Abstract
Pseudomonas aeruginosa is an opportunistic pathogen responsible for morbidity and mortality in humans, animals, and plants. This bacterium has been regarded to be widely present in terrestrial and freshwater environments, but not in open ocean environments. Our purpose was to clarify its presence in open ocean, and their genotypic and physiological characteristics were compared with those of isolates from clinical, animal, and freshwater sources. Water samples were collected from freshwater, bays, and offshore environments in Japan. Sixty-two isolates, including 26 from the open ocean, were identified as P. aeruginosa by phenotypic characteristics and the BD Phoenix System. Pulsed-field gel electrophoresis (PFGE) was performed on all strains, together with 21 clinical and 8 animal strains. The results showed that open ocean strains are composed of a few genotypes, which are separated from other strains. Although some clinical isolates made a cluster, other strains tended to mix together. Different antibiotypes were observed among marine isolates that had similar PFGE and serotyping patterns. Some were multidrug-resistant. Laboratory-based microcosm study were carried out to see the responses of P. aeruginosa toward increased NaCl concentrations in deionized water (DW). Marine strains showed better survival with the increase, whereas river and clinical strains were suppressed by the increase. These findings illustrate the potential significance of open ocean as a possible reservoir of P. aeruginosa, and there may be clones unique to this environment. To our knowledge, this is the first report on the presence and characterization of P. aeruginosa in the open ocean.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Since Pseudomonas aeruginosa was first described in 1872, it has been one of the most thoroughly investigated bacteria [44]. It is well known as a pathogen of human in association with cystic fibrosis [11, 46], and it is the quintessential opportunistic pathogen, causing a wide variety of infections in compromised hosts [20, 40]. P. aeruginosa also causes diseases in both plants [5, 56] and animals [41, 59]. Owing to its exceptionally high metabolic versatility in utilizing numerous organic compounds and its adaptability to various conditions, it can survive in terrestrial [49, 61], air [36], and freshwater environments [27, 38, 39, 42]. Although it has been isolated from river outfalls and shorelines in the sea, these isolates have been regarded as originating from freshwater or sewage [21, 22, 28, 33, 37, 54, 60]. In addition, the culture-independent methods have not yet indicated the presence of this bacterium in open ocean environments.
In 1995, marine chemists discovered the presence of certain dissolved proteins in the ocean [50, 51]. One 48-kDa protein was especially ubiquitous in the North Pacific, Indian Ocean, and Antarctic Ocean, from the surface to the deep layers of water [51, 52]. N-terminal amino acid sequencing [50] and immunochemical reaction [48] revealed that this protein was an outer membrane porin protein, OprP, of P. aeruginosa. This protein is specifically synthesized under phosphate-deficient conditions [18]. This finding raised questions on the origin of the protein, the release process of the protein, stability or turnover rate of the protein in the sea, the mechanism to specifically select OprP among various proteins, and so on. The final goal of the present investigation was to answer these questions. Because our recent microscopic study using fluorescent antibody suggested that P. aeruginosa is common in Tokyo Bay and coastal environments [28], it is expected that P. aeruginosa is present in marine environments including open ocean.
Recent study documented that the P. aeruginosa community in River Woluwe, Belgium, was almost as diverse as the global P. aeruginosa population, and the river harbored members of nearly all successful clonal complexes [39]. Römling et al. [42] reported that 19% of a collection of 573 P. aeruginosa strains isolated from various clinical cases and from the aquatic environment, especially rivers, belonged to the same clonal group. Similar homogeneity was found among P. aeruginosa strains isolated either from cases of clinical infection or from aquatic environments and from gasoline sources [6, 12, 43]. Although several recent studies indicated that environmental strains are homogeneous with clinical strains, the oceanic strains, if they exist, may be different from those of other environments because of the uniqueness of their habitat.
Our objectives were (1) to investigate the presence of P. aeruginosa in the open ocean and (2) to clarify the genotypic, phenotypic, and metabolic characteristics of the open ocean strains in comparison with those collected from clinical, animal, and freshwater sources. We succeeded in isolating a number of strains from the surface layer of the North Pacific Ocean. By applying pulsed-field gel electrophoresis (PFGE) [16], we confirmed that they have distinct genetic types. To our knowledge, this is the first report on P. aeruginosa from open ocean environments.
Materials and Methods
Water Sampling
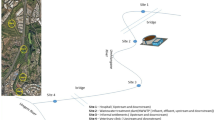
Water samples were collected from four stations in the Arakawa River (Sts. AK1, AK2, AK3, and AK4), two ponds (P1, Inokashira Pond, and P2, Zenpukujii Pond), and Lake Tamako in the Tokyo metropolitan area between September 2003 and April 2004. Samples from marine environments were collected from Tokyo Bay (Sts. T1, T2, T3, T4, and T5), Sagami Bay (St. S1), and open ocean environments (Sts. S2, S3, and S4). Marine samples were collected during cruises KT-03-05 and KT-03-07 on RV Tansei Maru of the Ocean Research Institute, The University of Tokyo, in 2003 (Fig. 1). Surface seawater was also collected from Kumamoto Bay in December 2003. The southernmost sampling site (St. S2) was about 300 km away from the Tokyo bay and is separated by the Kuroshio Current from the coastal area and mainland Japan. This current is analogous to the Gulf Stream in the Atlantic Ocean, transporting warm, tropical water northward toward the polar region [55, 57]. South of this current, there is virtually no possibility of contamination of freshwater or terrestrial influences.
Surface water samples were collected in a sterile bucket and kept in sterile Nalgene (Nalge Nunc International Corporation, Rochester, NY, USA) plastic bottles on ice until processing. During the cruises of RV Tansei Maru, water samples were collected with an ethanol-washed Niskin water sampler equipped with a conductivity, temperature, and depth (CTD) device (FSI, Cataumet, MA, USA). All samples were analyzed within 4 h after collection.
Measurement of Physicochemical Parameters
During the cruises, water temperature and salinity were measured with the CTD system. For other samples, the parameters were measured with a YSI Model 85 handheld oxygen, conductivity, salinity, and temperature system (YSI Incorporated, Yellow Springs, OH, USA). The pH of all samples was measured with a desktop Horiba pH meter F-21 (Horiba Ltd., Chiyoda-Ku, Tokyo, Japan).
Isolation and identification of P. aeruginosa
Selective and nonselective agar culture media were used for the isolation and identification of P. aeruginosa. Appropriate volumes of water samples were filtered through sterilized Nuclepore membrane filters (Whatman, Middlesex, UK) (pore size 0.2 μm and diameter 47 mm), which were placed on nalidixic acid cetrimide (NAC) agar [31] plates and Nutrient Broth agar (NA) (Difco Laboratories, Detroit, MI, USA) plates supplemented with 0.5% NaCl. After incubation of the plates at 20°C for 3–4 days, colonies appeared on the plates, were picked up, transferred to NAC agar, and incubated at 20°C for another 3 or 4 days to ensure the presence of the color unique to pyocyanine. Suspected colonies that showed the characteristic appearance and color were inoculated onto cetrimide kanamycin nalidixic acid agar (CKNA) [29] and incubated at 42°C overnight. Isolates that showed growth at 42°C were primarily identified as P. aeruginosa. In addition, eight animal and 21 clinical P. aeruginosa strains examined were from the collections of the Veterinary Assay Laboratory, Japan, and The Toho Medical University (Tokyo, Japan), respectively. All the isolated P. aeruginosa strains were preserved in 40% glycerol in nutrient broth at −85°C.
Identification by BD Phoenix System
The isolates were identified by the BD Phoenix Automated Microbiology System (Becton, Dickinson and Company, Sparks, MD, USA) according to the method described by Fahr et al. [10]. Briefly, the Phoenix system uses one ID (identification) and AST (antimicrobial susceptibility) combination panel, with the ID substrates on one side and the antimicrobial drugs on the other. Bacterial cells that had been precultured on Mueller-Hinton agar were inoculated into the Phoenix ID broth and were adjusted to 0.5–0.6 McFarland standards by using a Crystal Spec nephelometer (BD). After the transfer of 25 μL ID broth suspension to the Phoenix AST broth, the suspension was poured into the ID side of the Phoenix Combo panel. Once inoculated, the panel was logged and loaded into the Phoenix Automatic system, in which colorimetric and fluorometric signals were measured in every 20 min.
Serotyping
Serotyping for O-group specific antigen was carried out by using P. aeruginosa serotyping kit following the manufacturer's protocol (Denka Seiken Ltd., Japan). All isolates were tested for O-group-specific antigens. Three polyvalent (I, II, and III) and 14 serotypes (A, B, C, D, E, F, G, H, I, J, K, L, M, and N) were checked against all the isolates [23]. Briefly, individual colonies were emulsified in 20 μL PBS to which an equal volume of agglutination serum was added. Positive reactions were noted by the clumping of bacterial cells, under a light microscope as well as by the naked eyes.
Antibiotyping
The Phoenix AST method was used for antibiotyping [4, 10]. Briefly, Phoenix AST broth was supplemented with one drop of Phoenix AST indicator, an oxidation–reduction indicator based on resazurin (Alamar Blue). From the standardized ID suspension, 25 μL was transferred to the AST broth, to obtain about 5 × 105 CFU/mL solution. The broth was then poured into the AST side of the panel. As noted above, the panel was loaded into the Phoenix apparatus. For each antibiotic, a minimum of eight concentrations (serially doubled dilutions) were tested. In addition, specific detection of extended-spectrum beta-lactamase (ESBL) for Gram-negative bacteria was also performed on the AST side on respective Combo panels [34, 35]. Finally, the strains were typed depending on the number of antimicrobial substances to which they were resistant. The data were coded either as 0 (sensitive) or 1 (resistant). Following the method described by Martin-Kearly et al. [32], hierarchical cluster analysis was performed using the average linkage method with the rescaled distance measure. The dendrogram was produced using the program SPSS for Windows, Release 10.0 (SPSS Inc., Chicago, IL, USA).
Pulsed-field Gel Electrophoresis
To clarify the genetic relatedness among the strains, PFGE analyses were carried out by using the Genepath Group 5 reagent kit (Bio-Rad Laboratories, Richmond, CA, USA) by following the manufacturer's protocol with a slight modification, as reported by Ishii et al. [24]. In brief, 30–50 μL overnight cultures of Luria-Bertani (LB) broth (Tryptone 10 g/L, yeast extract 5.0 g/L, sodium chloride 10.0 g/L), in 1.5-mL microcentrifugation tubes, were centrifuged at 6700 g for 1 min at 4°C by a high-speed refrigerated microcentrifuge (MX-100, TOMY, Tokyo, Japan). The pellet was resuspended in 150 μL suspension buffer. Next, 150 μL of liquid 1.2% embedding agarose and 6 μL lysozyme were mixed with the suspension and cooled to 50°C. The mixture was poured into plug molds on ice. The plugs were then incubated for 30 min at 37°C in 500 μL lysis buffer containing 20 μL lysozyme. The plugs were treated with proteinase K overnight at 50°C, then each plug was washed with washing buffer for three or more times. Embedded DNA in each plug was digested with the restriction enzyme SpeI (5 U) in 300 μL SpeI buffer at 37°C for 2 h after treatment by 10 times diluted (0.1×) washing buffer. Fragments of DNA were separated at 14°C for 19.7 h on 1% SeaKem Gold agarose gel (FMC Bioproducts, Rockland, ME, USA) in 0.5× TBE buffer with 100 μM thiourea [62], with a switch ramped time from 5.3 to 34.9 s at a 120° angle, nonlinear 21%, on a CHEF Mapper apparatus (Bio-Rad Laboratories). The sizing ladder used for PFGE was a lambda ladder with a range of 0.05–1 MB (Bio-Rad Laboratories). The PFGE patterns were analyzed using the Molecular Analyst Fingerprinting Plus software package (version 1.2, Bio-Rad Laboratories). Levels of similarity between fingerprints were expressed as Dice coefficients, which were calculated by determining the ratio of twice the number of bands shared by two patterns to the total number of bands in both patterns. Isolates were clustered by using the unweighted pair group method with arithmetic averages (UPGMA) [45].
Survivability
Laboratory-based microcosms were prepared to see the comparative survival and growth responses of marine (strain 22), freshwater (strain 11), and clinical (strain 43) P. aeruginosa at 0.0–7.0% NaCl concentrations. Microcosms were prepared in deionized water. All microcosms were prepared in duplicates. Cells from the logarithmic phase of growth on LB broth were inoculated in the microcosms and were kept at room temperature (24±1°C). Samples were taken from each of the microcosms in sterile condition in a series of time intervals and were plated on LB and NAC agar medium with appropriate dilution and were grown at room temperature for 4–5 days. Based on preliminary experiments, the culturable cell counts and growth rate were calculated with the values after 24 h of incubation. Paired t test was performed on the data to estimate whether or not there was a significant difference between test series.
Results
P. aeruginosa Isolation
P. aeruginosa strains were isolated and identified after a series of examinations. As the first step in their isolation, 6700 colonies appeared on either NAC or NA plates with 0.5% NaCl. After their transfer to NAC agar plates to check their growth and the appearance of the color unique to P. aeruginosa, 1679 isolates were selected. Among them, 560 were able to grow at 42°C on NAC. They were transferred to CKNA agar plates and incubated at 42°C overnight. The isolates grown under this condition were tentatively regarded as P. aeruginosa, and further identified by using the BD Phoenix system. Finally, 62 strains were confirmed as P. aeruginosa; 26 from the open ocean, 3 from Tokyo Bay, 4 from Kumamoto Bay, 2 from Sagami Bay, 14 from the Arakawa River, and the remaining 13 from the ponds and lake in the Tokyo metropolitan area. A list of strains is given in Table 1, together with their sources, origins, and year of isolation.
The comparative numbers of colonies that appeared on selective agar plates and the numbers of culturable P. aeruginosa per liter of water are shown in Table 2. The number of P. aeruginosa per liter was ca. 1000 or more in the ponds and lake, and 3–30 in Arakawa River (Table 2). In the marine environment, the numbers were smaller and the appearance was sporadic with no clear trends among areas. Relatively high numbers of isolates were obtained at St. 2, the southernmost station during this cruise.
Physicochemical Parameters
Table 2 shows the physicochemical parameters, including salinity, water temperature, and pH. Salinity varied widely, depending on the source, and gradually increased from the river, the bay, to the open ocean. The values in Kumamoto Bay and four stations (Sts. T1 to T4) in Tokyo Bay were typical of those in coastal environments. Temperature ranged from 11.3°C in Kumamoto Bay at the end of December to 24.0°C in the open ocean at St. S3 in June. Water pH was higher in the coastal and open ocean environments ranging from 8.06 to 8.46.
Pulsed-field Gel Electrophoresis
To determine the genetic relatedness among marine, freshwater, animal, and clinical strains, PFGE analyses were performed on the genomic DNAs after SpeI digestion. A dendrogram was prepared including all strains from all sources (Fig. 2). Ninety strains from all sources resulted in 50 different restriction band patterns in the PFGE profiles at a 100% homogeneity level (Table 3). When 55% homogeneity level was taken as the criterion, six clusters appeared (Fig. 2). The first group (Group I) consisted of 14 strains, of which 13 were of clinical origin and one from Tokyo Bay (strain 25). The second one (Group II) contained oceanic strains and the Bio-Rad reference strains (strains 49, 50, and 51). With the exception of strain 26, which was closely related to those from Kumamoto Bay and Arakawa River, all open ocean isolates, and 1 strain from Sagami Bay (strain 12) were in this group. The open ocean strains also made a cluster at the 83% to 100% homogeneity level. The third group (Group III) was formed by 28 strains from almost all sources. Small clusters composed of river and pond strains were included. The fourth group (Group IV) consisted of 17 strains: 5 from the lake, 4 from animals, 3 from the river, 3 from the ponds, and the other 2 from Tokyo Bay and a clinical source. Two other groups were composed of only 2 strains each: a fifth group (Group V) containing a clinical strain and a river strain and a sixth group (Group VI) containing a river strain and a Tokyo Bay strain.
SpeI enzyme digested pulsed field gel electrophoresis band patterns of P. aeruginosa isolated from various sources. Strain numbers, their sources and groups are shown. A: animal; C: clinical; L: Lake Tamako; KB: Kumamoto Bay; O: open ocean; P: pond; R: Arakawa River; RC: reference strain provided by Bio-Rad; SB: Sagami Bay; TB: Tokyo Bay. Groups were made at 55% homogeneity level.
Table 3 shows the number of strains from each source and band types they produced. Among 50 band types obtained, only one type was found from multiple sources, i.e., from Sagami Bay and open ocean. All the rest was unique to each source or environment. Obviously, the numbers of band types were small in open ocean, suggesting the presence of strains with less genetic variations.
Antibiotyping
The antimicrobial resistance patterns of 90 P. aeruginosa toward 22 antimicrobial substances were examined by using the BD Phoenix AST panels. Resistance was judged based on growth at the maximum amount of respective substance used in the AST panel. Antibiotypes of all strains are shown in Table 1. Hierarchical cluster analyses revealed 29 different types among the strains (Fig. 3). The predominant type was XXIV; it was observed in 27.2% of the strains, including open ocean, coastal, freshwater, and animal (Fig. 3). This type is resistant to seven antimicrobial substances, i.e., cefazolin, cefuroxime-N, cefpodoxime, ampicillin, amoxicillin–clavulanate, ampicillin–sulbactam and tetracycline, whereas pattern XXV was shown by strains resistant to all the antimicrobial substances used in this study. The second predominant type (8.7%) was XXVII, which includes animal and pond strains. Types VIII, XVII, XXV, and XXIX were observed in 6.5% of strains. Type VII was restricted to animal strains, and XXV and XXIX were in Arakawa and Tamako Lake sharing with open ocean strains. Only one strain (1.1%) showed resistance corresponding to each of types I, V, VI, IX, X, XI, XII, XIII, XIV, XV, XVI, XVIII, XX, XXI, and XXII, the least common resistance patterns. Patterns I–VIII were strictly distributed among the clinical isolates (Fig. 1).
Dendrogram illustrating the clustering of antibiotyping patterns of P. aeruginosa. The dendrogram was produced by hierarchical cluster analysis using the average linkage method. The distance units are arbitrary, being based on the rescaled distance measure. Strains were arbitrarily grouped into different types. Numbers, sources, and antibiotypes are designated at the right side. A: animal; C: clinical; L: Lake Tamako; KB: Kumamoto Bay; O: open ocean; P: pond; AK: Arakawa River; R: reference strain provided by Bio-Rad; SB: Sagami Bay; TB: Tokyo Bay.
The maximum amounts of antimicrobial substances used in the BD Phoenix panels and the percentages of resistant strains from each source are shown in Table 4. Regardless of their origin, all isolates were resistant to six antimicrobial substances: ampicillin, cefazolin, cefuroxime sodium, cefpodoxime, ampicillin–sulbactam, and amoxicillin–clavulanate. Most of the clinical strains were resistant to almost all the antimicrobial substances monitored. Open ocean strains were resistant to 6–11 antimicrobial substances, whereas the animal isolates were resistant to 6–8 substances. All the open ocean isolates were at least resistant to the six antibiotics stated above and were sensitive to ceftazidime, meropenem, imipenem, amikacin, gentamicin, piperacillin–tazobactam, levofloxacin, cefepime, piperacillin, and ofloxacin.
Serotyping
The serotypes of all strains are presented in Table 1. Of the 26 open ocean strains, 25 belong to serotype E and only one (isolated from S4) belongs to serotype K. Type E was also found dominant among the clinical strains. Most of the freshwater strains belonged to serotype G. Out of the 7 strains from the ponds, 5 were of serotype G. Only 2 strains from the Arakawa River showed serotype K (Table 1). The greatest variation in the number of serotypes was observed among the clinical isolates. The animal strains belonged to serotypes E, F, G, and I.
Survivability of Marine Strains
To clarify their response to high sodium chloride concentration, three strains (marine-22, river-11, and clinical-43) were grown in LB with increase NaCl concentration. There was little difference among the three strains, indicating that P. aeruginosa is generally quite tolerant to high salt condition (Data not shown). Considering the natural environments, the survival of each strain was observed in deionized water with up to 7% concentration of NaCl. Figure 4a shows the percentage of culturable cells after 24 h, relative to the number at the onset of the incubation. As for marine strains, the percentage of culturable cells increases with NaCl concentration. At 7% NaCl, as much as 63% of the cells still retained culturability, whereas freshwater and clinical strains only retained 9% and 4%, respectively. The percentage of culturable cell counts decreased with the increase in NaCl concentration. Therefore, when regression lines were obtained only the marine strain showed positive slope, whereas the remaining strains showed a negative slope. Figure 4b shows the slopes of each strain during the course of incubations. Except for 4, 6, and 12 h, the marine isolate always showed a positive value, indicating that culturability increased with sodium concentration. The slopes for river and clinical strains were always negative, further showing that culturability consistently decreased with NaCl concentration.
(a) Survival of three P. aeruginosa strains in deionized water with different NaCl (0 to 7%) concentrations after 24 h incubation. Value represents the mean of two determinations from two independent experiments. Linear regression lines are for each of the strains. Straight, bigger dotted, and smaller dotted lines represent marine (strain 22), river (strain 11), and clinical (strain 43) strains respectively. Symbols ▪, ♦, and ▴ represent the marine (strain 22), river (strain 11), and clinical (strain 43) strains of P. aeruginosa respectively. Differences were significant among the marine and river (p=0.075), and marine and clinical (p=0.054) strains. (b) Survival of three P. aeruginosa strains in deionized water with different NaCl (0–7%) concentrations represented by slope against the time intervals. Value represents the mean of two determinations from two independent experiments. Symbols ▪, ♦, and ▴ represent the marine (strain 22), river (strain 11), and clinical (strain 43) strains of P. aeruginosa, respectively. Differences were significant among the marine and river (p≤0.01), and marine and clinical (p≤0.01) strains.
Discussion
To clarify the presence of P. aeruginosa in marine environments and to characterize them, we collected seawater samples from the open ocean where the influence of human activity was minimal or absent, and from coastal environments. Together with those from freshwater environments, 62 isolates were identified as P. aeruginosa by the BD Phoenix system. Considering the genetic distinctiveness clarified by PFGE analyses and the tendency toward high NaCl concentration, populations unique to marine environment seem to be present. However, the antibiotype was not specific to marine strains. In addition, the serotype of strains from open ocean was also observed among other strains. To our knowledge, this is the first report on the isolation and comparative analyses of P. aeruginosa isolated from open ocean environments.
In this work, phenotypic characteristics such as color of colonies and growth on selective media were used as the first screening. During the course of this study, we found that the growth at 42°C on selective medium was a simple and efficient way for P. aeruginosa, although a possibility to overlook some strains is not rejected. Thus strains selected were analyzed by the BD Phoenix system, which has been used for the identification of clinical isolates. Its reliability has been confirmed by several studies [10, 13]. We have sequenced the 16S rDNA of some strains identified as P. aeruginosa by this system, and found that the sequence agreed with that of P. aeruginosa PAO1. In addition, this system is known to be very accurate in detecting the antibiotic resistance in this group of microorganisms [4, 8, 10, 13]. AST performance with Gram-negative bacteria was equivalent to that of the standard broth microdilution method.
For genotypic analyses, several methods such as PFGE [16, 53], ribotyping [6], and arbitrary primer PCR-based fingerprinting methods [9] have been used. We used PFGE in this study, because we considered it suitable and more discriminating compared to other techniques for comparing P. aeruginosa isolated from various sources. DNA fingerprinting by PFGE of genomic DNA after digestion with an appropriate restriction endonuclease was considered as the “gold standard” for bacterial typing [53], and is the preferred method for P. aeruginosa because of its high discriminatory capacity, good reproducibility, and ease of interpretation [17]. Results showed that all the strains from various sources could be divided into six groups at the 55% homogeneity level. This indicates several aspects of the genetic relatedness among the strains. First, depending on the source, the strains tend to make clusters at different levels of homogeneity. For instance, among the 22 clinical isolates (including one from Bio-Rad), 13 made up Group I, together with 1 isolate from Tokyo Bay (strain 25). Except for strain 26, all open ocean isolates were in Group II. Because the open ocean strains showed different antibiotypes (Table 1), they may comprise multiple phenotypic groups. Compared with the oceanic strains, freshwater ones were genetically more divergent. Two strains from the Arakawa River (strains 72 and 79) and four strains from the ponds (strains 80, 81, 82, and 83) made one small cluster at the 82% homogeneity level in Group III. Other freshwater strains tended to make small clusters sharing the same genotype. On the other hand, the animal isolates were widely distributed in the dendrogram. These results indicate that, in nature, there may be some strains uniquely adapted to each environmental niche. Second, the present results also suggest frequent genetic exchanges among some strains and/or transfer of cells from one environment to another. The composition of Group III is a typical case. This group included strains from all sources, although there were small clusters at higher homogeneity levels. It should be noteworthy that if we were to analyze only the strains in Group III, we might have not found genetic differences among environmental and clinical strains. Some research groups have reported that clinical and environmental isolates of P. aeruginosa are genetically or phenotypically indistinguishable [3, 11, 12, 42]. A very recent study documented that the global population structure of P. aeruginosa is reflected in a relatively confined geographical area, a small river in Belgium [39]. Our present work indicates that, depending on the choice of strains and genes, apparent relatedness among strains may vary. Because marine strains inhabit in vast water mass with little influence of human activities, exchanges with those in other environments may be rare.
Although we succeeded in isolating marine P. aeruginosa, it is still not clear how commonly present this bacterium is in the open ocean. The distribution seems to be rather sporadic (Table 2), and the culture-independent approach still does not prove their wide distribution. Our findings support the previous article on the origin of the 48-kDa dissolved protein [50]. Application of the fluorescent antibody technique had demonstrated the presence of this bacterium in the sea [28]. This also supported the evidence of the detection of the 48-kDa dissolved protein by Western blotting [48]. The following findings might have been caused by the marine-type P. aeruginosa. P. aeruginosa has been found to cause bronchopneumonia and multiple large cutaneous ulcers in Atlantic bottlenose dolphins. Here, bacterial cells progress deep into the cutaneous tissue, causing serious damage to the animals [7]. Skin infections caused by this bacterium in occupational saturation divers have been also reported [1, 2]. Genome analysis of P. aeruginosa has clarified the presence of nqr, which encodes the primary sodium pump [47], giving an advantage for growth and survival in saline environments [19]. P. aeruginosa is well known for its high adaptability [19] and diverse phenotypic characteristics. This bacterium is able to use an extremely wide range of organic and inorganic compounds [14, 15, 58]. As various organic materials coexist at low concentrations in seawater, such physiological versatility should help the bacterium's growth and survival in marine environments [25]. Our results on survivability indicate that P. aeruginosa strains (including marine, river, and clinical strains) can grow and survive in deionized water and artificial seawater with high NaCl concentrations. Marine P. aeruginosa can survive better in higher NaCl in DW in comparison with freshwater and clinical isolates (Fig. 4), strongly indicating that they are forms specifically adapted to high salinity environments. We examined the presence of P. aeruginosa in tap water, sink, and laboratory facilities on the vessel, and inside and outside of the Niskin water samplers as possible contaminants. We could not isolate any P. aeruginosa. Considering all these factors, it seems reasonable to assume the presence of P. aeruginosa in open oceanic waters. We regard that the possible influence of contamination to be extremely small.
Why has the presence of this bacterium in the sea been overlooked for such a long time? First, when P. aeruginosa was isolated from coastal environments, it was regarded as being of terrestrial or freshwater origin [22, 28, 33, 37, 54, 60]. Second, its concentration in the open ocean is very small. We screened approximately 6700 colonies appearing on selective and nonselective agar medium from marine environments. Of these, less than 1% was confirmed as P. aeruginosa after a series of identification steps. Therefore, without focusing on this species, the chances of detection by using routine, nonselective marine media would be extremely small. Finally, the culture-independent methods also do not indicate the presence of this bacterium. Except in a few cases, sequences corresponding to those of P. aeruginosa have not been detected [26, 30]. However, these cannot be taken as proof for the absence of this bacterium in the open ocean.
The distribution of P. aeruginosa was rather sporadic, and it remains difficult to state any general trends from our data. There seem to be two possible explanations. First, P. aeruginosa in the sea may be associated with particulate matter or animals. As stated above, there are reports of its presence in dolphins [7]. Marine mammals may serve as reservoirs of this bacterium. Second, the cells may be present in viable but nonculturable (VBNC) state in marine environments. This is supported by the presence of their relatively high numbers, as enumerated by fluorescent-antibody technique, in Tokyo Bay [28]. However, as very limited information on VBNC is available for this bacterium, more work is required to address this issue.
We conclude that P. aeruginosa is present in the open ocean. PFGE analyses showed that oceanic strains are forming a distinct genetic cluster. The microcosm experiment indicates that a marine isolate survived better with higher concentrations of NaCl. This finding requires a change in our views on the origin, genetic exchanges, physiology, and ecology of this well-known bacterial species. Extensive studies on the phylogeny among strains from various sources and on the physiological characteristics at the molecular level are now being undertaken in our laboratory.
References
Ahlen, C, Mandal, LH, Iversen, QJ (1998) Identification of infectious Pseudomonas aeruginosa strains in an occupational saturation diving environment. Occup Environ Med 55: 480–484
Ahlen, C, Mandal, LH, Johannessen, LN, Iversen, QJ (2000) Survival of infectious Pseudomonas aeruginosa genotypes in occupational saturation diving environment and the significance of these genotypes for recurrent skin infections. Am J Ind Med 37: 493–500
Alonso, A, Rojo, F, Martínez, JL (1999) Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ Microbiol 1: 421–430
Brisse, S, Stefani, S, Verhoef, J, Van Belkum, A, Vandamme, P, Goessens, W (2002) Comparative evaluation of the BD Phoenix and VITEK 2 automated instruments for identification of isolates of the Burkholderia cepacia complex. J Clin Microbiol 40: 1743–1748
Buysens, S, Heungens, K, Poppe, J, Hofte, M (1996) Involvement of pyochelin and pyoverdin in suppression of pythium-induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Appl Environ Microbiol 62: 865–871
Denamur, E, Picard, B, Decoux, G, Denis, JB, Elion, J (1993) The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol Lett 110: 275–280
Diamond, SS, Ewing, DE, Cadwell, GA (1979) Fatal bronchopneumonia and dermatitis caused by Pseudomonas aeruginosa in an Atlantic bottle nosed dolphin. J Am Vet Med Assn 175: 984–987
Donay, JL, Mathieu, D, Fernandes, P, Prégermain, C, Bruel, P, Wargnier, A, Casin, I, Weill, FX, Lagrange, PH, Herrmann, JL (2004) Evaluation of the automated Phoenix system for potential routine use in the clinical microbiology laboratory. J Clin Microbiol 42: 1542–1546
Elaichouni, A, Vershraegen, A, Claeys, G, Devleeschouwer, M, Godard, C, Vaneechoute, M (1994) Pseudomonas aeruginosa serotype O12 outbreak studied by arbitrary primer PCR. J Clin Microbiol 32: 666–671
Fahr, AM, Eigner, U, Armbrust, M, Caganic, A, Dettori, G, Chezzi, C, Bertoncini, L, Benecchi, M, Menozzi, MG (2003) Two-center collaborative evaluation of the performance of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterococcus spp. and Staphylococcus spp. J Clin Microbiol 41: 1135–1142
Finnan, S, Morrissey, JP, O'Gara, F, Boyd, EF (2004) Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol 42: 5783–5792
Foght, JM, Westlake, DWS, Johnson, WM, Ridgway, HF (1996) Environmental gasoline-utilizing isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology 142: 2333–2340
Funke, G, Funke-Kissling, P (2004) Use of the BD PHOENIX automated microbiology system for direct identification and susceptibility testing of gram-negative rods from positive blood cultures in a three-phase trial. J Clin Microbiol 42: 1466–1470
Glazebrook, JS, Campbell, RS, Hutchinson, GW, Stallman, ND (1978) Rodent zoonoses in North Queensland: the occurrence and distribution of zoonotic infections in North Queensland rodents. Aust J Exp Biol Med Sci 56: 147–156
Green, SK, Schroth, MN, Cho, JJ, Kominos SK, Vitanza-Jack, VB (1974) Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl Microbiol 28: 987–991
Grothues, D, Koopmann, U, von der Hardt, H, Tümmler, B (1988) Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol 26: 1973–1977
Grundmann, H, Schneider, C, Hartung, D, Daschner, FD, Pitt, TL (1995) Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol 33: 528–534
Hancock, REW, Poole, K, Benz, R (1982) Outer membrane protein P of Pseudomonas aeruginosa: regulation by phosphate deficiency and formation of small anion-specific channels in lipid bilayer membranes. J Bacteriol 150: 730–738
Hase, CC, Fedorova, ND, Galperin, MY, Dibrov, PA (2001) Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev 65: 353–370
Head, NE, Yu, H (2004) Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect Immun 72: 133–144
Hoadley, AW (1968) On the significance of Pseudomonas aeruginosa in surface waters. J N Engl Water Works Assoc 82: 99–111
Hoadley, AW (1977) Potential health hazards associated with Pseudomonas aeruginosa in water. Am Soc Test Mater Spec Tech Publ 635: 80–114
Homma, JY (1982) Designation of the thirteen O-group antigens of Pseudomonas aeruginosa; an amendment for the tentative proposal in 1976. Japan J Exp Med 52: 317
Ishii, Y, Alba, J, Kimura, S, Nakashima, K, Abe, Y,Yamaguchi, K (2002) Rapid pulsed-field gel electrophoresis technique for determination of genetic diversity of Serratia marcescens. J Infect Chemother 8: 368–370
Karlowsky, JA, Deborah, CD, Mark, EJ, Thornsberry, C, Friedland, IR, Sahm, DF (2003) Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob Agents Chemother 47: 1681–1688
Kato, C, Li, L, Tamaoka, J, Horikoshi, K (1997) Molecular analyses of the sediment of the 11,000-m deep Mariana Trench. Extremophiles 1: 117–123
Khan, AA, Cerniglia, CE (1994) Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl Environ Microbiol 10: 3739–3745
Kimata, N, Nishino, T, Suzuki, S, Kogure, K (2004) Pseudomonas aeruginosa isolated from marine environments in Tokyo Bay. Microb Ecol 47: 41–47
Kodaka, H, Iwata, M, Yumoto, S, Kashitani, F (2003) Evaluation of a new agar medium containing cetrimide, kanamycin and nalidixic acid for isolation and enhancement of pigment production of Pseudomonas aeruginosa in clinical samples. J Basic Microbiol 43: 407–413
Li, L, Kato, C, Nogi, Y, Horikoshi, K (1998) Distribution of the pressure-regulated operons in deep-sea bacteria. FEMS Microbiol Lett 159: 159–166
Lilly, HA, Lowbury, EJL (1972) Cetrimide-nalidixic acid agar as a selective medium for Pseudomonas aeruginosa. J Med Microbiol 5: 151–153
Martin-Kearley, J, Gow, JA, Peloquin M, Greer, CW (1994) Numerical analyses and the application of random amplified polymorphic DNA polymerase chain reaction to the differentiation of Vibrio strains from a seasonally cold ocean. Can J Microbiol 40: 445–446
Mates, A (1992) The significance of testing for Pseudomonas aeruginosa in recreational seawater beaches. Microbios 71: 89–93
National Committee for Clinical Laboratory Standards (NCCLS) (1997) Methods for dilution in antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, PA
National Committee for Clinical Laboratory Standards (NCCLS) (1999) Performance standards for antimicrobial susceptibility testing. Ninth informational supplement M100-S9. National Committee for Clinical Laboratory Standards, Wayne, PA
Panagea, S, Winstanley, C, Walshaw, MJ, Ledson, MJ, Hart, CA (2004) Environmental contamination with an epidemic strain of Pseudomonas aeruginosa in a Liverpool cystic fibrosis centre, and study of its survival on dry surfaces. J Hosp Infect 59: 102–107
Papapetropourou, M, Rodopoulou, G (1994) Occurrence of enteric and non-enteric indicators in coastal waters of southern Greece. Bull Mar Sci 54: 63–70
Pellett, S, Bigley, DV, Grimes, DJ (1983) Distribution of Pseudomonas aeruginosa in a riverine ecosystem. Appl Environ Microb 45: 328–332
Pirnay, J-P, Matthijs, S, Colak, H, Chablain, P, Bilocq, F, Eldere, V, Vos, DD, Zizi, M, Triest, L, Cornelis, P (2005) Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ Microbiol 7: 969–980
Poirel, L, Lebessi, E, Castro, M, Fèvre, C, Foustoukou, M, Nordmann, P (2004) Nosocomial outbreak of extended-spectrum β-lactamase SHV-5-producing isolates of Pseudomonas aeruginosa in Athens, Greece. Antimicrob Agents Chemother 48: 2277–2279
Prevatt, AR, Sedwick, JD, Gajewski, BJ, Antonelli, PJ (2004) Hearing loss with semicircular canal transection and Pseudomonas aeruginosa otitis media. Otolaryngol Head Neck Surg 131: 248–252
Römling, U, Wingender, J, Müller, H, Tümmler, B (1994) A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol 60: 1734–1738
Ruimy, R, Genauzeau, E, Barnabe, C, Beaulieu, A, Tibayrenc, M, Andremont, A (2001) Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect Immun 69: 584–588
Schroeter, J (1872) Ueber einige durch Bacterien gebildete Pigmente. In: Cohn, FJU (Eds.) Beigrage zur Biologie der Pflanzen. Ern's Verlag, Breslau, pp 109–126
Sokal, RR, Sneath, PHA (1963) Principles of numerical taxonomy. Freeman, San Francisco, CA
Spilker, T, Coenye, T, Vandamme, P, LiPuma, JJ (2004) PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 42: 2074–2079
Stover, CK, Pham, XQ, Erwin, AL, Mizoguchi, SD, Warrener, P, Hickey, MJ, Brinkman, FSL, Hufnagle, WO, Kowalik, DJ, Lagrou, M, Garber, RL, Goltry, L, Tolentino, E, Westbrock-Wadman, S, Yuan, Y, Brody, LL, Coulter, SN, Folger, KR, Kas, A, Larbig, K, Lim, R, Smith, K, Spencer, D, Wong, GK-S, Wu, Z, Paulsen, IT, Reizer, J, Saier, MH, Hancock, REW, Lory, S, Olson, MV (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964
Suzuki, S, Kogure, K, Tanoue, E (1997) Immunochemical detection of dissolved proteins and their source bacteria in marine environments. Mar Ecol Prog Ser 158: 1–9
Szoboszlay, S, Atzel, B, Kriszt, B (2003) Comparative biodegradation examination of Pseudomonas aeruginosa (ATCC 27853) and other oil degraders on hydrocarbon contaminated soil. Commun Agric Appl Biol Sci 68: 207–210
Tanoue, E, Nishiyama, S, Kamo, M, Tsugita, A (1995) Bacterial membranes: possible source of a major dissolved protein in seawater. Geochim Cosmochim Acta 59: 2643–2648
Tanoue, E (1995) Detection of dissolved protein molecules in oceanic waters. Mar Chem 51: 239–252
Tanoue, E, Ishii, M, Midorikawa, T (1996) Discrete dissolved and particulate proteins in oceanic waters. Limnol Oceanogr 41: 1334–1343
Tenover, FC, Arbeit, RD, Goering, RV, Micklesen, PA, Murray, BE, Persing, DH, Swaminathan, B (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33: 2233–2239
Velammal, A, Aiyamperumal, B, Venugopalan, VK, Ajmalkhan, S (1994) Distribution of Pseudomonas aeruginosa in Pondicherry coastal environments. Ind J Mar Sci 23: 239–241
Vonder Haar, TH, Oort, AH (1973) New estimate of annual poleward energy transport by northern hemisphere oceans. J Phys Oceanogr 3: 169–172
Walker, TS, Bais, HP, Deziel, E, Schweizer, HP, Rahme, LG, Fall, R, Vivanco, JM (2004) Pseudomonas aeruginosa–plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol 34: 320–331
White, WB, McCreary, JP (1976) On the formation of the Kuroshio meander and its relationship to the large scale ocean circulation. Deep-Sea Res 23: 33–47
Williams, PA, Worsey, MJ (1976) Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol 125: 818–828
Yeruham, I, Elad, D, Avidar, Y, Goshen, T, Asis, E (2004) Four-year survey of urinary tract infections in calves in Israel. Vet Rec 154: 204–206
Yoshpe-Purer, Y, Golderman, S (1987) Occurrence of Staphylococcus aureus and Pseudomonas aeruginosa in Israeli coastal water. Appl Environ Microbiol 53: 1138–1141
Young, AL, Leung, AT, Cheng, LL, Law, RW, Wong, AK, Lam, DS (2004) Orthokeratology lens-related corneal ulcers in children: a case series. Ophthalmology 111(3): 590–595
Zhang, Y, Yakrus, MA, Graviss, EA, Williams-Bouyer, N, Turenne, C, Kabani, A, Wallace, RJ Jr (2004) Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J Clin Microbiol 42: 5582–5587
Acknowledgments
The study was supported by Grants-in-Aid for Creative Basic Research #12NP0201 (DOBIS), and #14208063 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. We are grateful to Nihon Becton Dickinson Co., Ltd., Japan, for providing many Phoenix panels for the analyses. Our special thanks go to Dr. Kumiko-Kita Tsukamoto and Ms. Katomi Yao (Ocean Research Institute, The University of Tokyo) for doing 16S rDNA analyses of randomly selected isolates. We are also thankful to Ms. Reiko Shimatsu (Toho University School of Medicine) for her assistance during the BD Phoenix analyses. We remain indebted to Dr. Minoru Wada (Ocean Research Institute, Tokyo University) for his valuable advice during the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khan, N.H., Ishii, Y., Kimata-Kino, N. et al. Isolation of Pseudomonas aeruginosa from Open Ocean and Comparison with Freshwater, Clinical, and Animal Isolates. Microb Ecol 53, 173–186 (2007). https://doi.org/10.1007/s00248-006-9059-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9059-3