Abstract
Kartchner Caverns in Benson, AZ, was opened for tourism in 1999 after a careful development protocol that was designed to maintain predevelopment conditions. As a part of an ongoing effort to determine the impact of humans on this limestone cave, samples were collected from cave rock surfaces along the cave trail traveled daily by tour groups (200,000 visitors year–1) and compared to samples taken from areas designated as having medium (30–40 visitors year–1) and low (2–3 visitors year–1) levels of human exposure. Samples were also taken from fiberglass moldings installed during cave development. Culturable bacteria were recovered from these samples and 90 unique isolates were identified by using 16S rRNA polymerase chain reaction and sequencing. Diversity generally decreased as human impact increased leading to the isolation of 32, 27, and 22 strains from the low, medium, and high impact areas, respectively. The degree of human impact was also reflected in the phylogeny of the isolates recovered. Although most isolates fell into one of three phyla: Actinobacteria, Firmicutes, or Proteobacteria, the Proteobacteria were most abundant along the cave trail (77% of the isolates), while Firmicutes predominated in the low (66%) and medium (52%) impact areas. Although the abundance of Proteobacteria along the cave trail seems to include microbes of environmental rather than of anthropogenic origin, it is likely that their presence is a consequence of increased organic matter availability due to lint and other organics brought in by cave visitors. Monitoring of the cave is still in progress to determine whether these bacterial community changes may impact the future development of cave formations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Kartchner Caverns was discovered in 1974, and purchased by the State of Arizona in 1988 [25]. This cave is widely acknowledged as a beautiful example of carbonate cave development and is considered to be one of the top 10 caves in the world in terms of mineralogical diversity. The diverse mineralogy of Kartchner Caverns is attributable to the presence of six different mineral classes: carbonates, nitrates, oxides, phosphates, silicates, and sulfates [10]. Carved into a down-dropped block of Mississippian Escabrosa limestone, the cave is situated between the Whetstone Mountains and San Pedro Valley, approximately 13 km south of Benson, AZ. Unlike most limestone caves, Kartchner Caverns is adjacent to an igneous and metamorphic terrain, which accounts for much of its unique mineralogical composition [11].
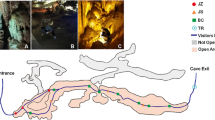
Kartchner Caverns has been slowly and carefully developed by the Arizona State Parks System to maintain its status as a living cave in this arid environment and to serve as a popular State Park attraction. Although some limited changes in the cave's climate have occurred, managed development has allowed maintenance of relatively stable cave conditions during the development process and the subsequent opening of the Rotunda–Throne Room complex to the public in 1999 and the Big Room complex in 2003 (Fig. 1) [24]. Tours are limited and closely supervised at all times to minimize touching of any surface. In addition, tour trail handrails and floors are washed down with water on a daily basis and the wash water is removed from the cave with a series of floor drains.
Map of Kartchner Caverns showing the low, medium, and high impact areas that were sampled. The high impact area is along the Rotunda–Throne room tour trail traveled daily by visitors (200,000 visitors year–1), the medium impact area is near an environmental monitoring station regularly checked by cave personnel (30–40 visitors year–1), and the low impact area is only visited for research purposes (2–3 visitors year–1).
Our work in Kartchner began in May 2001, in response to a slime formation problem that was discovered on painted fiberglass surfaces within the developed portion of the cave. These molded fiberglass blocks and fittings were mounted to disguise plumbing and electrical wiring as well as maintenance areas that might detract from the visitors' visual experience. In an effort to eliminate the slimy growths, the surfaces were washed with a 10% bleach solution. However, the biofilm growths returned within 1 month of treatment. Of primary concern was the fact that bacteria potentially introduced into the cave following the installation of synthetic surfaces or left behind from the skin, clothing, and shoes of people touring the cave might alter natural microbial communities found in the cave. Thus, the objective of this initial study was to determine the source of the observed slime and to compare the culturable heterotrophic populations isolated from the fiberglass surfaces to those present on adjacent natural rock surfaces.
After the initial study, a second study was designed to compare the community structure of bacteria isolated in the first study from the high human impact zone of the cave trail to that of more remote areas of the cave with varying degrees of human exposure. Microbial recovery was focused on the culturable fraction of the population because of the need to identify the slime-producing microbes associated with fiberglass moldings and to determine whether these organisms were a natural part of the indigenous cave microbiota. Indigenous cave bacteria are thought to originate from bacterial communities in soils hydrologically connected to the cave. The highly oligotrophic conditions in caves select for the portion of this community that can thrive under nutrient-limiting conditions. Thus, it was hypothesized that in Kartchner, surfaces in areas with little human traffic would harbor primarily highly oligotrophic bacteria, whereas surfaces exposed to higher levels of human activity (and thus organic matter) would either exhibit a community similar to that found in higher nutrient environments or a community based on microbiota brought into the cave by humans. The basis for this hypothesis is that human visitation brings with it bacteria that are associated with skin cells and lint that are left behind as people tour the cave, and lint from the clothing of human visitors is probably the largest anthropogenic source of organic matter deposition into Kartchner Caverns [14].
Materials and Methods
Cave Background
A microclimate study by Buecher [3] provides useful background information. The amount of water infiltrating the cave on a yearly basis is estimated to be 7.9 mm. Estimated water loss due to evaporation is 134,700 L year–1, representing over half of the 7.9 mm of infiltration water. Thus, during a dry year, when infiltration is low, the cave may undergo drying. The average relative humidity for the entire cave is 99.4%. Air temperatures in the cave range from 18.6°C in the back of the cave to 20.9°C near the main cave entrance (mean = 19.8°C). Temperatures within the cave are higher than the outside surface temperature (mean = 16.9°C). This difference is ascribed to regional geothermal heating. The elevated cave temperature impacts air flow in the cave whereby during the winter, colder outside air enters through the main cave opening, after which it warms and rises leaving out of small openings at higher elevations. During the summer months as the outside temperature increases, this induced airflow pattern weakens and finally reverses. Measures have been taken to maintain normal humidity levels in visited portions of the cave. Double airlock doors have been installed to prevent loss of humid air to the outside (or entry of dry outside air) as tourists enter the cave. Further, at the time of the study, a misting system was used in selected areas of the cave including the entrance where tourists walk under misters to dampen their clothing to prevent removal of existing moisture in the cave.
Sampling Site Information
Study samples were collected from Kartchner Caverns in August 2001 for the initial study designed to evaluate the slime production on painted fiberglass surfaces. The collection sites were located along the visitor trail for the Rotunda–Throne Room Complex (Fig. 1, Table 1), an area that has been exposed to 200,000 visitors year–1 since the opening of the cave in 1999. Sampling in this high human impact area of the cave included seven samples taken from two painted fiberglass barriers (2.1 m high × 1.4 m wide), and four samples taken from adjacent limestone cave rock surfaces in different areas of the cave. The fiberglass barriers were located adjacent to the visitor's path and each displayed a clear, slimy layer over their entire surface. The cave rock surface samples were obtained from each side of the fiberglass barriers and directly across the visitor's path.
Samples for the second study were collected in June 2003 from low and medium impact areas to evaluate the effect of human exposure on the natural bacterial communities of the cave. The culturable heterotrophic bacterial community was targeted to permit comparisons with populations identified in the initial study. Medium impact samples were collected from the Grand Central Station area exposed to 30–40 visitors year–1, whereas low impact samples were taken from the Subway Passage area exposed to only 2–3 visitors year–1 (Fig. 1, Table 1). In each area, triplicate samples were taken from adjacent limestone rock surfaces within a 1 m2 area.
The number of visits for the high and medium impact areas is constant and evenly spaced throughout the year as a result of tourism (limited to 500 tourists day–1 and accompanying docents) and routine monitoring, respectively. In contrast, the low impact area is not necessarily visited each year. The three sampling areas all experience similar relative humidity. In terms of temperature, the low impact area is in the colder back part of the cave, whereas the medium and high impact areas are closer to the main entrance. Air flow is greatest in the medium impact area that is nearest the cave entrance and is least in the low and high impact areas.
Sample Collection and Processing
Samples of all painted fiberglass and cave rock surfaces (30 cm2 surface area per swab) for both studies were collected in triplicate, using sterile swabs wetted with sterile tap water (pH = 7.2–7.8). The swabs were immersed in 3 mL of sterile tap water in test tubes, which were then capped, placed on ice, and transported back to the University of Arizona for same-day processing. Repetitive sampling of this cave has demonstrated that this sampling procedure provides reproducible, qualitative samples of bacterial populations on solid surfaces. In the laboratory, samples were vortexed for 5 min and then 0.1 mL volumes of five serial dilutions were plated on R2A medium (Difco Laboratories, Detroit, MI, USA) to enumerate heterotrophs and to allow visual assessment of morphological diversity. The plates were incubated at room temperature (∼23 C°) in a humidified chamber for 3 months. This temperature is close to average temperatures in the Throne Room, which are approximately 22°C.
Isolation and Grouping of Culturable Bacterial Isolates
The selection of unique bacteria for identification followed a two-step screening process. First, during the 3-month period of incubation, morphologically distinct colonies were selected for isolation from each dilution series (10–1 to 10–5) as they appeared. Colony size, shape, color, concavity, and growth rate served to characterize and group isolates morphologically. Representative colonies from each group were streaked repeatedly for isolation on R2A agar until pure colonies were obtained. Purity was confirmed microscopically after gram staining [16] by evaluating the homogeneity of cell shape, size, and gram stain. Replicate colonies from 10% of the groups were selected to confirm the accuracy of the first screening step. For these groups, two to six colonies of the same type were isolated to confirm that colonies assigned to the same morphological group were in fact of the same phylotype.
Repetitive extragenic palindromic-polymerase chain reaction (REP-PCR) was performed on each isolate as the second screening step [26]. We have shown in previous work [2] that REP-PCR profiles can differentiate between bacterial isolates at the strain level. Therefore, isolates with identical REP-PCR profiles were considered to be of the same genus and species. Isolates were prepared for REP-PCR analysis as follows. Each isolate was inoculated into 5 mL of R2B, and placed on a gyratory shaker (170 rpm) until turbidity was observed. One milliliter of turbid culture was transferred to a 1.7-mL microfuge tube, and centrifuged to pellet the cells. The supernatant was removed, and the cell pellet was resuspended in 1 mL of autoclaved and UV sterilized ddH2O. The cells were then subjected to three freeze–thaw cycles employing liquid nitrogen and boiling deionized water, which were followed by a 15-min boil and final freeze in liquid nitrogen. Cell lysates were stored at −20°C. In some instances, isolates did not grow when transferred into broth. We have previously encountered environmental isolates incapable of planktonic growth and have developed an alternative procedure. For these isolates, 2 mL sterile distilled water was added to a plate containing a pure culture. The plate was scraped with a cell scraper, aspirated for 30 s, and then the liquid was transferred to a microfuge tube. After centrifugation, the cells in the pellet were subjected to the freeze–thaw lysis procedure described above.
Lysates were subjected to REP-PCR by using a GeneAmp PCR System 2400 (PerkinElmer Inc., Boston, MA, USA). Primers used were REP1R-1 5′III ICG ICG ICA TCI GGC 3′ and REP2-1 5′ICG ICT TAT CIG GCC TAC 3′ [2]. A 25-μL REP-PCR reaction contained a 1-μM concentration of each primer, a 1.25-mM concentration each of dNTP, 1× buffer consisting of 10 mM Tris–HCl, 50 mM KCl, 2.5 mM MgCl2 (pH 8.9), 5% dimethyl sulfoxide (DMSO), 2.5 U of Taq DNA polymerase (Roche, Indianapolis, IN), and 2.5 μL of cell lysate. The amplification program run for REP-PCR was 95°C for 5 min, followed by 35 cycles of 95°C for 0.5 min, 43°C for 0.5 min, and 72°C for 4 min, and a final extension at 72°C for 16 min. PCR products were stored at −20°C. Amplified products were visualized after electrophoresis on 3% NuSieve 3:1 agarose gel (FMC Bioproducts, Rockland, ME, USA) following ethidium bromide (1 μg mL–1) staining. Isolates that could not be successfully amplified with the REP-PCR primers were amplified and sequenced as described below.
PCR Amplification of 16S rRNA and Sequencing
Each unique isolate as well as all isolates not successfully amplified by REP-PCR were subjected to 16S rRNA PCR by using primers 27f (5′ AGA GTT TGA TCC TGG CTC AG 3′) and 1492r (5′ TAC GGT TAC CTT GTT ACG ACT T 3′) [15]. Each 50-μL reaction contained 0.5 μM of each primer, 0.2 mM of each dNTP, 1× buffer consisting of 10 mM Tris–HCl, 50 mM KCl, 2.0 mM MgCl2 (pH 8.3), 5% DMSO, 1 U of Taq DNA polymerase, and 5.0 μL of cell lysate. The amplification program was 95°C for 5 min, followed by 30 cycles of 94°C for 1.0 min, 60°C for 1.0 min, and 72°C for 1.25 min, and a final extension at 72°C for 10 min. PCR products were stored at −20°C. Successfully amplified 16S rRNA products were submitted to The University of Arizona Research Labs Genomic Analysis and Technology Core for purification, quantification, and sequencing using their robotic 96-well plate program. Four primers, 27f, 518f (5′ CCA GCA GCC GCG GTA AT 3′), 1070r (5′ AGC TGA CGA CAG CCA T 3′), and 1492r, were used to obtain nearly full-length 16S rRNA gene sequences [15]. For some isolates the 27f primer did not work, in which case an alternative primer 518r (5′ ATT ACC GCG GCT GCT GG 3′) was used. Similarity searches were performed via BLAST analysis http://www.ncbi.nlm.nih.gov).
Phylogenetic Analysis
DNA sequences from all isolates from painted fiberglass and rock surfaces were combined and aligned with the PILEUP program of the GCG Sequence Analysis Software Package (ver. 10.2, Accelrys Inc., San Diego, CA, USA). Manual adjustments of sequence alignments were performed with the data editor program of MacClade Phylogenetic Software (ver. 4.0, Sinauer Associates Inc., Sunderland, MA, USA). Phylogenetic analyses were performed with programs contained in PAUP Phylogenetic Software (ver. 10.2 β, Sinauer Associates Inc.). Parsimony analysis heuristic searches for the most-parsimonious trees were conducted with random stepwise addition and branch swapping by tree bisection–reconnection (TBR). Sequence gaps were treated as missing data. To assess the effect of ambiguous regions in the alignment, analysis was repeated after removing such sections. Bootstrap analysis (1000 replicates) was performed to assess the statistical support for tree topology. All sequences have been submitted to NCBI GenBank (Tables 2, 3, 4, and 5).
Utilization of 100% Acrylic Paint as Sole Carbon Source
Unique isolates from the painted fiberglass were tested for the ability to grow on paint as a sole source of carbon and energy. Isolates were plated on mineral salts agar (MSA), R2A (normal strength) and 10% R2A media, all with a paint overlay. MSA is composed of (g L–1): KH2PO4, 1.0; Na2HPO4, 1.0; NH4NO3, 0.5; (NH4)2SO4, 0.5; MgSO4·7H2O, 0.2; CaCl2·2H2O, 0.02 (prepared as a 10× stock solution); FeCl3, 0.002 (prepared as a 100× stock solution); MnSO4·H2O, 0.002 (prepared as a 100× stock solution). Noble agar was added (15 g L–1), and the pH adjusted to 7.2 with 0.1 M NaOH. The paint overlay was prepared by dissolving 100% acrylic paint (Dunn Edwards Corporation, Los Angeles, CA, USA) in chloroform to final concentrations of 10,000 (1%), 1000 (0.1%), and 100 (0.01%) mg L–1. The paint was chosen by a Dunn Edwards expert as the best match for the paint on the fiberglass moldings. One milliliter of paint–chloroform solution was added to each plate. The plates were allowed to equilibrate under a fume hood for 30 min to evaporate the chloroform. The painted fiberglass isolates were precultured overnight in 5 mL R2B, pelleted and washed twice with saline, resuspended, and then 100 μL of the cell suspension was inoculated onto the plates in duplicate sets at a density of ∼1.0 × 106 CFU mL–1. Controls were performed by inoculating cultures onto Noble agar plates with no paint overlay. Plates were incubated at 23°C in a dark, humidified chamber for 1 month.
Results
Bacterial Communities in the High Impact Zone: Natural Rock vs Synthetic Surfaces
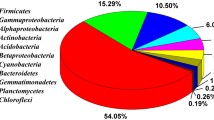
A total of nine unique isolates were obtained from painted fiberglass surfaces and 22 from adjacent natural rock surfaces (Tables 2 and 3). Replicate isolates sequenced from the same morphological group were found to have ≥97% identity, confirming that the morphological screening method did not prevent the identification of unique bacteria. The isolates obtained from the painted fiberglass slime biofilm represent the α-Proteobacteria, Bacteriodetes, and the Firmicutes (Table 2). The natural rock isolates were dominated by Proteobacteria with smaller numbers of Actinobacteria, Bacteriodetes, and Firmicutes (Fig. 2). Although the phylogenetic diversity of bacteria isolated from natural rock surfaces was greater than that of isolates from the fiberglass, strong similarities were observed between the two communities. Fiberglass isolate, PF-A (Bacillus), had 99.9% similarity to the natural rock isolate HI-G2, and both PF-D and PF-K (Sphingomonads) had 93% similarity to HI-D4. PF-M and HI-B13 were identified by BLAST as being most closely related to Sinorhizobium (Table 2, Fig. 3) and PF-I associated with the same Sphingomonas cluster (bootstrap = 100) as HI-D4, HI-K4, and HI-I1. These results suggest that the communities associated with the painted fiberglass surfaces are closely related to the organisms found on the adjacent natural rock surfaces.
Alignment of the 16S sequences resulted in a 1532-character dataset, of which 757 characters (49.4%) were variable and 691 characters (45.1%) were parsimony informative. Maximum parsimony analysis of the 16S dataset yielded 3049 equally most parsimonious trees (steps = 4482, CI = 0.326, RI = 0.788), which primarily differed in changes in relationships among taxa in terminal clades. Five principle clades were evident in all trees: Firmicutes, Actinobacteria, α-Proteobacteria, β-Proteobacteria, and γ-Proteobacteria. All principle clades were supported by bootstrap values >80%. Three isolates were identified as Bacteriodetes and composed the root of the tree. Analysis of the truncated datasets (ambiguous section removal) had only minor effects on the resulting tree topology compared to those resulting from analysis of the complete dataset, and the position and composition of major clades and high bootstrap support remained unchanged (data not shown).
The painted fiberglass isolates were evaluated for the ability to grow on acrylic paint as a sole source of carbon and energy to test the hypothesis that the paint coating is responsible for the observed biofilm formation on the fiberglass surfaces (Table 2). Of the nine painted fiberglass isolates, none displayed growth on Noble agar control plates (no added carbon source). Five isolates (PF-A, PF-H, PF-B, PF-I, and PF-D) were able to utilize the paint as a sole source of carbon at each of three tested concentrations. Three of these five isolates, PF-B (Dyadobacter), PF-H (Bacteriodetes) and PF-I, also displayed highly mucoid growth in vitro. Of the four remaining isolates, PF-K and PF-M demonstrated growth only at the lowest paint concentration (100 mg L–1), whereas PF-F (Staphylococcus) and PF-G (Brevibacillus) did not produce any colony growth during the 1-month incubation period. All isolates displayed growth on both 10% and regular-strength R2A with a paint overlay at all three concentrations. Therefore paint toxicity was excluded as a factor affecting the growth of isolates on plates with acrylic paint as the sole carbon source. Interestingly, five of the isolates capable of growth on paint were among those with similarities to isolates from the adjacent natural rock surfaces.
Comparison of Bacterial Communities in High and Minimally Impacted Areas
The second study was conducted to determine whether the culturable microbial diversity observed on the high impact rock surfaces was representative of bacterial communities in the less impacted areas of the cave. A total of 220 bacterial colonies were selected for isolation from the rock surfaces in the three regions with 76 from the high impact area, 73 from the medium impact, and 71 from the low impact. From these, 81 unique isolates were obtained including 22 from the high impact area, 27 from the medium, and 32 from the low impact rock surfaces (Tables 3, 4, and 5). Thus, the number of unique isolates obtained increased with decreasing human impact, suggesting that diversity increases as human impact decreases (Table 1).
Taxonomic and phylogenetic analyses of the sequences obtained for each impact designation reveals that the majority of isolates belong to three main phyla: Actinobacteria, Firmicutes, and Proteobacteria (Tables 2, 3, 4, and 5; Fig. 3). In addition, the high impact and painted fiberglass surfaces yielded sequences from the Bacteriodetes. A distinct pattern of recovered isolates emerged along the human impact gradient. In the high impact area, 77% of the isolates were Proteobacteria (evenly distributed among the α, β, and γ groups), whereas in the low and medium impact areas, Firmicutes predominated (66% and 52%, respectively) followed by Actinobacteria (19% and 22%, respectively) (Fig. 2). Only two isolates from the high impact area (HI-D3, HI-G2) were associated with the Firmicutes division. Both of these isolates were identified as Bacillus sp. and were found to have 100% identity with the low impact isolates L66 and L64, respectively. Likewise, a few of the medium impact Proteobacteria isolates were closely related to high impact isolates. Specifically, HI-G1 (Pseudomonas) and HI-B12 (Alcaligenes) had 99.8% and 99.7% identity to MI-o1 and MI-c1, respectively. Thus, although the community structure of the culturable bacteria is clearly impacted by the anthropogenic inputs, certain populations can be found throughout the cave.
One variable that must be considered when evaluating the impact of tourism on the cave is the fact that samples for the second study were taken 22 months after the initial study was conducted. Preliminary data from current monitoring of the portion of the cave opened to the public in November 2003 (using 16S rDNA/denaturing gradient gel electrophoresis analysis) suggest that changes to the bacterial community structure of low human impact regions over time is minimal (data not shown). This data strongly support the theory that the phylogenetic distribution of bacteria isolated from the low and medium impact areas of the cave is generally consistent over time. This observation does not necessarily apply to the high impact area exposed to a less stable environment as a result of human visitation.
Bacterial Numbers Recovered
A comparison of the numbers of bacteria recovered in the two studies shows that the culturable count from the natural rock in the high impact area was 2 orders of magnitude higher than those recovered from the lower impact zones or the adjacent painted fiberglass surfaces (Table 1). Although the swab sampling technique used in these studies is a qualitative one, we have found in subsequent sampling trips that the magnitude of the counts has remained fairly consistent as has the difference between the high impact zone and lower impact zones (data not shown). The count for the painted fiberglass surface was lower than the adjacent rock; however, the significance of this count is debatable because only one set of counts was performed and the fiberglass surfaces are periodically washed with 10% bleach to remove the accumulated biofilm. At the time of sampling for this study, a biofilm was present, but the impact of the washing on the magnitude of the population is unknown.
Discussion
Painted Fiberglass Isolates
Seven of the nine isolates from the painted fiberglass surfaces were capable of growth on acrylic paint as a sole source of carbon. We compared these results to a number of studies that have investigated biodeterioration of ancient wall paintings found in both churches and hypogean environments [7–9, 20–22]. These studies, as well as a study by Hunter [13] on slime formation on Tygon tubing (which leaches plasticizers) in Lechuguilla Cave, led us to hypothesize that paint used to coat the fiberglass was serving as an organic substrate for a microbial slime-producing consortium that colonized the surface. We further hypothesized that establishment of this selective niche would allow dominance by a few heterotrophs able to utilize the paint and/or not be affected by paint toxicity. This hypothesis is supported by the fact that the diversity of the microbial community on the painted fiberglass was much lower than that on the adjacent rock surfaces, and that the majority of these isolates were found to be closely related to bacteria isolated from adjacent rock surfaces. In addition, three paint-degrading isolates (PF-B, PF-H, and PF-I) produced mucoid colonies (Table 2). Isolate PF-B is most closely related to Dyadobacter fermentens, which was originally isolated from maize stems under nitrogen-limiting conditions. D. fermentens is known to lack flagella, but produces copious slime when grown on nitrogen-limited agar (nitrogen is likely limited in the cave as well) [6]. The nearest relative of isolate PF-I is a Sphingomonas sp. Sphingomonads are known for their production of the anionic polysaccharide gum, gellan, which is thought to have a role in attachment and adhesion to surfaces [1]. Isolate PF-H was classified only to the phylum level of Bacteriodetes, but like PF-B and PF-I exhibited copious slime production in vitro. The mucoid nature and physiological behavior of these three isolates suggest that they are critical for establishment and maintenance of the biofilm observed on the painted fiberglass surfaces.
Bacterial Diversity as a Function of Human Impact
The cultured natural cave bacterial community (low and medium impact areas) is dominated by Firmicutes, whereas Proteobacteria dominate in communities exposed to high levels of human contact (Fig. 2). Both of these dominant phyla can be described as versatile. Members of these phyla are saprotrophs that are commonly found in soil and water environments. Each encompasses a wide range of metabolic capabilities resulting in the ability to grow on a variety of organic substances ranging from complex organic materials to simple sugars. A major difference between these phyla is their relative ability to withstand environmental stress. The Proteobacteria are not commonly found in environments that are characterized by severe pH, temperature, nutrient, or water tension stresses [19]. In contrast, Firmicutes are often found under more extreme conditions and, in fact, they are among the Prokaryotes that are most resistant to dessication and nutrient stress, particularly the bacilli recovered in this study because of their ability to form endospores [23].
The different physiological characteristics of Proteobacteria and Firmicutes led us to explore the ecological differences between the impact zones studied. As pH, temperature, and relative humidity (> 98%) are relatively constant throughout the cave, we qualitatively examined differences in organic matter inputs among the sampling sites. Prior to development and tourism, organic matter probably entered the cave mainly through two sources. The first source is dissolved organic carbon (DOC) and colloidal organic matter transported by a water flow. This water enters the cave through either vertical infiltration (dripping) from surface rainfall or through lateral infiltration (flooding) from adjacent washes that periodically run during the winter rains and summer monsoons that characterize the climate of the Sonoran Desert environment.
The second source of organic matter comes from plants and animals; bats are likely the largest source, but plant roots, crickets, and woodrats also contribute. Periodically, over the last 50,000 years, parts of the cave have been used as the annual maternity roost for a Myotis velifer colony. This colony is probably the major source of organic matter that maintains the heterotrophic food chain in portions of Kartchner Caverns [3, 27]. The organic input from animals and plants tends to be localized (near the cave opening and near the maternity roost), and it is composed of larger and more complex organic matter components than DOC and colloids associated with water flow.
The balance of organic input from different sources determines the total input in any area. Prior to development of the cave, the Rotunda–Throne area (designated as high impact due to human visitation) is thought to have had the lowest organic input of the three areas tested. The reason for this is that no bats roost in this area, the ceiling is too thick for root penetration, and the area is not subject to flooding. Organic matter was probably solely introduced through percolating water. In contrast, the moderate impact zone is thought to have received the greatest amount of organic input. The reason for this is that the area is relatively shallow and close to the entrance. Because of its position, it receives a variety of biological inputs, including bat guano, cricket guano, and potentially some roots. It also receives organic matter from flowing water, both from infiltration and minor flooding. Finally, the low impact zone is thought to have received intermediate amounts of organic matter. Like the high impact zone, this area is far from entrances that allow bats and other animals into the area in large numbers. However, in addition to almost constant percolation of water from above, the low impact zone floods with waters containing dissolved organics from the outside.
Because development of the cave has taken place, this natural organic carbon input gradient has been confounded by anthropogenic inputs that primarily affect the high impact zone. These inputs have occurred for the past decade as a result of humans entering the cave on a daily basis. Human visitation brings with it a variety of foreign substances such as hair, sloughed skin cells, sweat, skin oils, fibers from clothing, and dust particles that could potentially provide nutrients for microbes [14]. These particles slough off workers and tourists as they travel through the cave and are deposited in the cave (especially along travel routes such as tour trails). A second source of increased organic matter in the high impact zones is localized algal growth in the proximity of cave lights known as lampenflora. The addition of lights in the cave allows photosynthetic algae and cyanobacteria to grow in almost ideal humidity conditions. In fact, algal growth can be observed on a number of natural rock surfaces in the developed section of the cave due to added lights. Arizona State Parks has attempted to minimize the growth of algae through use of dimmers to decrease light intensity, timers to reduce duration of lights, and physical means and directed light focusing to reduce localized light intensities; however, some algal growth is inevitable. Thus, the daily input of anthropogenic organic carbon due to lint and photoautotrophic carbon may very well have reversed the natural organic carbon gradient, making the high impact zone the most carbon-rich as opposed to its original position as least carbon-rich. The increased organic matter may serve as food for the types of metabolically diverse microbes recovered from Kartchner Caverns. Unfortunately, no quantitative data are available concerning the absolute contributions of the various anthropogenic organic matter sources. Arizona State Parks has an ongoing lint deposition study in the Big Room, but preliminary results are not yet available.
The enhanced nutrient availability in the high impact zone is one possible explanation for the dominance of Proteobacteria in this area compared to their lower diversity in the moderate and low impact areas. Moreover, the types of Proteobacteria recovered from the high impact zone support this hypothesis. Closest relatives for isolates recovered from the high impact sites include Pseudomonas, Sphingomonas, and Alcaligenes, all known for their ability to degrade a wide diversity of organic substrates including aromatic compounds (e.g., [17]). We also isolated a close relative of Methylobacterium that has been reported to utilize a wide variety of carbon substrates and to be resident in diverse environments [12]. Not surprisingly, given that reported levels of nitrogen in cave minerals are very low [18], close relatives of Rhizobium and Sinorhizobium, both root-associated nitrogen-fixing bacteria, were obtained as well.
In terms of culturable microbial diversity, the impact of humans in Kartchner appears to be more indirect than direct. A variety of microbes unique to the microbiota of humans (e.g., staphylococci) are undoubtedly transported into the cave on a daily basis. However, these organisms do not appear to be long-term components of the cave rock community. Rather, the Proteobacteria that are predominant on the painted fiberglass and high impact natural cave rock surfaces are ubiquitously found in soils and bodies of water around the world. These results suggest that alteration of the microbiota at Kartchner is more likely the result of changes in physicochemical (i.e., lighting and organic matter inputs) cave conditions due to development of the cave than to exposure to human-associated change to microbiota brought in by tourists. Microbial communities in oligotrophic environments are particularly susceptible to such changes, and continued study of the Kartchner communities may provide additional insights into human impacts on cave microbial ecology. Moreover, it is possible that the changes documented here could be a harbinger of other changes. For that reason, Arizona State Parks has embarked on a microbial monitoring program.
A limited number of studies have looked at culturable bacterial populations in caves environments. Chelius and Moore [5], using R2A agar, cultured Actinobacteria and Proteobacteria isolates from a little visited site in Wind Cave (South Dakota, USA), but did not recover any Firmicutes. A major difference between this study and ours is that it focused on sediment rather than rock wall (mineral) samples. A study by Northup et al. [18] examined samples of ferromanganese deposits and enrichment cultures from these deposits in two caves near Carlsbad, NM: Lechuguilla (a deep, oligotrophic, little visited cave) and Spider (a shallow, smaller cave that has frequent visitors). Representative enrichment culture clones from both Lechuguilla and Spider Caves were from the same bacterial phyla found in the present study, Proteobacteria, Actinobacteria, and Firmicutes, although the distribution among and within these phyla differed. And finally, a report by Cacchio et al. [4] showed a large relative abundance of calcifying bacteria among the total cultured community recovered from carbonate speleothems in Cervo Cave (L'Aquila, Italy), ranging from 8% to 100% of the isolates obtained from the speleothems tested. The majority of the calcifying microbes isolated came from the genus Kocuria. It should be noted that our study also recovered Kocuria isolates from all sites along the impact gradient. In addition to Kocuria, Cacchio et al. [4] report calcifying activity in Acinetobacter, Renibacterium, and Bacillus isolates from their site. Of these genera, only Bacillus isolates were recovered from Kartchner Caverns during our study.
In summary, although we do not have information concerning the precave development microbial community in the high impact area, the culturable population distribution is markedly different in this area than in parts of the cave that are less traversed and less impacted physically (i.e., lighting) and chemically (i.e., sloughed skin cells and oils). We speculate that originally the high impact zone microbial community would have been very similar to the low and medium impact zones. Little is known about the significance of these isolates to critical cave processes such as speleothem formation, but the culture collection created by this study provides the opportunity to evaluate the role of bacteria in speloethem formation and to determine whether the changes observed here could have a negative impact on the living status of the cave. Continued monitoring of the recently opened Big Room, as well as implementation of a polyphasic approach employing both culturable and molecular techniques, may allow for more definitive assertions about the actual impact of human presence on the microbial communities of Kartchner Caverns.
References
Azeredo, J, Oliveira, R (2000) The role of exopolymers in the attachment of Sphingomonas paucimobilis. Biofouling 16: 59–67
Bodour, AA, Drees, KP, Maier, RM (2003) Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl Environ Microbiol 69: 3280–3287
Buecher, RH (1999) Microclimate study of Kartchner Caverns, Arizona. J Caves Karst Stud 61(2): 108–120
Cacchio, P, Contento, R, Ercole, C, Cappuccio, G, Martinez, MP, Lepidi, A (2004) Involvement of microorganisms in the formation of carbonate speleothems in the Cervo Cave (L'Aquila-Italy). Geomicrobiol J 21: 497–509
Chelius, MK, Moore, JC (2004) Molecular phylogenetic analysis of Archaea and bacteria in Wind Cave, South Dakota. Geomicrobiol J 21: 123–124
Chelius, MK, Triplett, EW (2000) Dyadobacter fermentens gen. nov., sp. nov., a novel Gram-negative bacterium isolated from surface-sterilized Zea mays stems. Int J System Evol Microbiol 50: 751–758
Gonzalez, I, Laiz, L, Hermosin B, Caballero, B, Incerti, C, Saiz-Jiminez, C (1999) Bacteria isolated from rock art paintings: the case of Atlanterra shelter (south Spain). J Microbiol Methods 36: 123–127
Groth, I, Vetterman, R, Schuetze, B, Schumann, P, Saiz-Jiminez, C (1999) Actinomycetes in karstic caves of northern Spain (Altamira and Tito Bustillo). J Microbiol Methods 36: 115–122
Gurtner, C, Heyrman, J, Pinar, G, Lubitz, W, Swings, J, Rolleke, S (2000) Comparative analyses of the bacterial diversity on two different biodeteriorated wall paintings by DGGE and 16S rDNA sequence analysis. Int Biodeter Biodegrad 46: 229–239
Hill, C, Forti, P (1997) Cave Minerals of the World, 2nd ed. National Speleological Society, Huntsville, AL
Hill, CA (1999) Mineralogy of Kartchner Caverns, Arizona. J Cave Karst Stud 61: 73–78
Hiraishi, A, Furuhata, K, Matsumoto, A, Koike, KA, Fukuyama, M, Tabuchi, K (1995) Phenotypic and genetic diversity of chlorine-resistant Methylobacterium strains isolated from various environments. Appl Environ Microbiol 61: 2099–2107
Hunter, A (2004) Persistent coliform contamination in Lechuguilla Cave pools. J Cave Karst Stud 66: 102–110
Jablonsky, P, Kraemer, S, Yett, B (1993) Lint in caves. In: Pate, DL (Ed.) Proc. 1993 National Cave Management Symposium, Carlsbad, NM, American Cave Conservation Association, Horse Cave, Kentucky, pp 73–81
Lane, DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E, Goodfellow, M (Eds.) Nucleic Acid Techniques in Bacterial Systematics. Wiley, New York, NY, pp 115–175
Murray, RGE, Doetsch, RN, Robinow, CF (1994) Determinitive and cytological light microscopy. In: Gerhardt, P (Ed.) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, DC, pp 31–32
Nohynek, LJ, Nurmiaho-Lassila, EL, Suhonen, EL, Busse, HJ, Mohammadi, M, Hantula, J, Rainey, F, Salkinoja-Salonen, MS (1996) Description of chlorophenol-degrading Pseudomonas sp. strains KF1T, KF3, and NKF1 as a new species of the genus Sphingomonas, Sphingomonas subarctica sp. nov. Int J Syst Bacteriol 46: 1042–1055
Northup, DE, Barns, SM, Yu, LE, Spilde, MN, Schelble, RT, Dano, KE, Crossey LJ, Connolly, CA, Boston, PJ, Natvig, DO, Dahm, CN (2003) Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ Microbiol 5: 1071–1086
Palleroni, NJ (1992) Introduction to the family Pseudomonadaceae. In: Balows, A, Trüper, HG, Dworkin, M, Harder, W, Schliefer, K-H. (Eds.) The Prokaryotes, 2nd ed, Vol III. Springer Verlag, NY, pp 3071–3103
Rolleke S, Muyzer G, Wawer, C, Wanner, G, Lubitz, W (1996) Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 62: 2059–2065
Schabereiter-Gurtner, C, Saiz-Jiminez, C, Piñar G, Lubitz, W, Rölleke, S (2002) Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in the Tito Bustillo cave, Spain, and on its Paleolithic paintings. Environ Microbiol 4: 392–400
Schabereiter-Gurtner, C, Saiz-Jiminez, C, Piñar, G, Lubitz, W, Rölleke, S (2004) Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonic and La Garma). FEMS Microbiol Ecol 47: 235–247
Slepecky, RA, Hemphill, HE (1992) The genus Bacillus—nonmedical. In: Balows, A, Trüper, HG, Dworkin, M, Harder, W, Schliefer, K-H (Eds.) The Prokaryotes, 2nd ed, Vol III. Springer Verlag, NY, p 1663
Toomey, RS III, Nolan, G (2005) Environmental change at Kartchner Caverns: trying to separate natural and anthropogenic changes. In: Proceed Biodiversity and Management of the Madrean Archipelago II: Connecting Mountain Islands and Desert Seas. USDA-Forest Service, RMRS-P 36: 264–270
Tufts, R, Tenen, G (1999) Discovery and history of Kartchner caverns, Arizona. J Cave Karst Stud 61: 44–48
Versalovic, J, Koeuth T, Lupski, JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19: 6823–6831
Welbourn, W (1999) Invertebrate cave fauna of Kartchner Caverns, Kartchner Caverns, Arizona. J Cave Karst Stud 61: 93–101
Acknowledgments
This work was supported in part by grants MCB-0604300 and CHE-013237 from the National Science Foundation and in part by a grant from the Arizona State Parks Board.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ikner, L.A., Toomey, R.S., Nolan, G. et al. Culturable Microbial Diversity and the Impact of Tourism in Kartchner Caverns, Arizona. Microb Ecol 53, 30–42 (2007). https://doi.org/10.1007/s00248-006-9135-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9135-8