Abstract
Stable-isotope probing (SIP) was used to identify acetate- or methanol-assimilating bacteria under nitrate-reducing conditions in activated sludge. A sludge sample obtained from wastewater treatment systems was incubated in a denitrifying batch reactor fed with synthetic wastewater containing [13C]acetate or [13C]methanol as the main carbon source and nitrate as the electron acceptor. We analyzed how growth of bacterial populations was stimulated by acetate or methanol as the external carbon source in nitrogen-removal systems. Most of the acetate- or methanol-assimilating bacteria identified by SIP have been known as denitrifiers in wastewater treatment systems. When acetate was used as the carbon source, 16S rRNA gene sequences retrieved from 13C-labeled DNA were closely related to the 16S rRNA genes of Comamonadaceae (e.g., Comamonas and Acidovorax) and Rhodocyclaceae (e.g., Thauera and Dechloromonas) of the Betaproteobacteria, and Rhodobacteraceae (e.g., Paracoccus and Rhodobacter) of the Alphaproteobacteria. When methanol was used as the carbon source, 16S rRNA gene sequences retrieved from 13C-DNA were affiliated with Methylophilaceae (e.g., Methylophilus, Methylobacillus, and Aminomonas) and Hyphomicrobiaceae. Rarefaction curves for clones retrieved from 13C-DNA showed that the diversity levels for methanol-assimilating bacteria were considerably lower than those for acetate-assimilating bacteria. Furthermore, we characterized nitrite reductase genes (nirS and nirK) as functional marker genes for denitrifier communities in acetate- or methanol-assimilating populations and detected the nirS or nirK sequence related to that of some known pure cultures, such as Alcaligenes, Hyphomicrobium, and Thauera. However, most of the nirS or nirK sequences retrieved from 13C-DNA were clustered in some unidentified groups. On the basis of 16S rRNA gene clone libraries retrieved from 13C-DNA, these unidentified nir sequences might be identified by examining the nir gene in candidates for true denitrifiers (e.g., the families Comamonadaceae, Hyphomicrobiaceae, Methylophilaceae, and Rhodobacteraceae).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biological nitrate and/or nitrite removal from wastewater is achieved by denitrification, which involves the reduction of nitrate, via nitrite and nitric oxide, to nitrous oxide or dinitrogen gas [49]. Denitrification requires an oxidizing nitrogen compound as the electron acceptor and a carbon source. For wastewater with a low carbon/nitrogen ratio or which lacks readily biodegradable carbon sources, various organic compounds acting as external carbon sources, such as acetate, ethanol, glucose, and methanol, are added to achieve a satisfactory degree of denitrification. Methanol has often been chosen because of its relatively low cost and the small amounts of sludge produced compared with other organic compounds, although only some bacteria can utilize methanol as a carbon source [27].
In recent years, molecular biological analysis has been applied to understanding microbial ecology in wastewater treatment systems. For example, the 16S rRNA gene-based approach is the most widely used technique for organism identification and community analysis because it provides phylogenetic information based on a large database of sequence information.
Denitrification ability is found in many bacterial species in a variety of taxonomic groups [49]. These bacteria have been found using primer sets specific for functional genes involved in denitrification: genes encoding periplasmic and membrane-bound nitrate reductases (napA and narG) [5, 8, 9], genes encoding cytochrome cd 1 and copper-containing nitrite reductases (nirS and nirK, respectively) [2, 3, 10, 18, 26, 31, 38, 47], nitric oxide reductase (norB) [4], and a gene encoding a nitrous oxide reductase (nosZ) [35, 36]. Polymerase chain reaction (PCR)-based methods using these primers revealed the diversity and community structure of denitrifiers in activated sludge [10, 38], marine sediment [2, 18, 35], and soil [26, 31, 36]. However, active denitrifiers in various environments cannot be identified by these methods.
Recently, molecular techniques using radio isotopes or stable isotopes in biomarkers have been developed and applied to identifying active microbial populations [1, 12, 15, 20–23, 28, 32, 33, 44]. The combination of microautoradiography and fluorescence in situ hybridization (MAR-FISH) is used to simultaneously determine the phylogenetic composition and in situ substrate uptake patterns of complex microbial populations in wastewater treatment systems [1, 12, 15]. Radajewski et al. [32] established the stable-isotope probing (SIP) technique for linking the identity of active bacteria with their function in complex natural environments. DNA synthesized during microbial growth on a substrate enriched with a heavy, stable isotope is sufficiently labeled to be separated from unlabeled DNA by CsCl density gradient centrifugation. To date, the SIP technique has been successfully applied to identifying the active methylotroph populations in oak forest soil [32, 33], active ammonia oxidizers in lake sediment [44], active phenol-degrading microbial populations in an aerobic industrial bioreactor [21], and active methanol-assimilating denitrifiers in wastewater treatment systems [20].
In this study, SIP analysis was applied to clarify how growth of bacterial populations was stimulated by acetate or methanol as the external carbon source under nitrate-reducing conditions. Moreover, we characterized the community structure and diversity of functional genes (nitrite reductase genes, nirS and nirK) in acetate- or methanol-assimilating bacterial populations for a better understanding of denitrifying consortia.
Methods
Incubation with [13C] Substrates under Nitrate-Reducing Conditions
Activated sludge samples were collected from an anoxic–oxic activated sludge system for removing carbon and nitrogen from municipal sewage. To identify the microbial populations of the original sludge, some of the collected activated sludge was stored at −80°C. A sludge pellet (wet weight 8 g) was suspended in synthetic wastewater containing [13C] substrates. In the acetate experiment, 13CH3 13COONa (99% 13C; Cambridge Isotope Laboratories, Inc., Cambridge, MA, USA) was added, and 13CH3OH (99% 13C; Sigma, St. Louis, MO, USA) was added in the methanol experiment. The composition of the synthetic wastewater was as follows: NaCl (6.60 mg/L), MgSO4·7H2O (8.20 mg/L), KH2PO4 (18.6 mg/L), KCl (13.4 mg/L), dextrin (30.5 mg/L), bactopeptone (65.5 mg/L), yeast extract (65.4 mg/L), meat extract (74.6 mg/L), NaHCO3 (191 mg/L), NaNO3 (607 mg/L, i.e., 100 mg of NO3-N/L), 13CH3OH (550 mg/L, i.e., 216 mg of 13C/L), and 13CH3 13COONa (691 mg/L, i.e., 216 mg of 13C/L). The synthetic wastewaters used for both the acetate and methanol experiments were purged with pure N2 gas (>99.9%) to eliminate the dissolved oxygen while maintaining the temperature at 25°C. During incubation periods (6 days), it was confirmed by using a dissolved oxygen meter TOX-98E (Toko Chemical Laboratories Co., Ltd., Tokyo, Japan) that dissolved oxygen concentration was almost zero. Furthermore, NaNO3 (100 mg of NO3-N/L) and [13C] substrates (100 mg of 13C/L) were added every 24 h. Sludge samples were collected in the acetate and methanol experiments every 24 h and stored at −80°C. Nitrate, nitrite, and acetate concentrations were measured with a DX-100 ion chromatograph (Dionex, Sunnyvale, CA, USA), and methanol concentrations were measured with a GC380 gas chromatograph (GL Science Inc., Tokyo, Japan).
DNA Extraction and Ultracentrifugation
Total DNA was extracted from 0.15 g (wet weight) sludge pellets using Isoplant (Nippon Gene, Toyama, Japan) according to the manufacturer's instructions. The DNA was purified using a phenol/chloroform/isoamyl alcohol (25:24:1) solution and was precipitated by adding ethanol and sodium acetate.
CsCl–ethidium bromide density gradient centrifugation (270,000 × g, 20 h, 20°C) was used for the separation and collection of 13C-DNA from 12C-DNA fraction as described by Radajewski et al. [33]. Ethidium bromide was extracted from DNA fractions with an equal volume of isoamyl alcohol saturated with water. Purified DNA was dialyzed in a large volume of Tris–EDTA (TE) buffer to remove CsCl, precipitated by adding ethanol and sodium acetate overnight at −20°C, and dissolved in 30 μL of TE buffer.
PCR Characterization
The following primer sets were used for PCR amplification: (1) eub341f-univ907r [24] for the amplification of 16S rRNA gene fragments, (2) nirS1F-nirS6R [3] for the amplification of nirS fragments, and (3) nirK1F-nirK5R [3] for the amplification of nirK fragments. The PCR mixture contained 0.5 μM of each primer, 200 μM of dNTP, 2.0 mM MgCl2 for the 16S rRNA gene and nirK and 1.5 mM MgCl2 for nirS, 2.5 U of rTaq DNA polymerase (Toyobo, Osaka, Japan) for the 16S rRNA gene and nirS and 1.25 U of rTaq DNA polymerase for the nirK, 2 μL of 10× PCR buffer for rTaq, and sterile water added to a final volume of 20 μL. The PCR amplifications of the 16S rRNA gene, nirS, and nirK were conducted in a model 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA) using the following protocols: (1) 16S rRNA gene: 2 min at 94°C, 25 cycles (40 s at 94°C, 40 s at 54°C, 40 s at 72°C), 2 min at 72°C; (2) nirS: 5 min at 94°C, 30 cycles (1 min at 94°C, 1 min at 54°C, 1 min at 72°C), 5 min at 72°C; (3) nirK: 5 min at 94°C, 30 cycles (30 s at 94°C, 40 s at 48°C, 40 s at 72°C), 5 min at 72°C. The presence of PCR products was confirmed by 2% agarose gel electrophoresis and the subsequent staining of the gels with ethidium bromide.
Cloning, Sequencing, Rarefaction Analysis, and Phylogenetic Analysis
PCR products were purified by eluting the bands from 2% agarose gels using a Wizard SV gel and a PCR cleanup system (Promega Corp., Madison, WI, USA). The PCR amplicons were cloned using a QIAGEN PCR cloning kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Then, single colonies were picked up by using a toothpick, and the toothpick was immersed in Insert Check Ready Solution (Toyobo) for a few seconds. Amplified inserts were run on 2% agarose gels. Samples including inserts of estimated sizes were used for the subsequent sequencing. A DNA insert was amplified and used as template DNA in a cycle sequencing reaction with a DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Biosciences, Freiburg, Germany) according to the manufacturer's instructions. The 16S rRNA gene and nir gene fragments were sequenced with an ABI PRISM 3100-Avant DNA Sequencing System (Applied Biosystems). 16S rRNA gene sequences with more than 99% identity were considered to belong to the same operational taxonomic unit (OTU). The nir sequences exhibiting more than 97% identity were considered to belong to the same OTU. Rarefaction calculations were carried out using Analytic Rarefaction software [version 1.3, Stratigraphy Laboratory, University of Georgia (http://www.uga.edu/strata/software/Software.html)]. A database search was conducted using BLAST from the DNA Data Bank of Japan (DDBJ). Sequences determined in this study and those retrieved from the database were aligned using Clustal W [40]. Phylogenetic trees were constructed using a neighbor-joining algorithm [34] and were displayed using TreeView [29].
Nucleotide Sequence Accession Numbers
16S rRNA gene sequences determined in this study were deposited under accession numbers AB205646–AB206036, nirS sequences were deposited under accession numbers AB162225–AB162256, and nirK sequences were deposited under accession numbers AB162309–AB162341.
Results
SIP of Denitrifying Consortia
Nitrate reduction was observed from the beginning of incubation in both the methanol and acetate experiments (data not shown). Acetate was consumed rapidly from the beginning of the incubation. In contrast, methanol was consumed slowly at the beginning of incubation and then rapidly after 2 days of incubation. Total DNA was extracted from sludge samples after 1 to 6 days of incubation and then ultracentrifuged. Double DNA bands were detected for the sludge sample incubated for 3 days with the [13C] substrate, although a single DNA band was detected after 2 days of incubation with the [13C] substrate. When total DNA extracted from the original sludge before substrate addition was ultracentrifuged as a control, a single DNA band was detected. The position of the single DNA band, which was detected from the sludge sample after 2 days of incubation or before substrate addition, was almost the same as that of the upper DNA band. These results showed that the lower DNA band was generated due to an increase in buoyant density with the incorporation of 13C into the DNA. Therefore, it was judged that the upper band corresponded to 12C-DNA and the lower band to 13C-DNA.
Evaluation of Bacterial Populations Based on the 16S rRNA Gene Clone Library
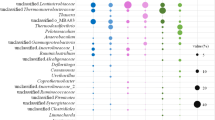
13C-DNA and 12C-DNA collected from sludge samples incubated for 3 days were chosen as the samples for the 16S rRNA gene clone library in both methanol and acetate experiments. Five 16S rRNA gene clone libraries were generated. The relative abundance of an individual clone obtained from each sample is shown as a histogram (Fig. 1). From acetate samples, 127 clones from 12C-DNA and 201 clones from 13C-DNA were obtained. In methanol samples, 130 clones from 12C-DNA and 137 clones from 13C-DNA were obtained. In addition, the 16S rRNA gene clone library (131 clones) was also constructed from the original sludge before substrate addition. In contrast to clone libraries retrieved from the 12C-DNA fraction and the original sludge before incubation, clone libraries from the 13C-DNA fraction showed a completely different bacterial composition and diversity. In these experiments, the identification of active population using SIP is a prerequisite for the incorporation of 13C into the DNA. Consequently, the clone libraries from the 12C-DNA fraction corresponded to organisms that did not grow with acetate or methanol under these conditions, i.e., those that were present in the original sludge and those that grew with carbon sources other than acetate and methanol.
Most of the clones were affiliated with the Proteobacteria in the clone libraries from 12C-DNA fractions and the original sludge. The remaining clones were affiliated with the Bacteroidetes, Chloroflexi, Actinobacteria, Firmicutes, Nitrospairae, Acidobacteria and Planctomycetes. In particular, many clones from the 12C-DNA were affiliated with the Actinobacteria, including high G + C Gram-positive bacteria. In both the methanol and acetate samples, bacterial community structures were obviously different between 12C-DNA and 13C-DNA libraries. These findings confirmed the successful separation of 12C-DNA and 13C-DNA corresponding to an increase in buoyant density with the incorporation of 13C into the DNA, and not to a difference in G + C content.
About 90% of the clones retrieved from the 13C-DNA fraction were affiliated with Proteobacteria. This showed that Proteobacteria actively consume acetate or methanol as a carbon source under nitrate-reducing conditions. The bacterial community structures in the 13C-DNA fraction were obviously different between acetate and methanol samples. The phylogenetic affiliations of all analyzed clones are summarized in Table 1.
Phylogenetic Analysis of Acetate-Assimilating Bacteria in Denitrifying Consortium
A total of 201 clones from the 13C-DNA fraction were assigned to 71 different OTUs (13C-A1 to 13C-A71). The cloned sequence types with more than 99% identity were considered to belong to the same OTU. The placements of OTUs within the 13C-DNA clone library are shown in phylogenetic trees (Figs. 2, 3, 4 and 5). The 13C-DNA clone library showed that acetate-assimilating bacteria were found in various phylogenetic groups. About 25% (50 clones) of all clones, including 11 different OTUs, were related to Comamonadaceae of the Betaproteobacteria, such as the genera Comamonas and Acidovorax. The OTU 13C-A7 contained many clones (18 clones) closely related to Comamonas denitrificans. A further 20.4% (41 clones) of all clones, including 17 OTUs, were clustered within Rhodocyclaceae of the Betaproteobacteria, such as the genera Thauera, Dechloromonas, Zoogloea and Azonexus. In particular, many clones related to the genus Thauera (e.g., Thauera aromatica, Thauera selenatis, and Thauera terpenica) were found (25 clones). Furthermore, 10 different OTUs (29 clones) were related to members of Rhodobacteraceae of the Alphaproteobacteria, such as the genera Paracoccus and Rhodobacter, and OTU 13C-A71 (22 clones) was very similar to Arcobacter cryaerophilus of the Epsilonproteobacteria. Nine different OTUs (23 clones) were clustered within the Gammaproteobacteria, including the genera Pseudomonas, Aeromonas, Thermomonas, and Stenotrophomonas. Furthermore, the remaining clones of this 13C-DNA clone library were seemingly distributed randomly throughout the bacterial domain, including the Actinobacteria, Bacteroidetes, Chlorflexi, and Firmicutes.
Phylogenetic affiliation of the alphaproteobacterial clones retrieved from each 13C-DNA by neighbor-joining analysis. The partial 16S rRNA gene sequences obtained from acetate and methanol samples are labeled as 13C-A and 13C-M, respectively. The number of clones assigned to each sequenced OTU with more than 99% identity is shown in parentheses. Geobacter metallireducens (accession no. L07834) are used as the outgroup. Bootstrap values >750 (closed circles) and in the range of 500 to 750 (open circles) are indicated at branch points. Scale bar = 5% nucleotide substitution.
Phylogenetic affiliation of the betaproteobacterial clones retrieved from each 13C-DNA by neighbor-joining analysis. The partial 16S rRNA gene sequences obtained from acetate and methanol samples are labeled as 13C-A and 13C-M, respectively. The number of clones assigned to each sequenced OTU with more than 99% identity is shown in parentheses. Geobacter metallireducens (accession no. L07834) are used as the outgroup. Bootstrap values >750 (closed circles) and in the range of 500 to 750 (open circles) are indicated at branch points. Scale bar = 5% nucleotide substitution.
Phylogenetic affiliation of the gamma-, delta-, and epsilonproteobacterial clones retrieved from each 13C-DNA by neighbor-joining analysis. The partial 16S rRNA gene sequences obtained from acetate and methanol samples are labeled as 13C-A and 13C-M, respectively. The number of clones assigned to sequenced OTU with more than 99% identity is shown in parentheses. Haloarcula vallismortis (accession no. D80851) are used as the outgroup. Bootstrap values >750 (closed circles) and in the range of 500 to 750 (open circles) are indicated at branch points. Scale bar = 10% nucleotide substitution.
Phylogenetic affiliation of the remaining bacterial clones except proteobacterial clones retrieved from each 13C-DNA by neighbor-joining analysis. The partial 16S rRNA gene sequences obtained from acetate and methanol samples are labeled 13C-A and 13C-M, respectively. The number of clones assigned to each sequenced OTU with more than 99% identity is shown in parentheses. Haloarcula vallismortis (accession no. D80851) are used as the outgroup. Bootstrap values >750 (closed circles) and in the range of 500 to 750 (open circles) are indicated at branch points. Scale bar = 10% nucleotide substitution.
Phylogenetic Analysis of Methanol-Assimilating Bacteria in Denitrifying Consortium
One hundred thirty-seven clones from the 13C-DNA were assigned to 22 different OTUs (OTU 13C-M1 to 13C-M22). The cloned sequence types with more than 99% identity were considered to belong to the same OTU. The phylogenetic positioning of OTUs obtained from the 13C-DNA clone library are shown in phylogenetic trees (Figs. 2 to 5). The 13C-DNA clone library showed two predominant groups related to known methylotrophs. Of all the clones, 63%, including six different OTUs, were related to Methylophilaceae of the Betaproteobacteria, such as Methylophilus, Methylobacillus, and Aminomonas. Most of the Methylophilaceae are obligate methylotrophs, which have the capacity to grow in the presence of methanol or methylamine but not in the presence of methane. A further 21% of all the clones, including four OTUs, were clustered within Hyphomicrobiaceae of the Alphaproteobacteria. The remaining clones in this library were clustered in Alphaproteobacteria (e.g., Blastobacter denitrificans), Betaproteobacteria (e.g., Ralstonia pickettii, Thauera aromatica, Comamonas sp.), Gammaproteobacteria (e.g., Ewingella americana), and Actinobacteria (e.g., Carnobacterium piscicola and Nocardioides sp.).
Nitrite Reductase Genes (nirS and nirK)
For the characterization of functional genes in acetate- and methanol-assimilating denitrifiers, the 13C-DNA fraction isolated in both experiments, in which activated sludge was incubated for 3 days, was used as a template for PCR amplification using the nirS- and nirK-specific primer pairs. Partial nirS and nirK sequences were determined and compared with those in the DDBJ database using BLAST. The phylogenetic analysis performed by the neighbor-joining method for all of the nirS and nirK clone sequences revealed that the denitrifying populations strongly depended on the type of organic carbon source (Figs. 6 and 7).
Neighbor-joining analysis of partial nirS gene products (261 amino acids) from the 13C-DNA retrieved from acetate samples (S-A) and methanol samples (S-M). The number of clones assigned to each nirS sequence with more than 97% identity is shown in parentheses. nirN from Pseudomonas aerginosa (accession no. D84475) was used as the outgroup. Bootstrap values >750 (closed circles) and in the range of 500 to 750 (open circles) are indicated at branch points. Scale bar = 10% nucleotide substitution.
Neighbor-joining analysis of partial nirK gene products (171 amino acids) from the 13C-DNA retrieved from acetate samples (K-A) and methanol samples (K-M). The number of clones assigned to each nirK sequence with more than 97% identity is shown in parentheses. aniA from Neisseria gonorrhoeae (accession no. M97926) was used as the outgroup. Bootstrap values >750 (closed circles) and in the range of 500 to 750 (open circles) are indicated at branch points. Scale bar = 10% nucleotide substitution.
(a) Nitrite Reductase Genes (nirS and nirK) in Acetate-Assimilating Populations
One hundred seventeen nirS clones retrieved from the 13C-DNA were assigned to 27 OTUs (S-A1 to S-A27). About 88% of the nirS clones belonged to cluster II and were related to a nirS mRNA clone detected from estuarine sediment [26]. In particular, OTU S-A24 (56 clones) was the most dominant in the 13C-DNA retrieved from acetate samples. Cluster I was predominantly composed of nirS clones from acetate samples, which were related to the nirS genes of the genus Thauera (e.g., T. aromatica and T. terpenica). The nirS clones in cluster V were related to the nirS clone detected in cyanobacterial aggregates (AJ457201). Only nirS clones from the incubation with acetate medium were found in clusters III and VI. The nirS clones in cluster III were related to the nirS genes of Azospirillum brasilense (AJ224912) and the nirS clones in cluster VI were related to the nirS genes of Ralstonia eutropha (X91394).
Eighty-seven nirK clones retrieved from the 13C-DNA were assigned to 27 OTUs (K-A1 to K-A29). Each of the nirK clones from the incubation with acetate medium belonged to all nirK clusters except for cluster V (I to IV and VI–XI). Most of the nirK clones showed less than 90% identity with nirK genes of isolated denitrifiers, such as Mesorhizobium, Pseudomonas, and Rhizobium. OTU K-A2 was highly related to the nirK genes of Alcaligenes faecalis (D13155). In the nirK clone library, the dominant OTUs were K-A3 (9 clones), K-A4 (14 clones), K-A17 (11 clones) and K-A19 (9 clones). OTUs K-A3 and K-A17 were related to the nirK clones detected in activated sludge and soil. OTU K-A4 was related to the nirK gene of Alcaligenes sp. (AB046603), and OTU K-A19 was related to the nirK gene of Pseudomonas sp. (M97294).
(b) Nitrite Reductase Genes (nirS and nirK) in Methanol-Assimilating Populations
Eighty-one nirS clones retrieved from the 13C-DNA were assigned to five OTUs (S-M1 to S-M5). About 96% of the total nirS clones belonged to cluster II and were related to a nirS mRNA clone detected from estuarine sediment [26]. In particular, OTU S-M4 (75 clones) was the most dominant in the 13C-DNA clone library generated from methanol samples. The nirS gene sequences of OTU S-M5 (1 clone) belonging to cluster I were related to the nirS genes of T. aromatica (AY078257). The remaining clones were related to the nirS clones detected in a cyanobacteria aggregate (AJ457201 and AJ457200).
Eighty nirK clones retrieved from the 13C-DNA were assigned to four OTUs (K-M1 to K-M4). The nirK clones from methanol samples were found in clusters III and V. OTU K-M4 (1 clone) was closely related to the nirK genes of Hyphomicrobium zavarzinii (AJ224902), and the remaining clones were related to the nirK genes of Rhizobium hedysari (U65658). In particular, K-M1 (71 clones) was the most dominant clone in the 13C-DNA clone library generated from methanol samples.
Rarefaction Analysis
The diversity of the 16S rRNA gene clones and nir clones isolated from acetate and methanol samples and the original sludge sample was evaluated by rarefaction analysis (Fig. 8). The 16S rRNA gene clones retrieved from the 13C-DNA indicated that the diversity level of methanol-assimilating bacteria was much lower than that of acetate-assimilating bacteria. Similarly, rarefaction analyses of nirS and nirK clones showed that the diversity level of methanol-assimilating denitrifiers was much lower than that of acetate-assimilating denitrifiers. There were no significant differences between the diversity levels of the 16S rRNA gene clones retrieved from the original sludge sample and the 12C-DNA in both methanol and acetate samples. Therefore, these results implied that the 3 days of incubation in the synthetic wastewater had no significant effects on the bacterial diversity.
Discussion
In this study, 16S rRNA gene- and functional gene (nitrite reductase gene)-targeted analysis combined with SIP revealed the identity and carbon uptake pattern of active denitrifying populations in activated sludge. Our results showed that the type of organic carbon source, such as acetate or methanol, had a strong effect on the active denitrifying populations. As for the community structure and ecological aspects of methanol-assimilating denitrifiers, some members of the genera Hyphomicrobium, Paracoccus, Rhodobacter, Blastobacter, and Hydrogenophaga have been reported as methanol-assimilating denitrifiers in freshwater environments [16, 17, 25]. However, active methanol-assimilating bacteria in a denitrifying consortium might have been missed because these members were identified by culture-dependent methods. Therefore, it is necessary to characterize active methanol-assimilating bacteria in a denitrifying consortium by molecular techniques linking microbial function with taxonomic identity [14]. Thus, we used SIP in the identification of active methanol-assimilating bacteria under nitrate-reducing conditions and found the existence of two predominant methanol-assimilating populations in the 13C-DNA clone library from methanol samples. One major phylotype was related to the members of the Methylophilaceae of the Betaproteobacteria, such as the genera Methyolphilus, Methylobacillus, and Aminomonas. Maneesha et al. also reported on a methanol-fed denitrifying community in activated sludge using the SIP and MAR-FISH techniques [20]. In their study, the dominant 16S rRNA gene phylotype in the 13C-DNA clone library was closely related to those of the obligate methylotroph of Methylophilaceae, most of which assimilated [14C]methanol in the MAR-FISH analysis. The other major phylotype was related to the genus Hyphomicrobium. The genus Hyphomicrobium (hyphomicrobia) consists of restricted facultative methylotrophs and has been found in soils, aquatic habitats, and a sewage treatment plant [30]. The diversity of hyphomicrobia in a sewage treatment plant and an adjacent receiving lake has been examined by Holm et al. [11]. An important characteristic of certain hyphomicrobia is their ability to grow with methanol as the carbon source and nitrate as the terminal electron acceptor. Moreover, we also found some clones related to B. denitrificans, which was reported as a methanol-assimilating denitrifier [41]. Hence, it was confirmed that SIP could specifically identify a methanol-assimilating population under nitrate-reducing conditions in a complex microbial community, such as activated sludge.
No clones related to the members of the family Rhodobacteraceae (e.g., Paracoccus and Rhodobacter), which were reported as methanol-assimilating denitrifiers, were detected in the 13C-DNA clone library generated from methanol samples. However, a number of clones related to the family Rhodobacteraceae were detected in the 13C-DNA clone library from acetate samples and in the 12C-DNA clone library from both acetate and methanol samples. Seven sequences obtained from the 12C-DNA of the acetate samples belonged to cluster A, and 13 sequences obtained from the 12C-DNA of the methanol samples belonged to cluster A (9 clones) and cluster C (4 clones) (Fig. 2). These results implied that these bacterial groups preferred other organic compounds (e.g., acetate, dextrin, and peptone) to methanol as carbon sources. Indeed, these bacterial groups can utilize various organic compounds for denitrification [13]. On the other hand, Claus and Kutzner [6] also reported that the members of Paracoccus grow poorly with methanol as the carbon source. Furthermore, they assumed that the growth of Paracoccus is supported by formate, which is produced by other bacteria (such as hyphomicrobia), or by organic compounds excreted by other bacteria. This assumption might lead to one limitation of SIP, that is, labeled intermediates and products might become incorporated into the DNA of other microbial communities when primary consumers metabolize the original substrates and excrete labeled metabolites. However, in this study, we considered this limitation unlikely because no clone obtained from the 13C-DNA isolated from methanol samples was related to the genus Paracoccus.
In denitrifying functional gene analysis of methanol-assimilating bacterial populations, a nirK clone highly related to the nirK gene of H. zavarzinii was detected only in the 13C-DNA retrieved from methanol samples. However, most of the nirS and the nirK clones were not closely related to the nir sequences of any known cultivated bacteria or nir clones found in various environments. Because the denitrification ability of methylotrophs was not previously characterized, there is a lack of nirS and/or nirK gene sequence data for most of the methylotrophs in the database. This might support the finding that many nirS and nirK clones found in this study, such as nirS clones S-M1 to S-M4 and nirK clones K-M1 to K-M3, were not related to any of the nirS and nirK gene sequences registered in the database. In future studies, we should obtain nir sequences from some pure cultures or isolates belonging to methanol-assimilating denitrifiers (e.g., Methylophilaceae and Hyphomicrobiaceae) identified by 16S rRNA gene clone libraries generated from the 13C-DNA clone library.
In contrast to methanol as an external carbon source, acetate was consumed quickly from the beginning of incubation. This implied that numerous heterotrophic bacteria in activated sludge can take up acetate under denitrifying conditions. MAR-FISH analysis conducted by Nielsen and Nielsen [22] revealed that wastewater treatment systems with nitrogen removal have relatively high numbers of acetate-assimilating bacteria (53–93%). The phylogeny of 16S rRNA gene sequences retrieved from the 13C-DNA indicated that a diverse range of bacteria in the activated sludge assimilated acetate under nitrate-reducing conditions. From the viewpoint of the number of clones retrieved from the 13C-DNA, the dominant acetate-assimilating bacteria were the Acidovorax-related groups (28 clones), Paracoccus-related groups (26 clones), Thauera-related groups (24 clones), Arcobacter-related groups (22 clones), Comamonas-related groups (19 clones), and Dechloromonas-related groups (8 clones). In previous studies, some denitrifying organisms in these genera, except for the genus Arcobacter, were isolated from activated sludge and were found to have the ability to degrade various xenobiotic compounds (e.g., aromatic compounds, hydrocarbons, and chlorinated compounds) under nitrate-reducing conditions [7, 13, 21, 45, 48]. Wagner and Loy reported that the Azoarcus–Thauera group of Betaproteobacteria represents numerous denitrifiers in wastewater treatment systems [43]. As for the genus Arcobacter, A. cryaerophilus was shown to be abundant in activated sludge [37]. Some Arcobacter spp. were shown to be sulfide oxidizers (with the production of sulfur), and it has been suggested that they play a role in the sulfur cycle by reoxidizing sulfide formed by microbial sulfate or sulfur reduction [39, 42, 46]. Teske et al. reported that Arcobacter spp. use acetate as the electron donor and nitrate as the electron acceptor [39]. However, to date, their denitrification ability in the environment has not been well documented.
Most of the acetate- or methanol-assimilating bacteria identified by SIP have been known as denitrifiers in wastewater treatment systems. There were interesting findings for 16S rRNA gene and nitrite reductase gene clone libraries in 13C-DNA clone libraries. The Thauera-related groups and Hyphomicrobium-related groups were detected in both 16S rRNA gene- and nitrite reductase gene clone libraries. However, no nirS or nirK clone retrieved from 13C-DNA had nir sequences related to the Paracoccus isolates, which were found to be dominant by 16S rRNA gene analysis, whereas nirK clones retrieved from 13C-DNA were highly related to A. faecalis (99% identity), which was not detected by 16S rRNA gene analysis. Although it may not be appropriate to compare the results obtained by the 16S rRNA gene- and the nitrite reductase gene-based approach, we discuss possible explanations for these findings below. First, the 16S rRNA gene approach detects all organisms that are active under conditions that are nitrate reducing or anaerobic (e.g., fermentation), not only under denitrifying conditions. Second, there is insufficient analysis of a statistically significant number of clones required for understanding complex communities. In this study, some rarefaction curves did not reach a plateau; reaching a plateau would indicate that the diversity was sufficiently analyzed. Third, there may be bias due to PCR amplification and degenerate primers. Lueders and Friedrich [19] reported that the degeneracy of primers has a considerable impact on bacterial diversity. They showed a decrease in bacterial diversity with increase in annealing temperature within a bacterial community model. However, highly degenerate primers often have to be used for functional genes to cover a wide phylogentic range because the sequence conservation of functional genes in comparison to 16S rRNA genes is constrained by the genetic code and its variability. Therefore, we should carefully characterize microbial community structures obtained by PCR-based analysis using highly degenerate primers, which might enhance the bias of PCR amplification. Other factors might be attributed to a lack of registered nir sequences of isolates with phylogenetic information from the database and to horizontal gene transfer. For instance, most of the nirS sequences registered in the database are retrieved from uncultured clones. Although only a part of the sequences are retrieved from pure cultures, most of these sequences are affiliated with limited bacterial groups (e.g., Alcaligenes, Marinobacter, Pseudomonas, Paracoccus, and Thauera). Thus, the primer sets used for detecting denitrifying bacteria, nirS1F–nirS6R and nirK1F–nirK5R, were designed on the basis of a limited number of sequences, mainly from laboratory strains. These primers were most commonly used, but this may need to be reassessed. Therefore, denitrifying gene sequences need to be obtained from many different isolates whose phylogenetic 16S rRNA positions have been identified. In particular, the denitrifying genes of candidates for true denitrifiers (e.g., the families Comamonadaceae, Hyphomicrobiaceae, Methylophilaceae, and Rhodobacteraceae) should be characterized because these bacteria are probably important populations in the nitrogen-removal system.
As for the horizontal gene transfer, Song and Ward reported an interesting correlation between 16S rRNA and nitrite reductase genes (nirS) and discussed the possibility of the horizontal gene transfer of nirS [38]. Most of the nirS genes from halobenzoate-degrading denitrifiers affiliated with the genera Thauera, Acidovorax, and Azoarcus of Betaproteobacteria are related to the nirS gene of Pseudomonas stutzeri of the Gammaproteobacteria, although phylogenetic relationships based on 16S rRNA genes from these genera are very different. The authors said that this might provide evidence for horizontal gene transfer of nirS among the genera Pseudomonas, Thauera, Acidovorax, and Azoarcus [38]. However, it was difficult to infer the horizontal gene transfer from our data sets at the community level.
This study showed how bacterial communities could be stimulated by external carbon sources (e.g., acetate and methanol) in denitrifying tanks. Moreover, SIP can be used for screening candidates for true denitrifiers from complex bacterial communities, such as activated sludge. Furthermore, the combination of SIP and other molecular techniques, such as MAR-FISH and real-time PCR, should significantly enhance our understanding of active denitrifying bacteria in activated sludge. Thus, in the future, the understanding of acetate- or methanol-assimilating bacteria under nitrate reducing conditions will be useful for the efficient and stable operation and suitable maintenance of nitrogen-removal systems.
References
Andreasen, K, Nielsen, PH (1997) Application of microautoradiography to the study of substrate uptake by filamentous microorganisms in activated sludge. Appl Environ Microbiol 63: 3662–3668
Braker, G, Ayala-del Rio, HL, Devol, AH, Fesefeldt, A, Tiedje, JM (2001) Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl Environ Microbiol 67: 1893–1901
Braker, G, Fesefeldt, A, Witzel, KP (1998) Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64: 3769–3775
Braker, G, Tiedje, JM (2003) Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl Environ Microbiol 69: 3476–3483
Cheneby, D, Hallet, S, Mondon, M, Martin-Laurent, F, Germon, JC, Philippot, L (2003) Genetic characterization of the nitrate reducing community based on narG nucleotide sequence analysis. Microb Ecol 46: 113–121
Claus, G, Kutzner, HJ (1985) Denitrification of nitrate and nitric acid with methanol as carbon source. Appl Microbiol Biotechnol 22: 378–381
Coates, JD, Chakraborty, R, Lack, JG, O'Connor, SM, Cole, KA, Bender, KS, Achenbach, LA (2001) Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411: 1039–1043
Flanagan, DA, Gregory, LG, Carter, JP, Karakas-Sen, A, Richardson, DJ, Spiro, S (1999) Detection of genes for periplasmic nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol Lett 177: 263–270
Gregory, LG, Karakas-Sen, A, Richardson, DJ, Spiro, S (2000) Detection of genes for membrane-bound nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol Lett 183: 275–279
Hallin, S, Lindgren, PE (1999) PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol 65: 1652–1657
Holm, NC, Gliesche, CG, Hirasch, P (1996) Diversity and structure of Hyphomicrobium populations in a sewage treatment plant and its adjacent receiving lake. Appl Environ Microbiol 62: 522–528
Ito, T, Nielsen, JL, Okabe, S, Watanabe, Y, Nielsen, PH (2002) Phylogenetic identification and substrate uptake patterns of sulfate reducing bacteria inhabiting an oxic–anoxic sewer biofilm determined by combining microautoradiography and fluorescence in situ hybridization. Appl Environ Microbiol 68: 356–364
Kelly, DP, Rainey, FA, Wood, AP (2001) The genus Paracoccus. In: Prokaryotes, An Evolving Electronic Resource for the Microbiological Community, Release 3.7, 3rd edn. Springer-Verlag, New York. http://link.springer-ny.com/link/service/books/10125/. 2 Nov.
Labbe, N, Juteau, P, Parent, S, Villemur, R (2003) Bacterial diversity in a marine methanol-fed denitrification reactor at the Montreal biodome, Canada. Microb Ecol 46: 12–21
Lee, N, Nielsen, PH, Andreasen, KH, Juretschko, S, Nielsen, JL, Schleifer, KH, Wagner, M (1999) Combination of fluorescence in situ hybridization and microautoradiography—a new tool for structure–function analyses in microbial ecology. Appl Environ Microbiol 65: 1289–1297
Lemmer, H, Zaglaure, A, Metzner, G (1997) Denitrification in a methanol-fed fixed-bed reactor. Part 1: Physico-chemical and biological characterization. Water Res 31: 1897–1902
Lemmer, H, Zaglaure, A, Neef, A, Meier, H, Amann, R (1997) Denitrification in a methanol-fed fixed-bed reactor. Part 2: Composition and ecology of the bacterial community in the biofilm. Water Res 31: 1903–1908
Liu, X, Tiquia, SM, Holguin, G, Wu, L, Nold, SC, Devol, AH, Luo, K, Palumbo, AV, Tiedje, JM, Zhou, J (2003) Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl Environ Microbiol 69: 3549–3560
Lueders, T, Friedrich, M (2003) Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69: 320–326
Maneesha, PG, Hugenholtz, P, Daimes, H, Wagner, M, Keller, J, Blackall, LL (2004) Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization–microautoradiography to study a methanol-fed denitrifying microbial community. Appl Environ Microbiol 70: 588–596
Manefield, M, Whiteley, AS, Griffiths, RI, Bailey, MJ (2002) RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol 68: 5367–5373
Nielsen, JP, Nielsen, PH (2002) Quantification of functional groups in activated sludge by microautoradiography. Water Sci Technol 46: 389–395
Morris, SA, Radajewski, S, Willison, TW, Murrel, JC (2002) Identification of the functionally active methanotroph population in a peat soil microcosm by stable isotope probing. Appl Environ Microbiol 68: 1446–1453
Muyzer, G, Brinkhoff, T, Nubel, U, Santegoeds, C, Schafer, H, Wawer, C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans, ADL, v. Elsas, JD, de Bruijin, FJ (Eds.) Molecular Microbial Ecology Manual, Kluwer Academic Publishers, Dordrecht, the Netherlands, 3.4.4., pp 1–27
Neef, A, Zaglaure, A, Meier, H, Amann, R, Lemmer, H, Schleifer, KH (1996) Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol 62: 4329–4339
Nogales, B, Timmis, KN, Nedwell, DB, Osborn, AM (2002) Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription–PCR amplification from mRNA. Appl Environ Microbiol 68: 5017–5025
Nyberg, U, Aspegren, H, Andersson, B, Jansen, J, Cour, L, Villadsen, IS (1992) Full-scale application of nitrogen removal with methanol as carbon source. Water Sci Technol 26(5–6): 1077
Padmanabhan, P, Padmanabhan, S, Derito, C, Gray, A, Gannon, D, Snape, JR, Tsai, CS, Park, W, Jeon, C, Madsen, EL (2003) Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol 69: 1614–1622
Page, RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358
Poindexter, JS (1992) Dimorphic prosthecate bacteria: the genera Caulobacter, Asticcacaulis, Hyphomicrobium, Pedomicrobium, Hyphomonas, and Thiodendron. In: Balows, A, Truper, HG, Dworkin, M, Harder, W, Schleifer, K-H (Eds.) The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Application, 2nd edn. Springer-Verlag, New York, pp 2176–2196
Prieme, A, Braker, G, Tiedje, JM (2002) Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl Environ Microbiol 68: 1893–1900
Radajewski, S, Ienson, P, Parekh, NR, Murrell, JC (2000) Stable-isotope probing as a tool in microbial ecology. Nature 403: 646–649
Radajewski, S, Webster, G, Reay, DS, Morris, SA, Ineson, P, Nedwell, DB, Prosser, JI, Murrel, JC (2002) Identification of active methylotroph populations in an acidic forest soil by stable isotope probing. Microbiology 148: 2331–2342
Saitou, N, Nei, M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425
Scala, D, Kerkhof, LJ (1998) Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett 162: 61–68
Scala D, Kerkhof, LJ (1999) Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol 65:1681–1687
Snaidr, J, Amann, R, Huber, I, Ludwig, W, Schleifer, KH (1997) Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol 63: 2884–2896
Song, B, Ward, BB (2003) Nitrite reductase genes in halobenzoate degradation denitrifier. FEMS Microbiol Ecol 43: 349–357
Teske, A, Sigalevich, P, Cohen, Y, Muyzer, G (1996) Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl Environ Microbiol 62: 4210–4215
Thompson, JD, Higgins, DG, Gibson, TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequencing weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680
Trotsenko, YA, Doronina, NV, Hirsch, P (1989) Genus Blastobacter Zavarzin 1961, 962AL. In: Staley, JT, Bryant, MP, Pfennig, N, Holt, JG (Eds.) Bergey's Manual of Systematic Bacteriology, Vol 3. Williams & Wilkins Co., Baltimore, pp 1963–1968
Voordouw, G, Armstrong, SM, Reimer, MF, Fouts, B, Telang, AJ, Shen, Y, Gevertz, D (1996) Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl Environ Microbiol 62: 1623–1629
Wagner, M, Loy, A (2002) Bacterial community composition and function in sewage treatment systems. Curr Opin Biotechnol 13: 218–227
Whitby, CB, Hall, G, Pickup, R, Saunders, JR, Ineson, P, Parekh, NR, McCarthy, A (2001) 13C incorporation into DNA as a means of identifying the active components of ammonia-oxidizer populations. Lett Appl Microbiol 32: 398–401
Willems, A, Vos, P (2001) The genus Comamonas. In: Prokaryotes, An Evolving Electronic Resource for the Microbiological Community, 3rd edn., release 3.7, Springer-Verlag, New York. http://link.springer-ny.com/link/service/books/10125/. Cited 2 Nov 2001
Wirsen, CO, Sievert, SM, Cavanaugh, CM, Molyneaux, SJ, Ahmad, A, Taylor, LT, DeLong, EF, Taylor, CD (2002) Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl Environ Microbiol 68: 316–325
Yoshie, S, Noda, N, Tsuneda, S, Hirata, A, Inamori, Y (2004) Salinity decreases nitrite reductase gene diversity in denitrifying bacteria of wastewater treatment systems. Appl Environ Microbiol 70: 3152–3157
Zhao JS, Ward, OP (1999) Microbial degradation of nitrobenzene and mono-nitrophenol by bacteria enriched from municipal activated sludge. Can J Microbiol 45: 427–432
Zumft, WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Osaka, T., Yoshie, S., Tsuneda, S. et al. Identification of Acetate- or Methanol-Assimilating Bacteria under Nitrate-Reducing Conditions by Stable-Isotope Probing. Microb Ecol 52, 253–266 (2006). https://doi.org/10.1007/s00248-006-9071-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9071-7