Abstract
In an anthropogenically disturbed soil (88% sand, 8% silt, 4% clay), 150-mg samples were studied to examine the fine-scale relationship of bacterial activity and community structure to heavy metal contaminants. The soils had been contaminated for over 40 years with aromatic solvents, Pb, and Cr. Samples from distances of <1, 5, 15, and 50 cm over a depth range of 40–90 cm underwent a sequential analysis to determine metabolic potential (from 14C glucose mineralization), bacterial community structure [using polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE)], and total extractable Pb and Cr levels. Metabolic potential varied by as much as 10,000-fold in samples <1 cm apart; log–log plots of metal concentration and microbial metabolic potential showed no correlation with each other. Overall, metal concentrations ranged from 9 to 29,000 mg kg−1 for Pb and from 3 to 8500 mg kg−1 for Cr with small zones of high contamination present. All regions exhibited variable metal concentrations, with some soil samples having 30-fold differences in metal concentration in sites <1 cm apart. Geostatistical analysis revealed a strong spatial dependence for all three parameters tested (metabolic activity, Pb, and Cr levels) with a range up to 30 cm. Kriging maps showed that in zones of high metal, the corresponding metabolic activity was low suggesting that metals negatively impacted the microbial community. PCR-DGGE analysis revealed that diverse communities were present in the soils with a random distribution of phylotypes throughout the sampling zones. These results suggest the presence of spatially isolated microbial communities within the soil profile.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
To date, several studies have shown the acute effects of heavy metals on the microbial community [7, 8, 12, 14, 18, 30, 42, 48, 49]. Results from these studies indicated that heavy metal amendments have a negative impact on the microbial community, resulting in a severe reduction of metabolic activity, microbial biomass, and bacterial abundance. A subsequent shift in the microbial community toward a more metal-tolerant or metal-resistant population has also been observed with increasing inputs of heavy metals [7, 13, 17, 39, 47]. These microbial responses tend to be dose-dependent, with increasing metal amendments causing a larger shift in the microbial response compared to the uncontaminated control [9, 50]. Few studies have attempted to address these same questions in chronically contaminated sites [26, 29], where natural selection should have selected for a metal-tolerant community. In addition, studies relating the issues of microbial activity and community structure to the spatial organization within the soil matrix are lacking in the literature.
Many of the studies performed on soil microbial communities are carried out with homogenized soil to disrupt the native spatial structure of the soil, allowing for bulk measurements to be performed. However, soil microorganisms are heterogeneously distributed in nature. The heterogeneity of soil is attributable to the combined biotic and abiotic factors present within the soil, such as competition, temperature, and available substrates [16]. Although bulk soil measures have helped in the understanding of large-scale dynamics, studies focusing on small-scale intricacies are rare. Furthermore, an understanding of microscale dynamics may lead to deeper understanding of factors limiting phenomena observed at larger scales.
Most bulk soil measurements do not adequately reflect what microbes are experiencing on a microscale level [37]. For example, fractionation studies have shown that the microbial community differs within a soil aggregate in the interior versus the exterior [49]. Other studies have also observed distribution patterns of microbes within the soil microarchitecture [23, 24, 32, 36, 40, 41] that would not have been seen using a bulk soil measure. Parkin [37] found that anaerobic microsites were responsible for most of the denitrification within a bulk soil sample. Thus, it is of interest to examine the microscale environment in soils where contaminant distribution is heterogeneous, to determine local effects on microbial activity and diversity.
Using geostatistical tools, spatial variation within a given area can be analyzed [16]. Unlike traditional statistics in which samples are assumed to be independent of one another, geostatistics provides a means to assess the spatial dependence of particular properties of microbes and their environment [45].
The purpose of the present study was to determine the spatial relationship between microbial community dynamics and local heavy metal contamination from a chronically contaminated site. A combination of traditional and geostatistical tools were used to determine if heavy metals negatively impacted the metabolic activity of the microbial communities in small, unsieved soil samples. Bacterial community analysis of these same samples was conducted to determine if particular phylotypes present within the communities varied with the level of local metal contamination.
Materials and Methods
Sampling Scheme and Study Site
Soil samples were gathered from an Indiana Department of Transportation (INDOT) site in Seymour, IN. The site was a former INDOT headquarters for road maintenance operations. In the late 1960s to early 1970s, excess road paint containing lead chromate as the pigment was dumped at thesite. In addition, painting instruments were cleaned with solvents (particularly toluene) at the site. A sandy backfill was brought in to cover the pit where the paint was dumped, where it then sat undisturbed for approximately 40 years.
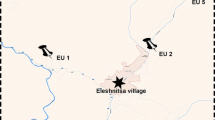
Soil samples for this study were collected in June 2001 from the Seymour INDOT property. A trench was dug in which samples were collected in nested hexagonal arrays with known distances between samples of 50, 15, and 5 cm (Fig. 1). A PbCrO4 paint layer, located 30 cm below the surface, ran the length of the sampling trench. Samples were taken below this paint layer and ranged in depth from 40 cm below the surface to 90 cm at the deepest point. Arrays 1 to 3 were located next to one another. The centers of these arrays were 50 cm from the next and spanned a distance of 1.35 m from the far end of array 1 to the farthest point of array 3. Array 4 was located directly opposite of array 1 on the opposing face of the sampling trench, 1.83 m to the north. Array 5 was taken from an area where no paint contamination was visible and no solvent odor could be detected. This array was 17.55 m away from the edge of array 3, and 18.90 m from the farthest sampling point in array 1. For each locus within the sampling array, three different 150 mg samples <1 cm apart from each other were analyzed. Individual 150-mg soil samples were subjected to sequential analyses for metabolic activity, bacterial community composition, and total metal concentrations.
(A) Schematic representation of arrays 1–3. Each array contains six subarrays with a center point located 15 cm from the center of the array as shown in array 3. For each subarray, samples were taken at the center point and at six locations 5 cm away from the center point. Sample locations were given a three-digit code, specified as array (ranging from 1 to 5), subarray (ranging from 1 to 6), and subarray location (ranging from 0 to 6). Subarrays and subarray location were numbered clockwise with “1” positioned 60° to the right of the vertical (as shown in array 3). Center points of each subarray were numbered with a “0”. (B) Representative results from subarrays 1 to 6 (circled in A). The three stacked numbers represent the three samples taken from within the same sampling point <1 cm distant. The two values for each point are the percent of glucose mineralized and lead concentration. A detailed list of all sample point metabolic potential and metal contents may be found on-line.
Metabolic Potential
The metabolic potential of microbes in soil was assessed by adding a commonly used substrate [uniformly labeled 14C glucose; 200 mCi mmol−1 (MP Biomedicals formerly ICN Biomedicals, Irvine, CA, USA)] and adding enough water so that water activity was not a limiting factor. A series of 25-mL serum vials were set up, each containing one 150-mg artificial aggregate along with a plastic trap containing 250 μL of 1 M KOH and a rubber stopper to seal the system. Soils were saturated with a water isotope mix of approximately 25 μCi of 14C glucose and 200 μL distilled water to ensure uniform distribution of the isotope; the concentration of glucose in the solution was 625 μM.
All vials were incubated at 22°C for 4 h. After 4 h, incubations were terminated by injecting 250 μL of 1 M HCl to the soil. Vials were allowed to sit for no more than 24 h, at which point potassium hydroxide present in the plastic cup trap was placed in scintillation vials containing 7 mL of Cytoscint™ scintillation cocktail. These vials were analyzed by scintillation counting (Tri-Carb, Model 1600 TR; Packard Instrument Company, Meriden, CT, USA). Prior to every run on the scintillation counter, the efficiency of the machine was tested with company standards to ensure that the machine was operating efficiently. Net cumulative disintegrations per minute (dpm) were calculated as the dpm from the aggregate minus a control consisting of only the isotope and water. Potential activity levels were expressed as the percent of added glucose metabolized.
Bacterial Community Analysis
Community structure of each artificial soil aggregate was determined using denaturing gradient gel electrophoresis (DGGE) to separate products from a polymerase chain reaction (PCR) of an amplified portion of the bacterial 16S rRNA gene. Total bacterial genomic DNA was extracted from soil samples using the QBioGENE (formerly BIO 101) FastDNA spin kit for soil (QBioGENE, Carlsbard, CA, USA). PCR amplification of the variable V3 region of 16S rDNA was performed using universal bacterial primers in a PTC100 Programmable Thermal Controller (MJ Research, Inc., Watertown, MA, USA). Primers were PRBA338F (5′-ACTCCTACGGGAGGCAGCAG) with a GC clamp needed to stabilize products in the DGGE [33], and PRUN518R (5′-ATTACCGCGGCTGCTGG). The PCR cycle consisted of a 92°C initial denaturation step for 5 min followed by 30 cycles of 30 s each at 92, 55, and 72°C, and ending with 15 min at 72°C. PCR product quality and quantity was confirmed by running 5 μL product on a 1% agarose gel.
Community structure was analyzed using the DCode™ Universal Mutation Detection system for DGGE gels (Bio-Rad Laboratories, Hercules, CA, USA). Polyacrylamide gels [8% (w/v) of 37.5:1 acrylamide/bisacrylamide] with a urea formamide denaturing gradient of 35–60% [where 100% denaturant contains 7 M urea, 40% (v/v) formamide] was utilized. Equal amounts of PCR product were loaded in each well of a preheated gel. Gels were run for 5.5 h at 60°C, with a constant voltage of 200 in 1× TAE buffer that was constantly stirred. SYBR Green I was used to stain the gels (1:10,000 dilution; Molecular Probes, Eugene, OR, USA), and gels were visualized on a UV transilluminator and photographed with a MP4 Land Camera (Cambridge, MA, USA).
Metal Analysis
Inductively coupled plasma atomic emission spectroscopy (ICP-AES) was used to determine the local level of metal contamination in each individual soil aggregate. Each sample was digested with nitric acid prior to ICP-AES analysis. Briefly, samples were predigested in 10 mL 50% nitric acid overnight, and subsequently heated at 140–150°C for 45 min. Five milliliters concentrated nitric acid was added to cooled samples. Samples were again heated for 45 min and 1-mL aliquots of 10% hydrogen peroxide were added to each sample until visible bubbling ceased. To remove particulate material, samples were filtered with 25 mm 0.45 μm Metricel® membrane filters (Pall Corporation, Ann Arbor, MI, USA).
Samples were run on an ICP–AES system (Atom Scan 16 Model, Thermo Jarrell Ash Corp Instrumentation Laboratory, Franklin, MA, USA) that uses an exogenous ultrasonic nebulizer (model number U-5000 AT+; CETAC Technologies, Omaha, NE, USA) to allow for equal distribution of the sample through the plasma flame to determine total extractable lead and chromium levels. Prior to each run, the system was calibrated using EPA standards (VWR International, Inc., West Chester, PA, USA) for lead and chromium and samples analyzed. For each run on the machine, the area under the curve was integrated five separate times and the average reported as the metal level for the sample.
Biomass Assessment by Direct Microscopic Counts
Thirty samples were randomly chosen from the five arrays to correlate metabolic potential analyses to cell numbers. We followed metabolic potential assays using 14C glucose as described above; bacteria were removed from the soil particles via shaking for 30 min at 125 rpm in the presence of 10 mM phosphate buffer. Cell suspensions were allowed to settle for 1–2 h, and the suspension was removed from the soil particulate with a pipette. One-milliliter samples were subsequently stained with 0.03% acridine orange, filtered onto 25-mm 0.22-μm black polycarbonate filters (Poretics Corporation, Livermore, CA, USA), and cells enumerated using an epifluorence standard research microscope (Carl Zeiss Inc., Thornwood, NY, USA) equipped with an HBO 200-W mercury burner, BG12 excitor filter and a barrier filter with a cutoff at 500 nm [5].
Statistical Analysis
Pairwise comparisons of individual DGGE gel fingerprints were made in the computer program BioNumerics (Applied Maths, Kortrijk, Belgium), for 178 samples. Using a set of eight amplicons run as two standard marker lanes run with each gel, DGGE gels profiles were normalized in BioNumerics to eliminate gel to gel variations. Only samples with high metal concentrations (>1500 mg kg−1 Pb), and high activity (≥1.6–40% glucose mineralized) were included in the analysis. Bands within individual profiles were visually confirmed and scored. Dice similarity coefficients were determined by comparing the portion of common bands present within the total number of bands between two discrete samples (SD = 2nAB/nA + nB, where nA is the number of bands in lane A, nB is the number of bands in lane B, and nAB is the number of common bands between lanes A and B) [12]. Dendrograms were constructed employing an unweighted pair group method with the arithmetic mean (UPGMA) algorithm. Statistically significant bands were identified using logistic regression to model the likelihood that a band was present in a sample as a function of each of (1) metabolic potential, (2) Cr concentration, and (3) Pb concentration. Because of the large number of statistical tests performed (i.e., 68 bands × 3 factors = 204 tests), p values were adjusted to control the false discovery rate (FDR) [3]. A significance cutoff of p < 0.05 was used. Statistical computations were performed in SAS (SAS Institute Inc, Cary, NC, USA).
Geostatistical analysis was performed using GS+: Geostatistics for the environmental sciences (Gamma Design Software, Plainwell, MI, USA). Isotropic and anisotropic analyses at 0, 45, 90, and 135° were performed on the data. As soon as the primary variate was calculated for each variable (activity, Pb, and Cr metal content levels) and the best-model fit, block kriging of the variates was performed. Block kriging was chosen over point kriging because this method allowed for slight differences between points to be smoothed; variate analysis performed using both point and block kriging showed no major differences. Spatial dependence (Q) was calculated as Q = C / (C + C0), where C is the sill variance (the maximum variance achieved with the model) and C0 is the nugget variance (the error unaccounted for by spatial autocorrelation) [20]. Q may range from 0 (no spatial structure) to 1 (strong spatial dependence).
Sequencing of Significant Bands
Bands found to be significant in the analysis described above were sequenced using an ALF express sequencer (Amersham Biosciences, Piscataway, NJ, USA). Significant bands were extracted from a DGGE gel (35–60% urea formamide gradient) using a sterile 1-mL pipette tip and placed in 100 μL RNase/DNase-free water to allow for the diffusion of the DNA from the gel into the water. PCR was then performed on the DNA from the gel plug using the same universal primers as those described above. If on additional DGGE gels the band of interest was not the only band in the profile, the band was again extracted and PCR repeated until this band was the only one present in the lane on subsequent DGGE gels. Purified bands were visually confirmed as that of interest by running a DGGE gel with the extracted band and the original sample side by side.
To sequence the bands, PCR was performed with the same primers used for DGGE analysis, but lacking the GC clamp. The amplicons were cloned into the pGEM T-EZ plasmid in DH5α. Clones were screened by DGGE analysis (using the same gradient as above) using whole-cell PCR. Those clones that migrated to the desired distance were sequenced using the Thermo Sequenase Primer Cycle Sequencing kit for ALF express sequencer (Amersham Biosciences). Both forward and reverse reactions were generated for each clone and multiple clones were sequenced for confirmation of the band. Using the BLASTn function [1], nucleotide sequences of excised bands were compared to sequences in the National Center for Biotechnology Information (NCBI) GenBank database and most similar sequences were noted.
Results
Metabolic Potential and Metal Contamination Levels Varied Within and Between Arrays
The four arrays that were contaminated with heavy metals showed highly variable levels of metabolic potential and metal content (Fig. 1B; a detailed list of all sample point metabolic potential and metal contents may be found on-line). Glucose mineralization was heterogeneous throughout the soil profile with levels varying by as much as 10,000-fold in samples less than 1 cm apart. Metabolic potential levels were not directly correlated with depth. Rather, a patchy distribution of isotope mineralization was evident, even atthe finest sampling scale (<1 cm distance). In array 5 (the uncontaminated site), samples exhibited high levels of metabolic potential; on average, 12 ± 5% of added glucose was mineralized. Metal levels were much lower than in the other arrays; the means and standard deviations were 17 ± 12 mg Pb kg−1 and 12 ± 8 mg Cr kg−1. The four contaminated arrays exhibited variable, heterogeneous metal concentrations, with much higher coefficients of variation than the uncontaminated samples (Table 1). Tenfold (or more) differences in metal level were found in samples separated by distances of <1, 5, 15, or 50 cm. The total metal concentrations ranged from9 to 29,000 mg kg−1 for lead and from 3 to 8500mg kg−1 for chromium.
To assess the relationship between microbial biomass and metabolic activity, 30 samples from the five arrays were assayed for metabolic potential using 14C glucose, followed by direct microscopic counting of bacterial cells extracted from the soil particles. Cell number and metabolic potential were not well correlated (r2 = 0.16), suggesting that there were differences in the physiological state of microbes among soil aggregations. Cell numbers were not that variable among the different samples tested, ranging from 0.5 to 3.2 × 107 cells g−1. Metabolic activity potentials were highly variable, ranging from no activity up to 40% mineralization of the added isotope.
Statistical Analysis of the Relationship Between Metabolic Potential and Metal Contamination
The direct relationship between microbial metabolic potential and metal contamination was evaluated in individual samples (Fig. 2); log–log correlations were poor when data from all samples were pooled. Lead and chromium levels were significantly correlated with one another (r = 0.95, p < 0.0001), whereas neither were significantly correlated with metabolic potential from arrays 1 to 4 (r= −0.04, p = 0.41 for Pb, r = 0.02, p = 0.67 for Cr).
Geostatistical analysis was then applied to evaluate spatial relationships. Semivariant and covariant analyses were performed on (1) the data for arrays 1–3, (2) array 4, and (3) array 5. Only arrays 1–3 could be grouped together for analysis as these arrays were physically located next to each other in a two-dimensional plane.
Analysis of arrays 1–3 indicated a spatial periodicity for net activity, Pb content, and Cr content. Both isotropic and anisotropic analyses at 0, 45, 90, and 135° were performed for each data set. The data showed strong anisotropy in the 45 and 135° directions as well as horizontally. In addition, the variograms indicated a spatial dependence range of 30 cm.
Block kriging was used to map the spatial relationships between the primary variate (activity) and its covariates (lead and chromium content). Kriging maps show that areas exist where increased metal concentrations occur with zones of decreased metabolic activity (Figs. 3A–C). This result is consistent with the idea that aggregations of high and low zones would be evident from a nested array sampling scheme [10].
Block kriging maps of (A) net metabolic activity, (B) Pb levels, and (C) Cr levels for arrays 1–3. The map portrays a plane that is 40 cm deep and 135 cm in width. The color coding is based on the natural log of microbial activity and metal content to better portray the large spatial differences that were found.
Variograms of arrays 4 and 5 indicated spatial ranges of 12 and 17 cm, respectively. In array 4, a heterogeneous distribution of heavy metals and metabolic potential was observed that was similar to that found in arrays 1–3. In contrast, array 5 had a homogeneous distribution of high metabolic potential and low concentrations of metals (data not shown).
Spearman rank order analysis for nonparametric data was used to evaluate cross correlations in order to determine the spatial relationships between the correlations of two variables. Cross-correlation coefficients were calculated for Pb to Cr levels as well as for either Pb or Cr to microbial metabolic potential (Fig. 4). The cross-correlation analysis showed that Pb and Cr were spatially structured and were thus spatially dependent. Cross correlations between the various metals and metabolic activity also exhibited spatial structure. At lags where the two metals were positively correlated (the metals cooccur), metabolic potential was negatively correlated to metal concentration; this confirms the view obtained from the kriging map (Fig. 3). In addition, the cross-correlation coefficients exhibit the same periodicity seen with the geostatistical variograms reinforcing the heterogeneous distribution of both the microorganisms and metal contaminants.
Using isotropic semivariograms, calculated values of spatial dependence showed that all three properties (activity, Pb content, and Cr content) were strongly spatially correlated for all arrays (Table 2). All of the calculated spatial dependence values were quite high, ranging from 0.77 to 0.99, indicating that there was a strong degree of spatial structure that best accounts for the observed differences in activity and metal contents at the individual sampling points.
Bacterial Community Analysis
Community analysis was performed to determine if particular phylotypes were selected under certain metal and activity conditions. The analysis concentrated on samples with both high metal contents and high metabolic activity. Cluster analysis of a group of 178 samples did not reveal any obvious pattern of association between community structure and particular metabolic potential levels or metal concentrations. A subset of those analyses is presented in Fig. 5. In addition, communities did not appear to group based on particular horizontal or vertical locations within the sampling space. For instance, array 4 samples did not form a distinct cluster. No dominant community fingerprint was found related to metabolic potential or metal content, suggesting that a diverse set of microorganisms is potentially responding to the metal stress. Furthermore, samples <1 cm distant had different banding patterns indicative of a high degree of phylogenetic heterogeneity among the communities. Community composition was heterogeneous on even the smallest spatial scales of <1 and 5 cm.
UPGMA comparisons of DGGE fingerprints from (A) samples within the same sampling point (<1 cm distant) all with high levels of metabolic potential and Pb contents, (B) samples within the same sampling point with high concentrations of Pb and variable levels of metabolic potential, and (C) samples within one subarray (5 cm distant) with high Pb contents and primarily high metabolic potential.
Of the 68 band classes generated in the community analysis, five were found to be unique to individual samples, and 44% were rarely found (in 1–10% of samples). Seventy-five percent of the total band classes were found in 20% or less of the 178 samples analyzed (Fig. 6). This suggests a high level of phylogenetic heterogeneity at this scale (150-mg aggregations of soil). No band class was found to occur at every sampling locus and only one band class occurred in over 50% of the samples. Further examination revealed that the band classes were randomly distributed throughout the soil. None of the bands appeared to localize with respect to depth. The majority of the bands were not localized to one particular array; rather, they occurred in all of the arrays or in arrays located at opposite ends of the sampling trench.
Frequency distribution of 68 band classes from the DGGE community profiles of 178 samples. The categories are defined as follows: Unique = found in only one sample, Rare = found in 1–10% of the samples, Infrequent = found in 10–20% of the samples, Less common = found in 20–35% of the samples, Common = found in 35–50% of the samples, Very common = found in more than 50% of the samples, Ubiquitous = found in every sample.
Two bands were identified that only occurred in samples with both high metal and high metabolic activity levels (mean Pb content = 5400 mg kg−1, mean metabolic activity = 22% metabolized. These phylotypes occurred in <15% of the 178 samples analyzed. These two bands were excised, cloned, and sequenced. Similarities of 100% to Actinobacteria sequences in the GenBank database were found for both 200-bp fragments. Specifically, one band had best matches to high G + C Gram-positive organisms including some in the families Actinobacteria (GenBank accession numbers AY167331.2 and AJ575541.1) and Microbacteriaceae (AY370615.1 andAF321022.1). For the other band, the best matches were to organisms in the genus Rhodococcus (GenBank accession numbers AF447392.1, AF447391.1, and AF420413.1).
Discussion
The negative acute effect of heavy metals on the microbial community has been well documented. Some of these acute responses include decreased microbial activity [7, 26–28, 39, 40, 42, 50], a decrease in microbial biomass [14, 38, 56], and a decrease in bacterial abundance [38] with increasing levels of metal inputs. In addition to studies that have examined the physiological response of the microbial community to metals, other studies have investigated how the microbial community is altered when a contaminant is added. Typically, metal is added to an uncontaminated soil and the resulting changes in the community are monitored. For instance, Díaz-Raviña et al. [9] added various concentrations of metals to soil microcosms and observed a shift toward a metal tolerant community. Changes in the microbial community toward one with more metal-tolerant or metal-resistant organisms, yet less overall phylogenetic diversity, has been widely observed in studies examining the acute effects of metals [7, 12, 13, 17, 27, 34, 38, 39, 42, 48, 54]. In contrast, only few studies have examined the effects of long-term, chronic exposure to particular contaminants [26, 29]. Communities from chronically exposed sites would have already undergone selection for a metal-tolerant community. We specifically chose a study site that had been contaminated for over 40 years to ascertain the effect of prolonged exposure to heavy metals on the microbial community. Specifically, we wanted to determine if the spatial distribution of the metal contaminant created a local microenvironment in the soil directly impacting the metabolic potential and community structure of small, 150-mg soil samples. In general, the results indicated thedegree of spatial heterogeneity within the site to be quite high.
The contaminated soils (arrays 1–4) exhibited a heterogeneous distribution of metabolic potential, whereas in uncontaminated soils from array 5 the distribution of activity was more homogeneous (Table 1). Heterogeneity was apparent in samples located less than 1 cm apart. It was possible that the metabolic potential assay merely reflected biomass size; however, metabolic activity was found to be poorly correlated to bacterial numbers. Of greater biological interest was the finding that metal content and activity were not significantly correlated. There are two possible explanations: (1) the metals were not bioavailable [50] or (2) there was selection for resistant microbial communities in some microsites with high metal concentrations.
Previous research had shown that a shift in the microbial community toward a metal-tolerant or metal-resistant community occurs in heavy metal contaminated soils [7, 12, 13, 17, 27, 34, 38, 39, 42, 48, 54]. We wanted to determine if particular phylotypes were present under different physical conditions and in particular if a dominant community fingerprint was evident in high activity/high metal situations. The sample size (150 mg) and sample handling (no sieving or pooling) used in the present study was chosen to permit detection of centimeter-scale spatial heterogeneity. Substantial heterogeneity in community composition was observed by using this approach; this implies either that the selective forces driving community structure vary at these spatial scales or that stochastic events such as immigration and extinction [25] are important forces structuring microbial communities in spatially isolated sites at these scales.
Results from the DGGE profiles indicated that contaminated samples were not populated by the same few phylotypes. Although the level of diversity was much lower than has been found in agricultural soil [52], the observed metacommunity diversity is consistent with reductions observed in soils dominated by a small amount of organic energy sources or contaminated with heavy metals [13, 21, 26, 43]. Of the 68 different phylotypes, the majority were found in less than 10% of the samples assayed. The horizontal transmission of plasmids with metal resistance determinants would be a mechanism to expand the number of successful phylotypes beyond a small number of metal-resistant strains that may have been initially present in the soils when they were originally contaminated over 40 years ago. Rasmussen and Sørensen [43] found a high degree of diversity within a soil community exposed to Hg, and concluded that diversity was attributable to horizontal gene transfer based on the large number of plasmids found within the recovered community. Some isolates from Seymour soils have been found to contain metal resistance mechanisms on mobile genetic elements [2, 6].
Because no clustering of metal or metabolic potential level was evident within the heterogeneous DGGE profiles, these factors do not appear to be driving the phylogenetic diversity seen in the different communities. A statistical approach [3] did find two Actinobacteria whose presence was restricted to those conditions. Other studies have also found a predominance of Gram-positive organisms in metal contaminated soils and in particular Actinobacteria [29, 32, 38]. In addition, the enrichment of Pb- or Cr-resistant bacterial cultures from these soils has yielded primarily Actinobacteria of the genera Arthrobacter and Rhodococcus ([1]; unpublished observations).
Because microorganisms are heterogeneously distributed within the soil, a variety of spatial scales of organization exist within the soil matrix. Geostatistics are a valuable tool used to assess spatial patterns within a given environment [37, 45]; traditional descriptive statistics do not provide this type of information. The sampling scheme (nested arrays) was designed to permit geostatistical analysis and represents one of the most complete data sets examining the spatial dependence of microbial distribution on multiple scales [10].
Studies attempting to address spatial heterogeneities in soils have fallen primarily into two groups. The first group is focused on spatial relationships at scales of many meters [11, 16, 46]. The second have focused on the microarchitecture of the soil [22–24, 32, 36, 40]. Few studies have addressed heterogeneities at an intermediate of centimeters to a few meters, as we have done in this study. In addition, the data set in this study was large enough to permit a significant number of comparisons to be made at a variety of scales.
Geostatistical analysis revealed that a spatial dependence was present in the sampling arrays. From the kriging maps for arrays 1–3 (which were contiguous to each other), a patchy distribution of metabolic activity and metal contamination was demonstrated. This is not an unexpected result, because it is known that soil is a nonuniform, heterogeneous environment in which the distribution of microorganisms is not ubiquitous [10]. Other studies have illustrated a similar trend in soil environments with regard to particular soil properties [19, 31, 36].
Inspection of the kriging maps suggested an inverse relationship between metal content and microbial activity. This result contradicts the analyses in Fig. 2. However, the two analyses have different underlying assumptions. In the log–log plots, samples are assumed to be independent of one another, whereas geostatistics was designed to analyze spatial relationships among samples. The analysis of individual samples indicated the forces operating at the microscale level, whereas geostatistical analysis integrated the effects that tend to dominate at a macroscopic scale. Taken together, the two analyses indicated that although there were active microbial communities in spatially isolated microsites with high metal concentrations, soil regions with high metal concentrations tended to be zones with lower metabolic activity.
Calculated cross correlations (Fig. 4) reinforce the geostatistical analysis by further illustrating the presence of spatial structure in the samples along with the inverse relationship between the metals and the metabolic activity potential. In addition, the cross-correlation analysis validates the need for samples to be gathered on a small scale, because a positive correlation between metals and metabolic activity would have been observed if samples were taken 30 cm apart, whereas a negative correlation between the metals and activity was observed using samples gathered a few centimeters apart. Cross correlation reveals the distances over which two variables are related to each other [35]. In this particular study (where gradients of Pb and Cr exist), it was of interest to determine the spatial relationships between these metals and microbial metabolic activity potential. As with any correlation, such relationships may be spurious, or they may indeed reveal information about the underlying process of interest. In this case, the periodicity observed in the cross correlation may be related to the applicationof the contaminant in the pit and subsequently redistributed.
Other studies have found a nonuniform distribution of microbes within the soil [4, 11, 15, 16, 44, 46, 51]. Although the Seymour community structure data was not analyzed for spatial dependence, the location of every community sample containing particular phylotypes was examined. The soil samples did not cluster by horizontal or vertical location. The 68 different phylotypes were randomly dispersed throughout the sampling trench. Ritz et al. [44] also observed a random distribution of diverse communities. Spatial isolation represents an explanation for high community heterogeneity [4, 55, 57, 58]. If samples are physically separated from one another, then interspecies resource competition is reduced, and rates of local immigration and extinction can result in higher diversity in the metapopulation than resource competition theory would predict [25]. Results consistent with this idea include experiments in microcosms with varying degrees of spatial isolation [53] and the high levels of microbial community diversity seen in sandy subsurface soils [4, 57, 58] and wetland sediment [4].
The heterogeneous community structures indicated by DGGE in this study are consistent with the idea that the soil matrix consists of a set of spatially isolated islands populated by microbes, particularly during the bulk of the year when the soils are not saturated with a continuous water layer. If the local (centimeter scale) communities are summed, the microbial metacommunity at this site is portrayed, and a higher level of diversity at the macroscopic level is revealed as a consequence of noncompetitive species dynamics [57] within the spatially isolated soil.
References
SF Altschul TL Madden AA Schaffer J Zhang Z Zhang W Miller DJ Lipman (1997) ArticleTitleGapped BLAST and PSI-BLAST: a new generation of protein database search programs Nucleic Acids Res 25 3389–3402 Occurrence Handle10.1093/nar/25.17.3389 Occurrence Handle1:CAS:528:DyaK2sXlvFyhu7w%3D Occurrence Handle9254694
Beasley, F (2004) Characterization of diversity, chromate resistance, and aromatic hydrocarbon degradation among Arthrobacter isolates from mixed waste soil. MS dissertation, Purdue University, West Lafayette
Y Benjamini Y Hochberg (1995) ArticleTitleControlling the false discovery rate—a practical and powerful approach to multiple testing J R Stat Soc, B 57 289–300
SJ Bent CL Gucker Y Oda LJ Forney (2003) ArticleTitleSpatial distribution of Rhodopseudomonas palustris ecotypes on a local scale Appl Environ Microbiol 69 5192–5197 Occurrence Handle10.1128/AEM.69.9.5192-5197.2003 Occurrence Handle1:CAS:528:DC%2BD3sXntlSjtLg%3D Occurrence Handle12957901
TD Brock (1978) ArticleTitleUse of fluorescence microscopy for quantifying phytoplankton, especially filamentous blue-green algae Limnol Oceanogr 23 158–160
JM Coombs T Barkay (2004) ArticleTitleMolecular evidence for the evolution of metal homeostasis genes by lateral gene transfer in bacteria from the deep terrestrial subsurface Appl Environ Microbiol 70 1698–1707 Occurrence Handle10.1128/AEM.70.3.1698-1707.2004 Occurrence Handle1:CAS:528:DC%2BD2cXisVKisL4%3D Occurrence Handle15006795
M Díaz-Raviña E Bååth (1996) ArticleTitleDevelopment of metal tolerance in soil bacterial communities exposed to experimentally increased metal levels Appl Environ Microbiol 62 2970–2977
M Díaz-Raviña E Bååth (2001) ArticleTitleResponse of soil bacterial communities pre-exposed to different metals and reinoculated in an unpolluted soil Soil Biol Biochem 33 241–248 Occurrence Handle10.1016/S0038-0717(00)00136-X
M Díaz-Raviña E Bååth Å Frostegård (1994) ArticleTitleMultiple heavy metal tolerance of soil bacterial communities and its measurement by a thymidine incorporation technique Appl Environ Microbiol 60 2238–2247 Occurrence Handle16349314
CH Ettema DA Wardle (2002) ArticleTitleSpatial soil ecology Trends Ecol Evol 17 177–183 Occurrence Handle10.1016/S0169-5347(02)02496-5
A Felske ADL Akkermans (1998) ArticleTitleSpatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils Microb Ecol 36 31–36 Occurrence Handle10.1007/s002489900090 Occurrence Handle1:CAS:528:DyaK1cXksVOrurs%3D Occurrence Handle9622562
K Feris P Ramsey C Frazar JN Moore JE Gannon WE Holben (2003) ArticleTitleDifferences in hyporheic-zone microbial community structure along a heavy-metal contamination gradient Appl Environ Microbiol 69 5563–5573 Occurrence Handle10.1128/AEM.69.9.5563-5573.2003 Occurrence Handle1:CAS:528:DC%2BD3sXntlSis7w%3D Occurrence Handle12957946
KP Feris PW Ramsey M Rillig JN Moore JE Gannon WE Holben (2004) ArticleTitleDetermining rates of change and evaluating group-level resiliency differences in hyporheic microbial communities in response to fluvial heavy-metal deposition Appl Environ Microbiol 70 4756–4765 Occurrence Handle1:CAS:528:DC%2BD2cXms1elu78%3D Occurrence Handle15294812
A Fließbach R Martens HH Reber (1994) ArticleTitleSoil microbial biomass and microbial activity in soils treated with heavy metal contaminated sewage sludge Soil Biol Biochem 26 1201–1205 Occurrence Handle10.1016/0038-0717(94)90144-9
RB Franklin LK Blum AC McComb AL Mills (2002) ArticleTitleA geostatistical analysis of small-scale spatial variability in bacterial abundance and community structure in salt marsh creek bank sediments FEMS Microbiol Ecol 42 71–80 Occurrence Handle1:CAS:528:DC%2BD38Xnt1KmtL4%3D Occurrence Handle12542032
RB Franklin AL Mills (2003) ArticleTitleMulti-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field FEMS Microbiol Ecol 44 335–346 Occurrence Handle1:CAS:528:DC%2BD3sXjs1Ckurk%3D Occurrence Handle12830827
Å Frostegård A Tunlid E Bååth (1996) ArticleTitleChanges in microbial community structure during long-term incubation in two soils experimentally contaminated with metals Soil Biol Biochem 28 55–63 Occurrence Handle10.1016/0038-0717(95)00100-X
Å Frostegård A Tunlid E Bååth (1993) ArticleTitlePhospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals Appl Environ Microbiol 59 3605–3617 Occurrence Handle16349080
LV Gonod C Chenu G Soulas (2003) ArticleTitleSpatial variability of 2,4-dichlorophenoxyacetic acid (2,4-D) mineralisation potential at a millimetre scale in soil Soil Biol Biochem 35 373–382 Occurrence Handle10.1016/S0038-0717(02)00287-0 Occurrence Handle1:CAS:528:DC%2BD3sXhvFGrtr0%3D
JH Görres MJ Dichiaro JB Lyons JA Amador (1998) ArticleTitleSpatial and temporal patterns of soil biological activity in a forest and an old field Soil Biol Biochem 30 219–230
BS Griffiths M Díaz-Raviña K Ritz JW McNicol N Ebblewhite N Bååth (1997) ArticleTitleCommunity DNA hybridisation and %G + C profiles of microbial communities from heavy metal polluted soils FEMS Microbiol Ecol 24 103–112 Occurrence Handle1:CAS:528:DyaK2sXmsl2ns78%3D
GL Grundmann D Debouzie (2000) ArticleTitleGeostatistical analysis of the distribution of NH4+ and NO2-oxidizing bacteria and serotypes at the millimeter scale along a soil transect FEMS Microbiol Ecol 34 57–62 Occurrence Handle1:CAS:528:DC%2BD3cXosVyrtLw%3D Occurrence Handle11053736
GL Grundmann A Dechesne F Bartoli JP Flandrois JL Chasse R Kizungu (2001) ArticleTitleSpatial modeling of nitrifier microhabitats in soil Soil Sci Soc Am J 65 1709–1716 Occurrence Handle1:CAS:528:DC%2BD38Xht1SmurY%3D
GL Grundmann P Normand (2000) ArticleTitleMicroscale diversity of the genus Nitrobacter in soil on the basis of analysis of genes encoding rRNA Appl Environ Microbiol 66 4543–4546 Occurrence Handle10.1128/AEM.66.10.4543-4546.2000 Occurrence Handle1:CAS:528:DC%2BD3cXnt1Cls78%3D Occurrence Handle11010914
SP Hubbell (2001) The Unified Theory of Biodiversity and Biogeography Princeton University Press Princeton, NJ
E Kandeler D Tscherko KD Bruce M Stemmer PJ Hobbs RD Bardgett W Amelung (2000) ArticleTitleStructure and function of the soil microbial community in microhabitats of a heavy metal polluted soil Biol Fertil Soils 32 390–400 Occurrence Handle10.1007/s003740000268 Occurrence Handle1:CAS:528:DC%2BD3cXosVSjtLY%3D
JJ Kelly M Häggblom RL Tate SuffixIII (1999) ArticleTitleChanges in soil microbial communities over time resulting from one time application of zinc: a laboratory microcosm study Soil Biol Biochem 31 1455–1465 Occurrence Handle1:CAS:528:DyaK1MXltVOks7Y%3D
M Khan J Scullion (2000) ArticleTitleEffect of soil on microbial responses to metal contamination Environ Pollut 110 115–125 Occurrence Handle10.1016/S0269-7491(99)00288-2 Occurrence Handle1:CAS:528:DC%2BD3cXksFyiur0%3D Occurrence Handle15092861
J Kozdrój JD Elsas Particlevan (2001) ArticleTitleStructural diversity of microbial communities in arable soils of a heavily industrialised area determined by PCR-DGGE fingerprinting and FAME profiling Appl Soil Ecol 17 31–42 Occurrence Handle10.1016/S0929-1393(00)00130-X
L Leita MD Nobili G Muhlbachova C Mondini L Marchiol G Zerbi (1995) ArticleTitleBioavailabilty and effects of heavy-metals on soil microbial biomass survival during laboratory incubation Biol Fertil Soils 19 103–108 Occurrence Handle10.1007/BF00336144 Occurrence Handle1:CAS:528:DyaK2MXkvVCkt7c%3D
SJ Morris (1999) ArticleTitleSpatial distribution of fungal and bacterial biomass in southern Ohio hardwood forest soils: fine scale variability and microscale patterns Soil Biol Biochem 31 1375–1386 Occurrence Handle1:CAS:528:DyaK1MXltVOks74%3D
DL Mummey PD Stahl (2004) ArticleTitleAnalysis of soil whole- and inner-microaggregate bacterial communities Microb Ecol 48 41–50 Occurrence Handle10.1007/s00248-003-1000-4 Occurrence Handle1:STN:280:DC%2BD2cvovVKgug%3D%3D Occurrence Handle15085298
G Muyzer EC Waal Particlede AG Uitterlinden (1993) ArticleTitleProfiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA Appl Environ Microbiol 59 695–700 Occurrence Handle1:CAS:528:DyaK3sXit1Kktrk%3D Occurrence Handle7683183
S Nazaret E Brothier L Ranjard (2003) ArticleTitleShifts in diversity and microscale distribution of the adapted bacterial phenotypes due to Hg(II) spiking in soil Microb Ecol 45 259–269 Occurrence Handle10.1007/s00248-002-2035-7 Occurrence Handle1:CAS:528:DC%2BD3sXkvFGls7Y%3D Occurrence Handle12658524
DR Nielsen O Wendroth (2003) Spatial and Temporal Statistics: sampling Field Soils and Their Vegetation Catena Verlag, GmbH Reiskirchen
N Nunan K Wu IM Young JW Crawford K Ritz (2002) ArticleTitleIn situ spatial patterns of soil bacterial populations, mapped at multiple scales, in an arable soil Microb Ecol 44 296–305 Occurrence Handle10.1007/s00248-002-2021-0 Occurrence Handle1:STN:280:DC%2BD38jhtFOmug%3D%3D Occurrence Handle12375095
TB Parkin (1993) ArticleTitleSpatial variability of microbial processes in soil—a review J Environ Qual 22 409–417
T Pennanen Å Frostegård H Fritze E Bååth (1996) ArticleTitlePhospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests Appl Environ Microbiol 62 420–428 Occurrence Handle1:CAS:528:DyaK28XovVCmug%3D%3D
L Ranjard E Brothier S Nazaret (2000) ArticleTitleSequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization andmicroscale distribution of soil bacterium populations responding to mercury spiking Appl Environ Microbiol 66 5334–5339 Occurrence Handle10.1128/AEM.66.12.5334-5339.2000 Occurrence Handle1:CAS:528:DC%2BD3MXjsVyhu7c%3D Occurrence Handle11097911
L Ranjard S Nazaret F Gourbière J Thioulouse P Linet A Richaume (2000) ArticleTitleA soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of community DNA fingerprints FEMS Microbiol Ecol 31 107–115 Occurrence Handle1:CAS:528:DC%2BD3cXltVCmug%3D%3D Occurrence Handle10640664
L Ranjard A Richaume (2001) ArticleTitleQuantitative and qualitative microscale distribution of bacteria in soil Res Microbiol 152 707–716 Occurrence Handle10.1016/S0923-2508(01)01251-7 Occurrence Handle1:STN:280:DC%2BD3Mnht1SqsQ%3D%3D Occurrence Handle11686384
L Ranjard A Richaume L Jocteur-Monrozier S Nazaret (1997) ArticleTitleResponse of soil bacteria to Hg(II) in relation to soil characteristics and cell location FEMS Microbiol Ecol 24 321–331 Occurrence Handle1:CAS:528:DyaK2sXotVyjtLk%3D
LD Rasmussen SJ Sørensen (1998) ArticleTitleThe effect of long-term exposure to mercury on the bacterial community in marine sediment Curr Microbiol 36 291–297 Occurrence Handle10.1007/s002849900312 Occurrence Handle1:CAS:528:DyaK1cXislWjsLc%3D Occurrence Handle9541566
K Ritz JW McNicol N Nunan S Grayston P Millard D Atkinson D Gollotte D Habeshaw B Boag CD Clegg BS Griffiths RE Wheatley LA Glover AE McCaig JI Prosser (2004) ArticleTitleSpatial structure in soil chemical and microbiological properties in an upland grassland FEMS Microbiol Ecol 49 191–205 Occurrence Handle1:CAS:528:DC%2BD2cXlsl2iur0%3D
RE Rossi DJ Mulla AG Journel EH Franz (1992) ArticleTitleGeostatistical tools for modeling and interpreting ecological spatial dependence Ecol Monogr 62 297–314
P Saetre E Bååth (2000) ArticleTitleSpatial variation and patterns of soil microbial community structure in a mixed spruce–birch stand Soil Biol Biochem 32 909–917 Occurrence Handle10.1016/S0038-0717(99)00215-1 Occurrence Handle1:CAS:528:DC%2BD3cXltVCrur4%3D
R-A Sandaa V Torsvik Ø Enger (2001) ArticleTitleInfluence of long-term heavy-metal contamination on microbial communities in soil Soil Biol Biochem 33 287–295 Occurrence Handle10.1016/S0038-0717(00)00139-5 Occurrence Handle1:CAS:528:DC%2BD3MXhslKls7s%3D
R-A Sandaa V Torsvik Ø Enger FL Daae T Castberg D Hahn (1999) ArticleTitleAnalysis of bacterial communities in heavy-metal contaminated soil at different levels of resolution FEMS Microbiol Ecol 30 237–251 Occurrence Handle1:CAS:528:DyaK1MXmsFahs78%3D Occurrence Handle10525180
A Sessitsch A Weilharter MH Gerzabek H Kirchmann E Kandeler (2001) ArticleTitleMicrobial population structures in soil particle size fractions of a long-term fertilizer field experiment Appl Environ Microbiol 67 4215–4224 Occurrence Handle10.1128/AEM.67.9.4215-4224.2001 Occurrence Handle1:CAS:528:DC%2BD3MXmslWisL0%3D Occurrence Handle11526026
W Shi M Bischoff R Turco A Konopka (2002) ArticleTitleLong-term effects of chromium and lead upon the activity of soil microbial communities Appl Soil Ecol 21 169–177 Occurrence Handle10.1016/S0929-1393(02)00062-8
MK Sliwinski RM Goodman (2004) ArticleTitleSpatial heterogeneity of Crenarchaeal assemblages within mesophilic soil ecosystems as revealed by PCR-single-stranded conformation polymorphism profiling Appl Environ Microbiol 70 1811–1820 Occurrence Handle1:CAS:528:DC%2BD2cXisVKisb0%3D Occurrence Handle15006808
V Torsvik L Øvreås TF Thingstad (2002) ArticleTitleProkaryotic diversity—magnitude, dynamics, and controlling factors Science 296 1064–1066 Occurrence Handle10.1126/science.1071698 Occurrence Handle1:CAS:528:DC%2BD38Xjslantrc%3D Occurrence Handle12004116
DS Treves B Xia J Zhou JM Tiedje (2003) ArticleTitleA two-species test of the hypothesis that spatial isolation influences microbial diversity in soil Microb Ecol 45 20–28 Occurrence Handle10.1007/s00248-002-1044-x Occurrence Handle1:STN:280:DC%2BD3s%2FmsVWltw%3D%3D Occurrence Handle12415419
R Turpeinen T Kairesalo M Häggblom (2004) ArticleTitleMicrobial community structure and activity in arsenic-, chromium- and copper-contaminated soils FEMS Microbiol Ecol 47 39–50 Occurrence Handle1:CAS:528:DC%2BD2cXltVyntg%3D%3D
B Xia DS Treves J Zhou JM Tiedje (2001) ArticleTitleSoil microbial community diversity and driving mechanisms Prog Nat Sci 11 818–824
H Yao J Xu C Huang (2003) ArticleTitleSubstrate utilization pattern, biomass and activity of microbial communities in a sequence of heavy metal-polluted paddy soils Geoderma 115 139–148 Occurrence Handle10.1016/S0016-7061(03)00083-1 Occurrence Handle1:CAS:528:DC%2BD3sXjvVCls7Y%3D
J Zhou B Xia H Huang AV Palumbo JM Tiedje (2004) ArticleTitleMicrobial diversity and heterogeneity in sandy subsurface soils Appl Environ Microbiol 70 1723–1734 Occurrence Handle1:CAS:528:DC%2BD2cXisVKisL0%3D Occurrence Handle15006798
J Zhou B Xia DS Treves L-Y Wu TL Marsh RV O'Neill AV Palumbo JM Tiedje (2002) ArticleTitleSpatial and resource factors influencing high microbial diversity in soil Appl Environ Microbiol 68 326–334 Occurrence Handle1:CAS:528:DC%2BD38Xjt1WmtQ%3D%3D Occurrence Handle11772642
Acknowledgments
This work was supported by a grant from the Department of Energy's Natural and Accelerated Bioremediation Research (NABIR) program (Grant no. DE-FG02-98ER62681). Our thanks to Indiana Department of Transport and, in particular, Bill Jarvis for giving us access to thissite.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Becker, J.M., Parkin, T., Nakatsu, C.H. et al. Bacterial Activity, Community Structure, and Centimeter-Scale Spatial Heterogeneity in Contaminated Soil. Microb Ecol 51, 220–231 (2006). https://doi.org/10.1007/s00248-005-0002-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-005-0002-9