Abstract
Inoculation with Azospirillum brasilense exerts beneficial effects on plant growth and crop yields. In this study, a comparative analysis of maize (Zea mays) root inoculated or not inoculated with A. brasilense strains was performed in two soils. Colonization dynamics of the rhizobacteria were tracked in various root compartments using 16S rRNA-targeted probes and 4′,6′diamidino-2-phenylindole staining, and the structure of bacterial populations in the same samples was analyzed by denaturing gradient gel electrophoresis (DGGE) of polymerase chain reaction products of the 16S rRNA gene. Based on whole cell hybridization, a large fraction of the bacterial community was found to be active in both the rhizoplane–endorhizosphere and rhizosphere soil compartments, in both soil types. A DGGE fingerprint analysis revealed that plant inoculation with A. brasilense had no effect on the structural composition of the bacterial communities, which were also found to be very similar at the root tip and at zones of root branching. However, rhizobacterial populations were strongly influenced by plant age, and their complexity decreased in the rhizoplane–endorhizosphere in comparison to rhizosphere soil. A clone library generated from rhizosphere DNA revealed a highly diverse community of soil and rhizosphere bacteria, including an indigenous Azospirillum-like organism. A large proportion of these clones was only distantly related to known species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The introduction of specific bacteria into the rhizosphere to promote plant growth and to antagonize soil-borne plant pathogens has been the subject of intensive research during the last decades, and this practice has become accepted in agriculture [50].
Plant-growth-promoting rhizobacteria (PGPR) are bacteria with plant-growth-stimulating activity, which may result from different mechanisms, such as the production of plant-stimulating growth substances (phytohormones) or the suppression of minor plant pathogens by various mechanisms [9, 14, 50]. Plant-growth-promoting effects of Azospirillum brasilense are well documented for crops of agronomic importance, with yield increases of cereals of up to 30% [7, 8, 36]. These effects are mainly derived from morphological and physiological changes of the inoculated plant roots, leading to an enhancement of water and mineral uptake [12, 35]. However, the ecological impacts that A. brasilense inoculation might have on the rhizosphere microbiota are not well known. Introduction of a large quantity of exogenous bacteria may bring about different outcomes: a marked perturbation in the rhizosphere-associated microbiota, disrupting the local ecosystem, transiently or permanently alteringthe composition of the indigenous microbial populations [31], or exert no effect on soil resident microbial populations [3]. Utilization of microorganisms in agriculture requires an evaluation of environmental risks associated with the introduction of indigenous or nonindigenous microorganisms into the plant rhizosphere for different purposes. Furthermore, the successful establishment of introduced microorganisms in the rhizosphere depends on the ability of the bacterium to colonize roots and to compete with the indigenous microbiota [6]. Many studies have shown the positive effects of Azospirillum inoculants on plant growth [36], but there is a scarcity of knowledge regarding their impact on the rhizosphere biota. Questions pertaining to root colonization dynamics by A. brasilense and the influence of its inoculation on natural bacterial communities should be addressed, as an increased knowledge of the ecology of plant-growth-promoting rhizobacteria is important for their successful implementation. This, in turn, may help promote sustainable agriculture.
In the present study, we applied molecular phylogenetic probes and primers targeting the 16S rRNA and rDNA of bacteria to assess the impact of A. brasilense on root and rhizosphere microbial colonization and on the structure of the microbial communities associated with various root compartments.
Materials and Methods
Bacterial Strain and Media
A. brasilense wild-type strains Cd [11] and Sp245 [2] were grown at 30°C in a low C/N medium adjusted to pH 6.8 and containing (in grams per liter) d-fructose, 6.67; MgSO4, 0.2; NaCl, 0.1; CaCl2, 0.02; K2HPO4, 6.0; KH2PO4, 4.0; yeast extract (Difco), 0.1; NH4Cl, 0.963; and microelements as described by Okon et al. [34]. Flasks were inoculated with about 109 cfu mL−1 of exponential-phase cultures of the bacterial strains inoculum (grown to an OD540 of approximately 0.2) and incubated on a rotary shaker (150 rpm) at 30°C for 24 h until late exponential phase.
Plants, Plant Growth, and Inoculation
Zea mays cv. Jubilee seeds (Hazera Co., Haifa, Israel) were surface sterilized with 1% sodium hypochlorite for 2 min at room temperature and 1 min in 70% ethyl alcohol. The seeds were washed extensively with sterile distilled water and allowed to germinate for 2 days in sterile dish plates with wet paper in the dark at 30°C. For inoculation purposes, A. brasilense strains Cd or Sp245 were grown as previously described, washed with a sterile NaCl solution (0.85%), and diluted to 5 × 107 cfu mL−1. Seedlings of approximately 1 cm in length were transferred to petri dishes containing the cell suspension and incubated for 1 h at room temperature. A sterile NaCl solution was used in control seedlings. The seedlings were then aseptically transferred (three seeds per pot) either to an Haploxeralfs brown-red sandy Rehovot soil (clay 0%, silt 4.65%, sand 95.3%, field capacity 5.5%, pH 7.23, conductivity 0.112 mS/cm, organic matter 0.9%) packed in 100 × 10 cm PVC tubes, amended with K2SO4 and super phosphate Ca(H2PO4) at 1 g/kg soil, or to a Chromoxererts brown alluvial Kfar Menahem soil (clay 6%, silt 8%, sand 86%, field capacity 27.9%, pH 8.4, organic matter 0.5%) in 30 × 25 cm 10-kg pots. One milliliter of cell suspension of strains Cd or 245 or Cd only was added to each seedling, directly into the soil, in the Rehovot and Kfar Menahem soils, respectively, thus combining seed and soil inoculation, insuring colonization of the seedling. Controls were mock-inoculated with a NaCl solution. Plants were grown either in a phytotron with a 28/22°C, 16/8 h day/night period at 75% humidity, watered once a week with modified Jensen nutrient solution [51] and twice a week with distilled water to near-field capacity (Rehovot soil), and sampled at different times [7 and 21 days postemergence in denaturing gradient gel electrophoresis (DGGE) analysis; 3, 7, and 21 days postemergence in fluorescence in situ hybridization (FISH) analysis], or in an acclimatized greenhouse with a 28/15°C day/night period and watered daily using a drip irrigation system to maintain near-field capacity. In that latter case, plants were harvested 7, 20, 29, and 40 days postemergence (Kfar Menahem soil).
In both soils, treatments were in triplicates, and the experiments were performed twice. As similar results were obtained, the results of only one experiment per soil are presented.
Preparation of Root Samples for DNA Extraction and Whole Cell Hybridization
At every sampling time point, roots were carefully separated from adhering soil particles and briefly washed in 0.05 M Phosphate buffer (PB) with 0.5% cetyltrimethylammonium bromide (CTAB). The soil suspension (root-adhering soil particles) constituted the rhizosphere soil. After fresh weight was measured and subsamples taken for dry shoot weight determination by oven dehydration at 70°C for 3–4 days, root tips and zones of branching roots of approximately 1 cm in length were aseptically separated and macerated in 0.05 M PB with 0.5% CTAB, 0.1% Na-cholate, 0.2 g chelex 100, and 0.25 g polyethylene glycol 6000 (PEG 6000) [45]. In the experiments with the KfarMenahem soil, the whole washed root system was used, with no separation between root tips and root branching zones. Macerated samples from root tips and branching zones included bacteria located inside the roots (endophytic bacteria) and at the root surface as well as tightly adhering soil particles and constituted the rhizoplane. Bacterial cells were extracted from the rhizosphere soil and from roots by chemical dispersion and centrifugation as described by Schallmach et al. [44], with some modifications. Only three centrifugation stepswere used, two at 700 rpm, 4°C for 10 min, and a final one at 4500 rpm, 4°C for 10 min. Subsamples weretaken for formaldehyde fixation and nucleic acid extraction (one subsample for every procedure). Fixation was performed overnight using 4% paraformaldehyde inPBS at 4°C. The subsamples were then washed andstored in 50% ethyl alcohol at −20°C until hybridization.
Fluorescence In Situ Hybridization and Epifluorescence Microscopy
Fluorescence in situ hybridization (FISH) was performed as described by Stoffels et al. [47] on Teflon-coated glass with eight wells for independent positioning of the samples, following the method of Manz et al. [25]. Formamide and NaCl were added to hybridization and washing buffers, respectively, according to the probes used for the hybridization. The following oligonucleotide probes were used: (1) a mixture of the oligonucleotide probes EUB338 [1], EUB788c [21], EUB927c [13], EUB1055c [21], and EUB1088c [21], complementary to regions of the 16S rDNA specific for the domain Bacteria and referred to as EUBmix; (2) AZO440b, complementary to a region of the 16S rDNA conserved in the Azospirillum–Rhodocista–Skermanella cluster [47]; (3) AZOI-655, complementary to a region of the 16S rDNA conserved in the Azospirillum cluster [47]; and (4) Abras1420, complementary to a region of the 16S rDNA conserved in A. brasilense—applied only in confocal microscopy [47]. Probes AZO440b and AZOI-655, when used together in combination, were referred to as AZOmix. Oligonucleotide probes labeled at the 5′-end with the fluorescent dyes 6-carboxytetramethylrhodamine (TAMRA, used for labeling the EUBmix) and fluorescein (FLUOS, used for labeling the AZOmix) were purchased from MWG Biotech, Ebersberg, Germany. Following hybridization, 4′,6′diamidino-2-phenylindole (DAPI, Sigma, Israel) staining was performed. Epifluorescence microscopy was performed with a BX51 Olympus upright microscope (Tokyo, Japan). Cells were counted under oil immersion using a 100 × 1.3 objective and a 10/10 grid ocular with an area of 10,000 μm2. When AZOmix-labeled probes were used, the sample was cohybridized with the EUBmix and counterstained with DAPI, thus allowing triple staining of the sample. Fractions of EUBmix hybridizing to DAPI-stained cells, and of cells hybridizing with specific probes (AZOmix) to EUBmix positive cells, were always related to the same fields.
Confocal Laser Scanning Microscopy
Confocal microscopy was performed on samples stained by FISH with Cy3- or TAMRA-labeled EUBmix and Cy5-labeled AZOI-655 or Cy5-labeled Abras1420 oligonucleotide probes in combination. Visualization was performed using a Zeiss 510LSM confocal laser scanning microscope (Zeiss, Oberkochen, Germany) with two helium neon lasers for excitation of Cy3 and TAMRA at 543 nm and for Cy5 at 633 nm. Cells were observed under water immersion using a 63× objective. In hybridizations using Cy5- and TAMRA-labeled oligonucleotide probes, fluorescent signals were assigned a red color and a green color, respectively. In hybridizations using Cy3- and Cy5-labeled oligonucleotide probes, fluorescent signals were assigned red and blue colors, respectively. When Cy5- and TAMRA-labeled oligonucleotide probes were used, the combination of the two single color images into one picture identified A. brasilense cells as yellow. Alternatively, a magenta coloring identified A. brasilense cells when the Cy3-labeled EUBmix (red fluorescence) and Cy5-labeled Abras1420 (blue fluorescence) oligonucleotide were applied.
DNA Extraction, PCR Conditions, and DGGE Analysis
The procedure used for DNA extraction was based on Tsai and Olson [48], with slight modifications as described by Schallmach et al. [44]. The forward GC-clamped GM5F and reverse 907R primers, yielding c. 580-bp-long products of the 16S rRNA gene of the members of the Bacteria domain, were used [30]. Polymerase chain reaction (PCR) amplifications of 16S rDNA fragments were performed on an automated PCR thermoblock (Eppendorf Mastercycler Gradient, Brinkmann Instruments, USA). PCR mixtures contained 5 μL of 10× magnesium-free buffer (Promega, Wisconsin, USA), 0.4 mg/mL bovine serum albumin, 3.75 mM MgCl2, 300 μM of each deoxynucleoside triphosphate, 800 nM of each primer, 0.03 U μL−1 of Red Taq DNA polymerase (Sigma), and 1 μL of template DNA in a final volume of 50 μL using sterile 0.2 μm filtered double-distilled water. The PCR included denaturation at 95°C for 1 min, 35 thermal cycles of 20 s at 95°C, 25 s of annealing at 57°C, and 30 s at 72°C, ending with a 1-min extension step at 72°C. Products were checked by electrophoresis in 1% (w/v) agarose and stained with ethidium bromide (0.2 mg/mL). Denaturing gradient gels were prepared according to Muyzer et al. [29] and ran in a Phor-U2 system (Ingeny, Goes, Holland). The PCR products were separated in a 6% (w/v) polyacrylamide gel [acrylamide/bisacrylamide (37:1)] in a 1× Tris–acetate–EDTA (TAE) buffer with a 30–60% denaturing gradient [80% denaturant corresponding to 7 M urea and 32% (v/v) formamide]. Electrophoresis was performed in 1× TAE at 60°C, 85 V, for 20 h. The gels were stained for 30 min in 0.5 mg L−1 ethidium bromide in a 1× TAE and rinsed for 15 min in a 1× TAE buffer. Visualization was performed with an AlphaImager™ System (Alpha Innotech Corporation, California, USA) using the AlphaEaseFC™ standard software package (Labtrade Inc., Florida, USA). Analysis was performed using the Gel-Pro Analyzer software (Media Cybernetics, Maryland, USA) that enables quantification of the number of bands in each lane and determines their relative position. Pictures were digitized and analyzed with the Quantity One software (Bio-Rad, Rishon Le Zion, Israel). The analysis of the DGGE fingerprints was performed using the Dice algorithm (Dice coefficient of similarity) taking into account the presence of a band and its position when comparing samples for similarity. The unweighted pair group method with mathematical averages option was used for cluster analysis and for the construction of complete linkage dendrograms.
Clone Library Construction and Phylogenetic Analysis
Polymerase chain reaction products generated using the GM5-907R primer set were excised from agarose gels using sterile razor blades and purified with the High pure PCR product purification kit (Boehringer Mannheim, Germany). Competent Escherichia coli DH 5a cells were prepared as described by Sambrook et al. [43]. The pGEM-T® Easy Vector System I (Promega) was used for the cloning of reamplified excised bands according to the manufacturer's instructions, with an additional 5-min incubation step at 42°C followed by 5 min on ice, repeated five times. Plasmids from selected colonies were purified with the QIAprep Spin Miniprep Kit (Qiagen, Germany). Clones were sequenced (in the forward direction) at the Center for Genomic Technologies of the Hebrew University (Jerusalem, Israel). Sequences were analyzed by CHIMERA_CHECK at the Ribosomal Database Project II (http://rdp.cme.msu.edu/html) [24], and suspected chimera sequences were removed from the analysis. A phylogenetic tree was constructed using the ARB package [23]. The ARB-ALIGN tool was used for sequence alignment. The resulting alignments were checked and corrected manually. Phylogenetic trees were constructed based on E. coli numbering of 16S rDNA sequences, using the ARB package software neighbor joining, parsimony, and maximum-likelihood methods in combinations with different filters and different corrections. The consistency of the tree was also verified by bootstrapping (n = 1000) for neighbor joining and parsimony.
Nucleotide Sequence Accession Numbers
Sequences of clones obtained in this work were deposited atthe GenBank under accession numbers AY669090–AY669116.
Statistical Analysis
A two-way analysis of variancewas applied using ANOVA in JMP (SAS Institute Inc., North Carolina, USA). The F test was applied to the whole experimental model, and significant differences (p = 0.05) were determined. In root colonization, a lognormal distribution was assumed [22]. A sample was defined as bulked root compartments (rhizosphere, tip, or branching zone) of a plant replicate in a specific treatment and time point of sampling following emergence. Because data for the different root zones were collected from the same plant, the individual plant was added as a factor in the model, and the variance between plants within treatment was taken as the error term when comparing treatments. Triplicate plants per treatment and sampling time were analyzed. In each triple-stained sample, i.e., DAPI stained cells, EUBmix hybridizable cells, and the combined specific probes AZOmix were counted in 20 different fields.
Results
Identification and Visualization of A. brasilense Cells on Root Segments
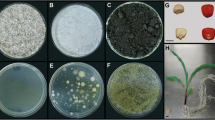
A. brasilense bacterial cells colonizing the rhizosphere and rhizoplane of young maize seedlings were visualized by confocal laser scanning microscopy (CLSM). CLSM enabled the localization andexamination of the spatial distribution of the inoculated bacteria. The inoculated strains were abundant in the rhizoplane, colonizing primary and secondary root surfaces (Figs. 1A–C) and appeared mainly as microcolonies, whereas in the rhizosphere, the cells appeared to cluster with other rhizobacteria (Fig. 1D). Similar results were obtained in both Rehovot sand and Kfar Menahem loamy sand. A low background signal and an enhanced contrast resulted in improved image quality. However, quantitative counting of fluorescence-labeled bacteria and statistical analysis of root colonization in the CLSM images was not possible. Therefore, cell counts were performed using epifluorescence microscopy.
In situ identification and localization of A. brasilense colonizing maize roots grown in Rehovot and Kfar Menahem soils using confocal imaging. (A)–(C). Branching zone of a maize root inoculated with A. brasilense strain Sp245 grown in Rehovot soil. Simultaneous hybridization with Cy3-labeled consensus probe EUBmix and Cy5-labeled probe Abras1420 identified numerous A. brasilense (marked by white arrows) on the root surface. Other colonizing bacteria are also visible (hollow arrows). (D) Rhizosphere of a maize root inoculated with A. brasilense strain Cd grown in a Kfar Menahem soil. Simultaneous hybridization with TAMRA-labeled consensus probe EUBmix and Cy5-labeled probe AZOI-655. A. brasilense (marked by white arrows) was identified by the overlay of TAMRA and Cy5-conferred fluorescence. Several other rhizobacteria are shown (hollow arrows). Bar = 10 μm.
Quantification of Spatial and Temporal Patterns of Root Colonization
Quantification and comparison of bacterial cell numbers obtained by DAPI staining in the loamy Kfar Menahem soil revealed that the total number of rhizosphere soil and rhizoplane (whole root) bacteria did not differ significantly between inoculated and control plants throughout the course of the experiments (Table 1A). A high proportion of the DAPI-stained cells also hybridized with the EUBmix probes. Because DAPI stains dead, inactive, and active bacteria, the high ratio of EUBmix (hybridizable)-stained cells to DAPI-stained cells indicated that the large majority of the bacterial communities at the rhizoplane–endorhizosphere interface was composed of metabolically active cells. This was also observed in rhizosphere soil bacterial communities.
The use of probes AZOI-655 and AZO440b (AZOmix) with the EUBmix probe in a top to bottom approach yielded high numbers (>7 × 107 cells g−1) of hybridizing cells in the rhizoplane, both in the control and in the inoculated treatments. However, control experiments with a nonsense probe (a nucleotide probe having the nucleotide sequence complementary to the nucleotide sequence of the Azospirillum specific sense probe) revealed that about 35% of the signal was due to nonspecific binding by the fluorescein fluorochrome part of the probe interacting with hydrophobic cell components (data not shown).
Similar trends were observed when maize was grown in a sandy Rehovot soil under the controlled conditions of a phytotron. The total cell number, as counted by DAPI staining, did not differ significantly between inoculated and control plants (Table 1B). Significantly more cells were counted in the rhizoplane, particularly in the root tip zone, than in the rhizosphere soil. In the root tip (T) and branching zone (B), virtually all DAPI-stained cells hybridized with the Bacteria (EUBmix) domain-specific probes indicating an active bacterial community. Significantly more AZOmix hybridizing cells were found on the root tips of inoculated than in control plants during the first week following emergence. However, as was previously observed, this population was overestimated, in part due to nonspecific binding of the fluorescein fluorochrome of the probes. No differences between the treatments (control vs strains Cd and Sp245 as well as Cd vs Sp245) were found in the branching zone at any time point. At 21 days postemergence, the number of EUB-hybridizable cells declined in all treatments, although this trend was not statistically significant.
DGGE Analysis
Total community DNA was extracted from the same root samples used in whole cell hybridization. PCR products obtained with eubacterial primers GM5F-907R were analyzed in different combinations on DGGE gels to test both inoculation of A. brasilense and plant growth stage effects on bacterial populations in the rhizosphere soil and in the rhizoplane of maize.
Cluster analysis of the rhizosphere soil fingerprints of plants grown in Kfar Menahem loamy sandy soil revealed a high degree of similarity between A. brasilense strain Cd-inoculated and control plants. No significant differences were observed between the treatments throughout the course of the experiments. Although inoculation had no apparent effect on the structure of the rhizobacterial community, a clear temporal, i.e. plant age effect was detected: samples from the two earliest sampling dates (7 and 20 days post emergence) were more similar to each other than samples from the two latter sampling dates (29 and 40 days post emergence) (Fig. 2). Bacterial communities of the rhizoplane–endorhizosphere fraction of whole root samples (excluding the rhizosphere soil) were more temporally stable, as the DGGE profiles for both inoculated and control plants were highly similar among sampling dates (not shown).
Dendrogram representing the similarity of bacterial PCR-DGGE profiles of rhizosphere soil bacterial communities of control maize plants and of plants inoculated with A. brasilense strain Cd, grown in a Kfar Menahem loamy soil. Control (1, 2, 3, 7, 8, 9, 13, 14, 15, 19 20, 21), Cd-inoculated (4, 5, 6, 10, 11, 12, 16, 17, 18, 22, 23, 24) plants from the 7 (1 to 6), 20 (7 to 12), 29 (13 to 18) and 40 (19 to 24) days post emergence. 7d, 20d, 29d, 40d numbering indicate days post emergence. Scale indicates similarity. Similarity is expressed as a value of the Dice correlation coefficient with a value of zero indicating the samples are completely different, whereas a value of 1.00 indicates that they are identical.
In a Rehovot sandy soil, the DGGE patterns generated in the rhizosphere soil of A. brasilense Cd- and Sp245-inoculated plants one and three week post emergence were similar and clustered together with those inferred for the control treatment, but the two sampling dates were clearly separated (Fig. 3), implying that plant age was a major factor influencing rhizobacterial community structure. The composition of the rhizoplane–endorhizosphere communities appeared not to be affected by the application of the PGPR treatments. However, the community structure was influenced by plant age, both at the root tips and in the zones of emerging secondary roots (not shown).
Dendrogram representing the similarity of bacterial PCR-DGGE profiles of rhizosphere soil bacterial communities of control maize plants and of plants inoculated with A. brasilense strains Cd and Sp245, and grown in a Rehovot sandy soil. Control (lanes 1, 2, 3, 10, 11, 12), Cd-inoculated (lanes 4, 5, 6, 13, 14, 15) and Sp245-inoculated (lanes 7, 8, 9, 16, 17, 18) plants from the first (lanes 1 to 9) and third (lanes 10 to 18) week post emergence. Scale indicates similarity. Similarity is expressed as a value of the Dice correlation coefficient with a value of zero indicating the samples are completely different, whereas a value of 1.00 indicates that they are identical.
In Rehovot soil, the DGGE profiles of root tips and zones of root branching were highly similar at each sampling point. Population complexity (determined using the Gel-Pro software) in these samples was significantly lower (p < 0.05) than in samples of rhizosphere soil (Fig. 4). A similar, though nonsignificant trend was observed with plants grown in the Kfar Menahem soil, for which the analysis of rhizoplane–endorhizosphere was performed using whole root systems.
DGGE fingerprints of rhizoplane–endorhizosphere and rhizosphere soil bacterial communities of control maize plants and of plants inoculated with A. brasilense strains Cd and Sp245 grown in Rehovot sandy soil. (A) Rhizosphere soil. Control (lanes 1, 2, and 3), Cd-inoculated (lanes 4, 5, and 6), and Sp245-inoculted (lanes 6, 7, and 8), plants from the third week post emergence. (B) Branching zones. Control (lanes 1 and 2), Cd-inoculated (lanes 3, 4, and 5) and Sp245-inoculted (6, 7, and 8) plants from the third week post emergence.
Rhizosphere Clone Library Analysis
To identify components of the rhizobacterial community, a clone library was generated from the rhizosphere of plants grown in Rehovot soil. Although the sequences obtained were relatively short (c. 580 bp), clear affiliations could be defined by phylogenetic analysis of the twenty-nine cloned sequences (Fig. 5). Five clones were related to Ralstonia pickettii but yet formed a distinct group. Other sequences exhibited similarity to known soil and rhizosphere bacteria from the Actinobacteria (98% similarity), Bacterioidetes (94% similarity), β-Proteobacteria (97% similarity) and α-Proteobacteria (97% similarity). Among the latter, two clones appeared to be closely related to Azospirillum lipoferum. Seven sequences were only very distantly related to known organisms or clustered with sequences originating from uncultured bacteria present in the GenBank database. When run in parallel in a DGGE gel using the conditions previously described, bands from rhizosphere samples migrated exactly as the ones from the Ralstonia, Chryseobacterium, Leptothrix, and Massilia clones.
Unrooted phylogenetic tree showing relatedness of rhizosphere soil 16S rDNA clones from maize plants grown in a Rehovot soil. The tree is based on a maximum-likelihood (FastDNAml) analysis, using a 50% conservation filter and the Olsen correction method. Branch points supported by parsimony bootstrap values (1,000 replicates) of >90%, >75% and >50% are indicated by open, filled gray, and filled black circles, respectively, while branch points without circles were not resolved (bootstrap values of <50%). Scale bar indicates 10% estimated sequence divergence.
Plant-Growth-Promoting Effect of A. brasilense Inoculation
In a Rehovot soil, inoculation with Sp245 resulted in a significant (P = 0.05) increase in the fresh weight of roots (1.92 ± 0.2, 2.66 ± 0.3, 2.28 ± 0.3 g mean and standard deviation for Cd., Sp245 and control, respectively) in the first week following emergence. At three weeks following emergence a similar trend (though non significant) was observed, as inoculated plants exhibited higher fresh root weights. Although no significant differences between A. brasilense Cd-inoculated plants and control plants grown in a Kfar Menahem soil were found, in all experiments and in both soil types, inoculation with Azospirillum resulted in a visible increase in root and shoot development, especially during plant establishment.
Discussion
Azospirilla, and more specifically A. brasilense are one of the best-studied PGPRs. The plant-growth-promoting effects of these bacteria have been demonstrated in cereals, legumes and other crops, in widely different soils, under various climatic and agrotechnical conditions [36]. The main mechanism underlying the PGPR properties of A. brasilense is probably phytohormone excretion [5] but to what extent this affects bacterial communities in the rhizosphere is not well known. Azospirilla colonize the root surface, and rarely, the root interior [49]. Therefore, in this study no attempt was made to differentiate between rhizoplane stricto sensu and endorhizosphere. Growth promotion due to inoculation with A. brasilense was seen in all experiments in both soil types to a greater or to a lesser extent. This was in accordance with field and greenhouse experiments (reviewed by [8]). Following inoculation, A. brasilense affects the rhizosphere environment indirectly through enhanced root development, increased root branching and a higher density of root hair, providing increased surfaces for nutrient absorption and rhizobacterial colonization [37]. Inoculation with azospirilla also leads to an increase in plant root exudation [16, 20, 52], and both changes in root structure and exudation are potential factors influencing the type of microorganisms colonizing the radicular environment.
Inoculation with two strains of A. brasilense had no impact, neither on the concentration of rhizosphere and rhizoplane bacteria nor on their state of activity, as assessed by whole cell hybridization. The relatively high availability of carbon in the rhizosphere supports highly active bacterial communities, which are readily detectable with rRNA-targeted fluorescent probes, as the amount of ribosomes present per cell, the intensity of fluorescence detected and the cell growth capacity are related [39, 42, 44]. In both soils, the rhizoplane–endorhizosphere (root tip in Rehovot sandy soil) sustained the highest bacterial population and the rhizosphere soil the lowest, consistent with the presence of more easily metabolizable substrates (exudates, sloughed cells) at this interface than further at a distance from the root surface [4, 15].
However, a more developed root system, as obtained in Azospirillum inoculated plants means that the total number of rhizosphere bacteria per soil unit volume increases as the plant output of readily assimilable organic substrates also increases. Growing roots are considered a significant source of carbon—the factor most limiting to microbial growth—and support a milieu of high microbial activity and biomass where elemental turnover is enhanced, as was recently shown for root-associated Cytophaga-like bacteria [18]. Therefore, a secondary impact of a more developed root system may be the stimulation of C, N, and P turnover per unit of soil volume in an inoculated field.
DGGE analysis showed that inoculation with A. brasilense did not alter bacterial community structures in the rhizoplane or in the rhizosphere of plants grown in the two soils. Inoculation with PGPRs may lead to different outcomes, and no relationship between mechanisms of growth promotion and changes in rhizobacterial communities following inoculation can be drawn: while in this work, no effect of inoculation was detected, Pandey et al. [38] found that A. brasilense stimulated certain rhizopopulations of beneficial bacteria. Inoculation with Sinorhizobium meliloti had only minor to no influence on the rhizobacterial community structure [28] while a trifolitoxin antibiotic-producing Rhizobium etli strain reduced the complexity of RISA patterns in the rhizosphere of common bean plants [41], to cite a few previous works. Although many factors may stand behind this apparent lack of consistency, the interaction between plant and bacterial genotypes appear to be very important in determining the fate of rhizobacterial communities [19].
DGGE analysis of rhizophere soil and rhizoplane samples indicated that the bacterial populations inhabiting the root environment were strongly influenced by plant age, which is also an important factor in determining the quality and quantity of root exudates [15]. No temporal influence was detected in the rhizoplane–endorhizosphere from whole roots samples of plants from Kfar Menahem. The most active bacterial communities are found at the root tips and at root branching points so shifts in these regions may have been blurred in total root extracts.
Rhizoplane–endorhizoplane samples from root tips and from root branching points yielded very similar DGGE patterns. The complexity of rhizoplane samples was also lower than that of rhizosphere samples. These results indicate the formation of distinct bacterial communities in different locations of the maize phytosphere, maybe resulting from selective effects of plant roots toward certain bacterial groups [27, 32]. Marilley and Aragno [26] found that in Trifolium repens and Lolium perenne bacterial communities in the rhizoplane–endorhizoplane differed from those of rhizosphere soil and bulk soil and that the phylogenetic diversity decreased in the proximity of plant roots.
While some variability was seen between triplicate samples in the DGGE, recurrent analysis of the same samples yielded identical results. In this study, soils were not sieved, homogenized or mixed with other growth media. This suggests that while specific root compartments are mostly colonized by common or very closely related organisms, biological variability still occurs. Whether this results from de novo colonization of the growing root as proposed in the moving wave model [46] or from primary colonizers of the root surface influencing the succession and species composition of bacterial communities that develop on this root part [53] remains to be shown.
Biological variety was reflected in the diversity of the sequences identified in a clone library. The library was dominated by the α and β Proteobacteria, and of the latter, sequences clustering with Ralstonia spp. represented the largest group of clones. In a recent analysis of wheat rhizosphere, Ralstonia spp. were found to be the most dominant species, followed by Microbacterium, Arthrobacter, Chryseobacterium, Massilia and Flavobacterium [19], which were also detected in this work, with the exception of Flavobacterium. A relatively large proportion of the clones was only very distantly related to known species, or was related to environmental clones. Four clones clustered with the Chloroflexi (the green nonsulfur bacteria) a phylum detected in 25–75% of the clone libraries of soil bacterial communities [17], but comprising few cultivated members of diverse phenotypes [40]. Although the primers used were the same, only part of the sequences obtained in the library were detected in the DGGE gels, suggesting that the cloning procedure may introduce biases. These sequences most certainly represent very dominant groups.
Two Azospirillum clones were also identified in the library, indicating the presence of indigenous Azospirillum-like organisms in Rehovot soil. While both clones were very similar, one was identical to an endophytic nitrogen-fixing isolate from rice [10]. Also, Nur and co-workers [33] isolated an organism resembling Azospirillum in a survey of nitrogen-fixing bacteria associated with grasses in Israel over 20 years ago, but whether this bacterium belonged to the genus Azospirillum was not determined.
In conclusion, under our conditions, inoculation of maize seedlings with A. brasilense appears not to affect dominant rhizosphere bacteria populations of maize plants. Whether an effect occurs on less dominant populations or the genetic diversity of specific populations is affected without causing significant changes in population levels remains to be evaluated.
References
RI Amann BJ Binder RJ Olson SW Chisholm R Devereux DA Stahl (1990) ArticleTitleCombination of 16S rRNA-targeted olinucleotide probes with flow cytometry for analyzing mixed microbial populations Appl Environ Microbiol 56 1919–1925 Occurrence Handle2200342 Occurrence Handle1:CAS:528:DyaK3cXkvFCgsrs%3D
VLD Baldani JI Baldani J Dobereiner (1987) ArticleTitleInoculation of field grown wheat (Triticum aestivum) with Azospirillum spp. in Brasil Biol Fertil Soils 4 37–40
M Basaglia S Casella U Peruch S Poggilini T Vamerali G Mosca J Vanderleyden P Troch ParticleDe MP Nuti (2003) ArticleTitleField release of genetically marked Azospirillum brasilense in association with Sorghum bicolor L Plant Soil 256 281–290 Occurrence Handle10.1023/A:1026198522123 Occurrence Handle1:CAS:528:DC%2BD3sXotV2jtr8%3D
S Burdman E Jurkevitch Y Okon (2000) Involvement of extracellular components in the aggregation of Azospirillum brasilense FO Pedrosa M Hungria G Yates WE Newton (Eds) Nitrogen Fixation: From Molecules Crop Productivity Kluwer Academic Publishers Dordrecht, The Netherlands 415–416
S Burdman D Kadouri E Jurkevitch Y Okon (2001) Bacterial phytostimulators in the rhizosphere: from research to application G Bitton (Eds) The Encyclopedia of Environmental Microbiology. Vol. Soil Microbiolgy John Wiley & Sons New York, USA
F Cello ParticleDi A Bevivino L Chiarini R Fani D Paffetti S Tabacchioni C Dalmastri (1997) ArticleTitleBiodiversity of a Burkholderia cepecia population isolated from the maize rhizosphere at different plant growth stages Appl Environ Microbiol 63 4485–4493 Occurrence Handle9361434
S Dobbelaere A Croonenborghs A Thys D Ptacek Y Okon J Vanderleyden (2002) ArticleTitleEffects of inoculation with wild type Azospirillum brasilense and A. irakese strains on development and nitrogen uptake of spring wheat and grain maize Biol Fertil Soils 36 284–297 Occurrence Handle10.1007/s00374-002-0534-9 Occurrence Handle1:CAS:528:DC%2BD38Xns1OgtrY%3D
S Dobbelaere A Croonenborghs A Thys D Ptacek J Vanderleyden P Dutto C Labandera-Gonzalez J MelladoCaballero JF Aguirre Y Kapulnik S Berner S Burdman D Kadouri S Sarig Y Okon (2001) ArticleTitleResponses of agronomically important crops to inoculation with Azospirillum Aust J Plant Physiol 28 871–879
S Dobbelaere J Vanderleyden Y Okon (2003) ArticleTitlePlant growth-promoting effects of diazotrophs in the rhizosphere CRC Crit Rev Plant Sci 22 107–149 Occurrence Handle1:CAS:528:DC%2BD3sXjt1yktLY%3D Occurrence Handle10.1080/713610853
A Elbeltagy K Nishioka T Sato H Suzuki B Ye T Hamada T Isawa H Mitsui K Minamisawa (2001) ArticleTitleEndophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolate d from wild rice species Appl Environ Micrbiol 67 5285–5293 Occurrence Handle1:CAS:528:DC%2BD3MXotlSnsLs%3D
DL Eskew DD Focht IP Ting (1977) ArticleTitleNitrogen fixation denitrification and pleomorphic growth in highly pigmented Spirillum lipoferum Can J Microbiol 34 582–585 Occurrence Handle1:CAS:528:DyaE1cXjsVentg%3D%3D
E Fallik S Sarig Y Okon (1994) Morphology and physiology of plant roots associated with Azospirillum Y Okon (Eds) Azospirillum/Plant Associations CRC Press Boca Raton 77–87
SJ Giovannoni EF Long ParticleDe GJ Olsen NR Pace (1988) ArticleTitlePhylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells J Bacteriol 170 720–726 Occurrence Handle2448289 Occurrence Handle1:CAS:528:DyaL1cXktF2lsbY%3D
BR Glick (1995) ArticleTitleThe enhancement of plant growth by free-living bacteria Can J Microbiol 41 109–117 Occurrence Handle1:CAS:528:DyaK2MXks1ygsrY%3D
SJ Grayston D Vaughan D Jones (1996) ArticleTitleRhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability Appl Soil Ecol 5 29–56
T Heulin A Gukert J Balandreau (1987) ArticleTitleStimulation of root exudation of rice seedlings by Azospirillum strains: carbon budget under gnotobiotic conditions Biol Fertil Soils 4 9–14
P Hugenholtz BM Goebel NR Pace (1998) ArticleTitleImpact of culture-independent studies on the emerging phylogenetic view of bacterial diversity J Bacteriol 180 4765–4774 Occurrence Handle9733676 Occurrence Handle1:CAS:528:DyaK1cXmt1Sgu7k%3D
JE Johansen SJ Binnerup (2002) ArticleTitleContribution of Cytophaga-like bacteria to the potential of turnover of carbon, nitrogen and phosphorus by bacteria in the rhizosphere of barley (Hordeum vulgare L.) Microb Ecol 43 298–306 Occurrence Handle10.1007/s00248-002-2006-z Occurrence Handle12037608 Occurrence Handle1:CAS:528:DC%2BD38XkvFSnsr8%3D
BB Landa DM Mavrodi LS Thomashow DM Weller (2003) ArticleTitleInteractions between strains of 2,4-diacetylphloroglucinol-producing. Pseudomonas fluorescens in the rhizosphere of wheat Phytopathology 93 982–994 Occurrence Handle1:CAS:528:DC%2BD3sXmsFGjur8%3D Occurrence Handle18943865
KJ Lee MH Gaskins (1982) ArticleTitleIncreased root exudation of 14C-compounds by sorghum seedlings inoculated with nitrogen-fixing bacteria Plant Soil 69 391–399 Occurrence Handle1:CAS:528:DyaL3sXhsVSmurs%3D
S Lee C Malone PF Kemp (1993) ArticleTitleUse of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria Mar Ecol Prog Ser 101 193–201 Occurrence Handle1:CAS:528:DyaK2cXivFSqsL0%3D
JE Loper TV Suslow MN Schroth (1984) ArticleTitleLognormal distribution of bacterial populations in the rhizoshpere Phytopathology 74 1454–1460 Occurrence Handle10.1094/Phyto-74-1454
W Ludwig O Strunk R Westram L Richter H Meier BA Yadhukumar T Lai S Steppi G Jobb W Förster I Brettske S Gerber AW Ginhart O Gross S Grumann S Hermann R Jost A König T Liss R Lüβmann M May B Nonhoff B Reichel R Strehlow A Stamatakis N Stuckmann A Vilbig M Lenke T Ludwig A Bode KH Schleifer (2004) ArticleTitleARB: a software environment for sequence data Nucleic Acids Res 32 1363–1371 Occurrence Handle10.1093/nar/gkh293 Occurrence Handle14985472 Occurrence Handle1:CAS:528:DC%2BD2cXhvFSqu7k%3D
BL Maidak JR Cole JCR Parker GM Garrity N Larsen B Li TG Lilburn MJ McCaughhey GJ Olsen R Overbeek S Pramanik TM Schmidt JM Tiedje CR Woese (1999) ArticleTitleA new version of the RDP (Ribosomal Data Project) Nucleic Acids Res 27 171–173 Occurrence Handle10.1093/nar/27.1.171 Occurrence Handle9847171 Occurrence Handle1:CAS:528:DyaK1MXpsVKjsw%3D%3D
W Manz R Amann W Ludwig M Wagner KH Schleifer (1992) ArticleTitlePhylogenetic oligodeoxynucleotide probes for the major subclass of Proteobacteria: problems and solutions Syst Appl Microbiol 15 593–600
L Marilley M Aragno (1999) ArticleTitlePhylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots Appl Soil Ecol 13 127–136 Occurrence Handle10.1016/S0929-1393(99)00028-1
P Mavingui G Lauerre O Berge T Heulin (1992) ArticleTitleGenetic and phenotypic diversity of Bacillus polymyxa in soil and in wheat rhizosphere Appl Environ Microbiol 58 1894–1903 Occurrence Handle1:CAS:528:DyaK38XksVCgtLw%3D Occurrence Handle16348720
R Miethling G Wieland H Backhaus CC Tebbe (2000) ArticleTitleVariation of microbial communities in response to crop species, soil origin and inoculation with Sinorhizobium meliloti L 33 Microb Ecol 40 43–56 Occurrence Handle10977876 Occurrence Handle1:CAS:528:DC%2BD3cXnt12mu7w%3D
G Muyzer EC Waal ParticleDe AG Uitterlinden (1993) ArticleTitleProfiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA Appl Environ Microbiol 59 695–700 Occurrence Handle7683183 Occurrence Handle1:CAS:528:DyaK3sXit1Kktrk%3D
G Muyzer A Teske CO Wirsen HW Jannash (1995) ArticleTitlePhylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments Arch Microbiol 164 165–172 Occurrence Handle10.1007/s002030050250 Occurrence Handle7545384 Occurrence Handle1:CAS:528:DyaK2MXovFyjsrk%3D
C Nacamulli A Bevivino C Dalmastri S Tabacchioni L Chiarini (1997) ArticleTitlePerturbation of maize rhizosphere following seed bacterization with Burkholderia cepacia MCI 7 FEMS Microbiol Ecol 23 183–193 Occurrence Handle1:CAS:528:DyaK2sXlt1Ohtr4%3D
B Normander JI Prosser (2000) ArticleTitleBacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions Appl Environ Microbiol 66 4372–4377 Occurrence Handle10.1128/AEM.66.10.4372-4377.2000 Occurrence Handle11010885 Occurrence Handle1:CAS:528:DC%2BD3cXnt1Cmurw%3D
I Nur Y Okon Y Henis (1980) ArticleTitleComparative studies of nitrogen-fixing bacteria associated with grasses in Israel with Azospirillum brasilense Can J Microbiol 24 967–980
Y Okon SL Albrecht RH Burris (1977) ArticleTitleMethods for growing Spirillum lipoferum and for counting it in pure culture and in association with plants Appl Environ Microbiol 33 85–88 Occurrence Handle16345192
Y Okon Y Kapulnik (1986) ArticleTitleDevelopment and function of Azospirilluminoculated root Plant Soil 90 3–16 Occurrence Handle1:CAS:528:DyaL28Xhs1Ghur4%3D
Y Okon CA Labandera-Gonzalez (1994) ArticleTitleAgronomic applications of Azospirillum: an evaluation of 20 years of worldwide field inoculation Soil Biol Biochem 26 1591–1601 Occurrence Handle10.1016/0038-0717(94)90311-5 Occurrence Handle1:CAS:528:DyaK2MXisFWrtbY%3D
Y Okon J Vanderleyden (1997) ArticleTitleRoot-associated Azospirillum species can stimulate plants ASM News 63 366–370
A Pandey E Sharma LMS Palni (1998) ArticleTitleInfluence of bacterial inoculation on maize in upland farming systems of the Sikkim Himalaya Soil Biol Biochem 30 379–384 Occurrence Handle10.1016/S0038-0717(97)00121-1 Occurrence Handle1:CAS:528:DyaK1cXhvFWrs70%3D
LK Poulsen G Ballard DA Stahl (1993) ArticleTitleUse of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms Appl Environ Microbiol 59 1354–1360 Occurrence Handle7685999 Occurrence Handle1:CAS:528:DyaK3sXks1Chs7k%3D
MS Rappe SJ Giovannoni (2003) ArticleTitleThe uncultured microbial majority Annu Rev Microbiol 57 369–394 Occurrence Handle10.1146/annurev.micro.57.030502.090759 Occurrence Handle14527284 Occurrence Handle1:CAS:528:DC%2BD3sXptFWlsr8%3D
EA Robleto J Borneman EW Triplett (1998) ArticleTitleEffects of bacterial antibiotic production on rhizosphere microbial communities from a culture independent perspective Appl Environ Microbiol 64 5020–5022 Occurrence Handle9835600 Occurrence Handle1:CAS:528:DyaK1cXnvF2htLc%3D
R Ruimy B Breittmayer V Boivin R Christen (1994) ArticleTitleAssessment of the state of activity of individual cells by hybridization with a ribosomal RNA targeted fluorescently labeled oligonucelotide probe FEMS Microbiol Ecol 15 207–214 Occurrence Handle1:CAS:528:DyaK2MXhsVais78%3D
J Sambrook EF Fritsch T Maniatis (1989) Molecular Cloning: A Laboratory Manual Cold Spring Harbor Laboratory Press New York
E Schallmach D Minz E Jurkevitch (2000) ArticleTitleCulture-independent detection of changes in root-associated bacterial populations of common bean (Phaseolus vulgaris L.) following nitrogen depletion Microb Ecol 40 309–316 Occurrence Handle12035089 Occurrence Handle1:CAS:528:DC%2BD3MXitV2qug%3D%3D
M Schloter A Hartmann (1998) ArticleTitleColonization of wheat roots (Triticum eastivum) and endophytic potential of different Azospirillum brasilense strains studied with strain-specific monoclonal antibodies Symbiosis 25 159–179
AM Semenov AHC Bruggen Particlevan VV Zelenev (1999) ArticleTitleMoving waves of bacterial populations and total organic carbon along roots of wheat Microb Ecol 37 116–128 Occurrence Handle10.1007/s002489900136 Occurrence Handle9929400 Occurrence Handle1:CAS:528:DyaK1MXotVyltQ%3D%3D
M Stoffels T Castellanos A Hartmann (2001) ArticleTitleDesign and application of new 16 rRNA-targeted oligonucleotide probes for Azospirillum–Skermanella–Rhodocista-cluster System Appl Microbiol 24 83–97 Occurrence Handle1:CAS:528:DC%2BD3MXltFert7k%3D
YL Tsai BH Olson (1992) ArticleTitleDetection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction Appl Environ Microbiol 58 754–757 Occurrence Handle1610201 Occurrence Handle1:CAS:528:DyaK38XhsVCqtb0%3D
M Umali-garcia DH Hubell M Gaskins F Dazzo (1980) ArticleTitleAssociations of Azospirillum with grass roots Appl Environ Microbiol 39 219–226 Occurrence Handle16345490
J Veen Particlevan LS Overbeek Particlevan JD Elsas Particlevan (1997) ArticleTitleFate and activity of microorganisms introduced into the soil Microbiol Mol Biol Rev 61 121–135 Occurrence Handle9184007
JM Vincent (1970) A Manual for the Practical Study of Root-Nodule Bacteria. International Biological Programme. vol. 15 Blackwell Scientific Publishers Oxford
H Volpin S Burdman S Castro-Sowinski Y Kapulnik Y Okon (1996) ArticleTitleInoculation with Azospirillum increased exudation of rhizobial nod—gene inducers by alfalfa roots Molecular Plant–Microbe Interactions 9 388–394 Occurrence Handle1:CAS:528:DyaK28XjsFCiurc%3D
CH Yang DE Crowley (2000) ArticleTitleRhizosphere microbial community structure in relation to root location and plant iron nutritional status Appl Environ Microbiol 66 345–351 Occurrence Handle10618246 Occurrence Handle1:CAS:528:DC%2BD3cXktlSrsg%3D%3D Occurrence Handle10.1128/AEM.66.1.345-351.2000
Acknowledgements
This work was supported by the German Israeli Foundation (586-095) and by the European Union-5th Framework contract QLK3-CT-2000-31759-ECO-SAFE.
Author information
Authors and Affiliations
Corresponding author
Additional information
Herschkovitz and Lerner contributed equally to this work.
Rights and permissions
About this article
Cite this article
Herschkovitz, Y., Lerner, A., Davidov, Y. et al. Inoculation with the Plant-Growth-Promoting Rhizobacterium Azospirillum brasilense Causes Little Disturbance in the Rhizosphere and Rhizoplane of Maize (Zea mays). Microb Ecol 50, 277–288 (2005). https://doi.org/10.1007/s00248-004-0148-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-004-0148-x