from the brood chamber sometimes carry more spores compared with adult bees sampled in the super [8, 13], the differences are slight. Thus, the increased amount of work to sample from the brood chamber, when samples are collected from colonies with honey supers, is hardly justified.
It is interesting to note that there is a strong correlation between the number of colony-forming units and the proportion of positive bees (Fig. 4). Goodwin et al. [8] reported the proportion of positive bees in their samples but did not relate this proportion to the level of clinically diseased cells or to amount of spores carried by each individual bee. Goodwin et al. [7] stated “that most bees in an AFB infected colony carry B. larvae (later renamed to P. larvae [6]) spores, even those with low level of clinical infection.” Unfortunately, they did not give any data or reference to support their claim. Figure 3 clearly shows that as more bees become spore carriers, each bee also tends to be contaminated with increasing number of spores. Increased number of spores per bee may possibly translate to more efficient transmission of spores to susceptible larvae, and additional larvae can potentially be infected by each contaminated bee [2]. The spores are stored in the intestines of the bees for several days and may be accumulated there if the bee constantly ingests more spores [16], as would probably be the case in a heavily infected colony.
There was a strong relationship between the number of clinically diseased cells in a colony and the mean number of bacterial colonies per bee that grew on the agar plates (Fig. 4). This is congruent with data from composite samples of adult bees [13]. The spore load of the adult bees rise fast as the number of clinically diseased cells increase. If the infection level is low and kept under control by the colony, then clinically diseased cells are probably only manifested at irregular intervals. Then, the proportion of adult bees that carry spores will indeed be small, and samples from such colonies may suggest that the disease disappears, only to reappear, although it is a continuously ongoing low-grade infection.
Microbiological methods was used to measure the spore loads in the samples. Although molecular methods have a lower detection threshold, the culturing of P. larvae spores is sufficiently accurate for the purposes of this study. Moreover, for molecular methods to be applicable spore walls need to be broken, which requires special equipment.
The results suggest that false-negative culturing results from composite samples of adult bees from individual colonies with clinical symptoms of AFB are highly improbable. At the apiary level, the outcome is strongly dependant on the number of healthy and clinically diseased colonies in the apiary.
References
Colton T (1974) Statistics in medicine. Little Brown, Boston, p 372
Dieckman U et al (ed) (2002) In: Adaptive dynamics of infectious disease. In Pursuit of virulence management. Cambridge University Press, Cambridge, UK
Dingman DW, Stahly DP (1983) Medium promoting sporulation of Bacillus larvae and metabolism of medium components. Appl Environ Microbiol 46(4):860–869
Ellis JD, Munn PA (2005) The worldwide health status of honey bees. Bee World 86:88–101
Fries I, Lindström A, Korpela S (2006) Vertical transmission of American foulbrood (Paenibacillus larvae) in honey bees (Apis mellifera). Vet Microbiol 114:269–274
Genersch E et al (2006) Reclassification of Paenibacillus larvae subsp. larvae and Paenibacillus larvae subsp. pulvifaciens as Paenibacillus larvae without subspecies differentiation. Int J Syst Evol Microbiol 56:501–511
Goodwin RM, Perry JH, Brown P (1993) American foulbrood disease part III: spread. NZ Beekeeper 219:7–10
Goodwin RM, Perry JH, Haine HM (1996) A study on the presence of Bacillus larvae spores carried by adult honey bees to identify colonies with clinical symptoms of American foulbrood disease. J Apic Res 35(3/4):118–120
Hansen H (1984) Methods for determining the presence of the foulbrood bacterium Bacillus larvae in honey. Dan J Plant Soil Sci 88:325–328
Hansen H, Brödsgaard CJ (1999) American foulbrood: a review of its biology, diagnosis and control. Bee World 80(1):5–23
Hornitzky MAZ (1998) The spread of Paenibacillus larvae subsp larvae infections in an apiary. J Apic Res 37(4):261–265
Hornitzky MAZ, Karlovskis S (1989) A culture technique for the detection of Bacillus larvae in honeybees. J Apic Res 28(2):118–120
Lindström A, Fries I (2005) Sampling of adult bees for detection of American foulbrood (Paenibacillus larvae subsp. larvae) spores in honey bee (Apis mellifera) colonies. J Apic Res 44(2):82–86
Nordström S, Forsgren E, Fries I (2002) Comparative diagnosis of American foulbrood using samples of adult honey bees and honey. J Apic Sci 46(2):5–12
Nordström S, Fries I (1995) A comparison of media and cultural conditions for identification of Bacillus larvae in honey. J Apic Res 34(2):97–103
Wilson WT (1971) Resistance to American foulbrood in honey bees. XI. Fate of Bacillus larvae spores ingested by adults. J Invertebr Pathol 17:247–255
Winston M (1987) The biology of the honey bee. Harvard University Press, Cambridge, MA, p 281
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lindström, A. Distribution of Paenibacillus larvae Spores Among Adult Honey Bees (Apis mellifera) and the Relationship with Clinical Symptoms of American Foulbrood. Microb Ecol 56, 253–259 (2008). https://doi.org/10.1007/s00248-007-9342-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9342-y