Abstract
Plants harbor diverse communities of fungi and other microorganisms. Fungi are known to occur both on plant surfaces (epiphytes) and inside plant tissues (endophytes), but the two communities have rarely been compared. We compared epiphytic and endophytic fungal communities associated with leaves of coffee (Coffea arabica) in Puerto Rico. We asked whether the dominant fungi are the same in both communities, whether endophyte and epiphyte communities are equally diverse, and whether epiphytes and endophytes exhibit similar patterns of spatial heterogeneity among sites. Leaves of naturalized coffee plants were collected from six sites in Puerto Rico. Epiphytic and endophytic fungi were isolated by placing leaf pieces on potato dextrose agar without and with surface sterilization, respectively. A total of 821 colonies were isolated and grouped into 131 morphospecies. The taxonomic affinities of the four most common nonsporulating fungi were determined by sequencing the nuclear ribosomal internal transcribed spacer (ITS) region: two grouped with Xylaria and one each with Botryosphaeria and Guignardia. Of the most common genera, Pestalotia and Botryosphaeria were significantly more common as epiphytes; Colletotrichum, Xylaria, and Guignardia were significantly more common as endophytes. Suprisingly, more morphospecies occurred as endophytes than as epiphytes. Differences among sites in number of fungi per plant were significant. Thus epiphytic and endophytic communities differed greatly on a single leaf, despite living only millimeters apart, and both communities differed from site to site. Significant correlations between occurrence of fungal morphospecies suggested that fungi may have positive or negative effects on their neighbors. This is the first quantitative comparison of epiphytic and endophytic fungal floras in any plant, and the first to examine endophytic fungi or epiphytic fungi in leaves of coffee, one of the world’s most valuable crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaves have epiphytic microorganisms on them and endophytic microorganisms inside them. The two floras coexist within millimeters of each other, but are usually studied separately. Few studies on endophytes even mention epiphytes, and vice versa. Microbial epiphytes and endophytes may have important implications for plant health and plant protection [2, 7, 51, 52], microbial biodiversity [5, 16, 25, 28, 41], and drug discovery [51].
Relationships between epiphytes and endophytes have important implications for fungal biodiversity and plant health. It is unclear to what extent plants control which endophytes are able to enter the leaf, and to what extent epiphytes may affect this process [30]. Comparison of endophytic and epiphytic floras may help to determine the basis for selectivity.
In this study we asked whether the fungal community on the surface of a leaf differs from the community within the leaf. Because plants are presumably able to exert more control over colonization of internal tissues than over exterior surfaces, we predicted that the composition of epiphyte and endophyte communities would be different, and that epiphyte communities would be more diverse. We also asked whether communities of epiphytes and endophytes differ among sites. Because endophytes are sheltered from environmental conditions by plant tissues and presumably live in a more constant environment, we predicted that endophyte abundance, diversity and species composition would vary less among sites than epiphyte abundance, diversity, and species composition. We also asked whether occurrence of certain fungi was correlated with that of other fungi. Because fungi may inhibit growth of other fungi by secondary metabolite production, parasitism, or competition for resources, we predicted that the presence of some common fungi would be negatively correlated with the presence of others.
These questions would be valid and novel for any plant. There were three reasons to use coffee (Coffea arabica L.). First, it is naturalized in Puerto Rico and is common as an understory plant in many areas [33]. Because few cultivars are planted in Puerto Rico and those are closely related [14], the different populations we studied are presumably genetically uniform. Second, there are no published studies about the epiphyte or endophyte floras of coffee leaves, despite the fact that coffee is one of the world’s most valuable crops. Third, epiphytes and endophytes may potentially be useful for biocontrol of pathogens [2, 7, 52], and several coffee pathogens are very destructive [53].
Methods
Collection of Leaves and Isolation of Fungi
Healthy, mature leaves were collected from coffee plants at six sites in Puerto Rico; all sites were >5 miles and <75 miles apart. The sites (with municipality, approximate altitude in m and average annual precipitation in mm) were: E1 Verde (Río Grande municipality, 300 m, 3640 mm), Hacienda Buena Vista (Ponce, 300 m, 1400 mm), Ciales (Ciales, 291 m, 2111 mm), Barranquitas (Barranquitas, 627 m, 1416 mm), Río Chiquito (Luquillo, 110 m, 2694 mm), and UPR’s Jardín Botánico (San Juan, 28 m, 1707 mm). All sites except the Jardín Botánico were abandoned coffee plantations that are now secondary forests. Three plants per site were chosen and five healthy, mature leaves were sampled per plant. (Only two plants were available in Río Chiquito and Jardín Botánico). From each leaf, four pieces were sampled for epiphytes and four for endophytes; a total of 640 pieces were sampled.
For epiphytes, 5 × 5 mm leaf pieces were placed on plates (adaxial surface down) for 1 h and then removed. For endophytes, pieces were surface-sterilized in 70% EtOH (1 min), 2.6% NaClO2 (50% Clorox) (3 min), and 70% EtOH (1 min) [9, 12, 24]. 2 × 2 mm pieces were used, since small leaf pieces are recommended for studies of endophyte biodiversity [16, 25, 35]. The culture medium was potato dextrose agar (PDA, half strength; Difco, Inc., Detroit, MI) with 50 ppm each tetracycline, penicillin, and streptomycin added after autoclaving; PDA is a good medium for isolating a variety of endophytes [4, 24, 25]. Plates were incubated at 22°C and observed weekly for development of fungal colonies.
Fungi that did not sporulate or could not be readily identified were transferred to other media. In some cases, these media induced sporulation or formation of distinctive structures that could be used to identify fungi to genus. In other cases, fungi could not be identified but could be grouped into morphospecies based on similar colony morphology and growth rate in culture. Four media were used for this purpose: half-strength PDA with 35 ppm benomyl, half-strength PDA with 35 ppm rose bengal, half-strength cornmeal agar (Difco), and Czapek-Dox agar (Difco) with 25 gL−1 KC1O3, pH = 7.0 [12]. Nonsporulating fungi are common among endophytes of tropical plants [6, 9, 13, 26, 35]; previous studies have shown that most morphospecies defined by morphology in culture correspond to putative taxa as determined by DNA sequencing [6, 26]. Because leaf pieces were not contiguous, it is unlikely that the same individual colony was sampled twice. However, we refer to them as colonies rather than as individuals because fungal growth is usually indeterminate and the boundaries of each genet were not known.
DNA Sequencing
One isolate each from the four most common unidentified morphospecies was identified by DNA sequencing. Fungi were grown in 50 mL PD broth and mycelia were removed and washed. DNA was extracted by a standard miniprep method [32]. The internal transcribed spacer (ITS) and 5.8S gene of the nuclear ribosomal gene repeat were amplified by polymerase chain reaction (PCR) with the universal primers ITS1 and ITS4 [54]. The PCR products were cleaned on QIAquick columns (Qiagen, Inc.), cloned with the TA cloning kit (Invitrogen Corp.) and sequenced in both directions on a LiCor automatic sequencer. The six most similar sequences in GenBank were found for each sequence by mean of BLAST searches. Sequences were aligned using CLUSTAL and BioEdit programs and edited manually; gaps were excluded. The most informative sequences were used to construct phylogenetic trees using PAUP. Appropriate outgroups were chosen based on published studies of each group; trees were rooted to the outgroup. Trees presented had identical topologies by maximum likelihood and parsimony criteria. Parsimony bootstrap values were based on 1000 replicates.
Data Analysis
Number of colonies of endophytes and epiphytes were compared for each of the five most common genera by Mann-Whitney tests; data from all sites were combined. Differences among sites in number of colonies of the five most common genera were compared with MANOVA tests. (Parametric tests were not used because their assumptions of normality and homogeneity of variance were not fulfilled.) For data analysis, each leaf was treated as a separate unit, because studies have established that different leaves of a single plant are independent in terms of endophytic fungal flora [13, 24, 35].
Richness of morphospecies in epiphyte and endophyte populations was compared using the Chao1 and jackknife1 estimators with the program EstimatesS, version 6.0b1 [17]. The Chao1 formula estimates number of species based on number of singletons and doubletons in a sample (that is, species represented by one or two individual colonies), and the jackknife is based on the number of species that occur in only one sample [18]. Diversity indices were not calculated because the epiphyte and endophyte communities appeared to be undersampled.
Pairwise comparisons were made to determine associations between fungi. If two genera occurred independently of each other, the proportion of leaves having both genera should be the product of the frequencies of each genus considered alone [10, 12], Fisher’s exact tests were used to compare these expected frequencies with the observed frequencies of pairs of species. (Chi squared tests were not used because many values were ≤5.) The null hypothesis in each case was that occurrence of the two genera was independent. Each leaf fragment was treated as a unit for these tests, because the size of each fungal colony was presumably small and two colonies would have to be in close contact to interact. These tests were only done among epiphytes or among endophytes; epiphytes could not be compared with endophytes because different fragments were used. For each pair of fungi, data from all sites where the fungi occurred in ≥10% of fragments were pooled.
Results
Number and Diversity of Fungi
A total of 821 fungi were isolated and assigned to 131 morphospecies (including both identified genera and morphologically distinct but unidentified types). The distribution of isolates among the 131 morphospecies approximated a log-normal pattern, with a few common taxa and many rare taxa (Fig. 1).
Five genera were common in more than one site: Pestalotia (20% of all isolates), Botryosphaeria (19%), Xylaria (18%), Colletotrichum (10%), and Guignardia (3%). The other identifed genera were rare, each ≤ 1% of isolates. These genera were Aspergillus, Cladosporium, Coprinus, Fusarium, Penicillium, Mucor, Rhizopus, and Trichoderma. In most cases these isolates were not identified to species; in the tropics many fungal genera include cryptic species that have not been identified [28]. The remaining 117 morphospecies comprised 25% of isolates. These did not sporulate in culture and could not be identified, a common problem with endophytic fungi [6, 9, 24, 26].
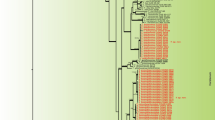
The four most common unidentified morphospecies were identified by BLAST searches and phylogeny. Two grouped within the Botryosphaeriaceae, isolate J1 with Botryosphaeria rhodina and its anamorph Lasiodiplodia theobromae (= Botryodiplodia theobromae), and J30 with Guignardia endophyllicola (Fig. 2a). (These are included in Botryosphaeria and Guignardia in the paragraph above.) Two sequences grouped within Xylaria, J26 with X. enteroleuca and J48 with X. hypoxylon and X. arbuscula, all common endophytes [9, 13, 35, 47], and were combined with the Xylaria isolates identified by morphology for purposes of analysis (Fig. 2b).
(a) Phylogenetic affinities of isolates J1 and J30. The tree was produced with PAUP 4.0b10. Tree length is 173 steps with 64 informative characters, CI = 0.89, RI = 0.91. Venturia inaequalis AF065839 was defined as the outgroup [56]. Bootstrap values above 60% are shown to the upper left of the node. Bar at lower left shows length of 1 change. The most similar sequences in GenBank, with source, E-value, and percent similarity in area of overlap, were for J1, AY612337, Australia, E = 0.0, 99%; for J30, AF532314, India, E = 0.0, 100%. (b) Phylogenetic affinities of isolates J48 and J46. The tree was produced with PAUP 4.0b10. Tree length is 245 steps with 96 informative characters, CI =0.77, RI = 0.79. Diatrype disciformis AJ390410 was defined as the outgroup [31, 43]. Bootstrap values above 60% are shown to the left of the node. The most similar sequences in GenBank, with source, E-value, and percent similarity in areas of overlap, were: for J26, AY315405, Jamaica, E = 0.0, 96% [20]; for J30, for J48, AF502739, unidentified leaf litter ascomycete, Puerto Rico, E = e−132, 97%.
Epiphytes vs Endophytes
Two of the most common genera were significantly more common as epiphytes than as endophytes: Botryosphaeria (Mann-Whitney, U = 784, P < 0.0001) and Pestalotia (U = 513, P < 0.0001) (Fig. 3). Three common genera were significantly more common as endophytes: Xylaria (U = 1588, P < 0.0001), Colletotrichum (U = 1425, P < 0.0001), and Guignardia (U = 2325, P < 0.001) (Fig. 3). These differences were also significant when number of fungal colonies per unit area, instead of per leaf fragment, was used (data not shown).
Epiphytes were more abundant than endophytes—481 epiphyte colonies were isolated vs 340 endophyte colonies. However, epiphyte communities were not noticeably richer than endophyte communities: total number of morphospecies were similar (63 vs 66), and species accumulation curves had similar shapes (Fig. 4). Jackknife estimates of total number of morphospecies were very similar for epiphytes and endophytes. The Chao1 estimator suggested greater morphospecies richness for endophytes, despite the higher number of epiphytes, which was contrary to our hypothesis. Both these measures suggested that the expected number of morphospecies was much greater than the number observed, implying that both communities were undersampled in this study.
Differences among Sites
Frequency of all five common genera differed significantly among sites (Fig. 5). For example, Botryosphaeria was a common epiphyte in all sites except Barranquitas, Xylaria was the only common endophyte in Ciales, and was entirely absent in Jardín Botánico. Total number of isolates and total numbers of morphospecies also differed significantly among sites.
Associations among Fungi
There was a significant, positive correlation between presence of Xylaria and presence of Guignardia in leaf fragments (FET, P = 0.005). There were significant, negative correlations between Xylaria and Colletotrichum (P < 0.0001) and between Guignardia and Colletotrichum (P = 0.004). Correlation between the epiphytes Botryosphaeria and Pestalotia was not significant (P = 0.54).
Discussion
Epiphytes vs Endophytes
Results supported the hypothesis that different fungi would predominate in epiphyte and endophyte communities: each of the five most common genera was more common either outside or inside the leaf, and differences were highly significant in all cases (Figs. 1 and 3). Although these communities live <1 mm apart, they are distinct. The fact that a single leaf simultaneously supports two distinct fungal floras has been overlooked in most studies of endophyte biodiversity.
Surprisingly, endophyte communities were not noticeably less speciose than epiphyte communities. We had predicted that many fungi would land on the leaf, but relatively few would be able to penetrate the leaf. However, the data did not support this idea: in most sites the number of species found was similar between epiphytes and endophytes (Figs. 1 and 4), Abundance of species in both communities approximated a log-normal distribution, as is typical for fungal communities [21, 24].
Very few studies have compared endophytic and epiphytic fungi, especially in tropical plants. However, one such study examined coconut leaves in Brazil [45]; 45 species were exclusively endophytic and 44 were exclusively epiphytic, with 29 species found in both floras. The most common endophyte was Pestalotiopsis palmarum and the most common epiphyte was Cladosporium spp. (isolated from 33% and 20% of leaves, respectively. In a study of dead beech leaves in Japan, Pestalotiopsis and Trichoderma were common as epiphytes, Xylaria as endophytes, and Ascochyta as both—similar to the present study, at least in terms of Xylaria and Pestalotiopsis [40]. In contrast, comparison of fungi isolated from washings of tomato fruits with fungi isolated from homogenized fruits found that Cladosporium, Penicillium, and Aspergillus were the most common genera in both cases [46]. However, because the fruits were bought at retail rather than collected in the field, comparisons with the present study are difficult; some epiphytes may have been removed by handling and processing.
It is likely that some epiphytes and endophytes were underrepresented because they did not grow well on PDA under the conditions used. For example, Mucor and Cladosporium were uncommon, although they are very common fungi in the tropics. Other fungi such as rusts and members of the Capnodiaceae were likely present but do not grow in culture. Methods based on direct detection of nucleic acids (rather than culturing) could reveal such fungi, although those methods also have limitations [6, 24]. Also, some of the fungi isolated may have been propagules that landed on or in the leaf but were not able to grow there. Furthermore, not all epiphytes on the surface of the leaves were likely to have been left on the agar when the leaf fragments were removed. Leaf washing and dilution plating is another method for sampling epiphytes, but it would underrepresent nonsporulating fungi that adhere tightly to the leaf surface. Also, spatial heterogeneity of endophytic fungi within a leaf means that sampling strategy may strongly affect results [24, 25, 35, 50]. Although these problems complicate the comparison of epiphytes and endophytes, the differences between the two communities were so pronounced that it is unlikely that different isolation methods would significantly change the results.
The Common Fungi
Both Botryosphaeria and Guignardia are widely distributed in the tropics. Both include pathogens of many plants, and both include plant pathogens and endophytes [8, 38, 44, 49, 55]. In fact, the most closely related sequences to our Guignardia sequence came from endophytes of Rhododendron (G. endophyllicola, Fig. 4; [38]) and an unidentified endophyte (AF413039) (Fig. 2a). In most cases it is unclear if endophytic strains are potential pathogens or if they constitute genetically distinct, nonpathogenic lineages [8]. Given that these fungi are important pathogens, however, and that endophytes may convert to pathogenicity and vice versa the relationship of endophytic and pathogenic populations should be studied [2, 23, 39]. Surprisingly, there are few reports of Botryosphaeria as epiphytes, whereas we found the Botryosphaeria populations on coffee leaves to be 99% epiphytic.
Our initial searches with isolate J30 produced AF218262 as the sequence with the highest BLAST score. This sequence was deposited in GenBank as Psilotum nudum, a plant. Its authors apparently amplified the ITS of an endophytic fungus believing that they had amplified that of P. nudum itself. This sequence has subsequently been withdrawn from GenBank, and is thus not included in Fig. 4. However, it shows the extent to which endophytic fungi are overlooked by plant biologists, and how sequence databases are limited by the quality of the sequences deposited. Other workers have made similar mistakes when trying to amplify genes from bamboo [55] and from pines [34].
Xylaria is a common wood decomposer worldwide and a common endophyte in the neotropics [20, 41, 47, 48]. In this study, two of the most common morphospecies were revealed to be Xylaria, on the basis of DNA sequences, even though their cultural morphology was not typical of Xylaria. The unidentified morphospecies probably included additional Xylaria isolates. There is a clear need for a selective culture medium on which Xylaria can be distinguished. This need is supported not only by their importance as endophytes and saprotrophs, and by their contribution to fungal biodiversity [41, 48], but also by their importance as producers of secondary metabolites of pharmaceutical interest [1, 37, 43].
Colletotrichum includes many tropical plant pathogens and endophytes [22, 42, 53]. Several species are endophytes of coffee, one of which, C. coffeanum, is a serious pathogen of fruits and flowers [29]. Pestalotia also includes common pathogens [42, 53] and has been found as an endophyte in Puerto Rico [14, 24, 35]; it has not been widely reported as a tropical epiphyte, perhaps because fewer studies have focused on epiphytic fungi than on endophytes.
Given that sequences from epiphytes and endophytes are poorly represented in GenBank relative to pathogens, it is impressive that many of the sequences most closely related to ours come from endophytes (Fig. 2 a, b). If this represents a general pattern, it may suggest that some groups are specialized for endophytism, rather than being latent pathogens [20, 48]. However, relationships between endophytism, saprotrophy, and pathogenicity are complex [8, 16, 20, 23, 39, 48],
Differences among Sites
Fungi that were dominant epiphytes or endophytes in some sites were often absent in others (Fig. 5). Differences among sites may be due to differences in environmental conditions or presence of inoculum [2, 47]. Studies of endophytic floras should include plants from different populations, because single populations may not be representative [25, 35, 36].
Associations among Fungi
Why were leaf fragments with Xylaria more likely to contain Guignardia than fragments without Xylaria, and why were fragments with Colletotrichum less likely to contain Xylaria and Guignardia than those without Colletotrichum? Our data suggest the possibility of competition or antagonism between endophytes. This explanation is attractive because it suggests that Xylaria may be able to restrict growth of Colletotrichum, which includes important pathogens of many plants [29, 42, 53]. However, it is equally plausible that conditions that favor growth of Xylaria and Guignardia do not favor Colletotrichum, and vice versa, and that interactions among fungi are not involved. These tests show correlations and not causality [11, 13]. The significant, positive association between Xylaria and Guignardia may reflect that by inhibiting Colletotrichum, they promote the growth of each other, or it may simply reflect that their colonization or growth is favored by the same environmental conditions. It is also not clear to what exent colonies of endophytes or epiphytes come in physical contact with other colonies; some endophytes have been shown to form very small colonies, limited to a single host cell [16, 50]. This suggests that differences between endophyte floras of different leaf tissues may be as great as the differences between epiphytes and endophytes shown here—a level of endophyte biodiversity that is virtually unexplored.
Significant negative correlations between the presence of Colletotrichum and Xylaria and between that of Colletotrichum and Guignardia were also found in leaves and roots of the orchid Lepanthes [13]. Because coffee and Lepanthes have very different growth habits and habitats, the associations observed here may be generally applicable. More plants should be studied in this regard, given the implications for disease biocontrol.
Colonizations of Leaves by Endophytic Fungi
It is unclear how most endophytic fungi colonize plants in the tropics—or as Lebrón et al. put it, “Where is the gate to the party?” [30]. To establish whether fungal endophytes are transmitted horizontally or vertically, fungal flora of seeds and leaves were compared in four tropical woody plants: Manilkara bidentata, Casuarina equisitifolia, Theobroma cacao, and coffee [3, 9, 30]. These studies suggested that endophytes of these plants are horizontally transmitted, with airborne or rainborne inoculum colonizing leaves. If this is true, then the epiphytic flora present on leaves may be a factor in determining which potential endophytes become established. In the case of coffee, spores of coffee leaf rust (Hemileia vastatrix) and brown-eye leaf spot (Cercospora coffeicola)—both important pathogens—must land on the leaf and germinate before infection. Interactions with epiphytic or endophytic fungi are likely to affect the establishment of these pathogens, but neither the communities or their interactions have been characterized in coffee.
Epiphytic and endophytic fungi presumably interact and engage in cross-talk in ways that affect the host plant. Interactions within each community are poorly understood, and interactions between endophytes and epiphytes are completely unexplored. To complicate matters, there are other communities of endophytes and epiphytes (notably bacteria), and their interactions with fungi are also poorly understood. Recent studies have shown that bacterial flora may influence development of human digestive systems and other aspects of animal development [27]. It is possible that endophyte and epiphyte communities influence not only plant function and health, but also plant development. Understanding these communities and interactions, and manipulating them to improve plant health, represents one of the most promising and poorly understood areas of agricultural biotechnology [19, 52].
References
Abate D, Abraham WR, Meyer H (1997) Cytochalasins and Phytotoxins from the fungus Xylaria obovata. Phytocheinistry 44: 1443–1448
Andrews JH, Harris RF (2000) The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol 38:145–80
Arnold AE, Herre EA (2003) Canopy cover and leaf age affect colonization by tropical fungal Endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia 95:388–398
Arnold AE, Lutzon F (2003) Foliar endophytes of Magnolia grandiflora: morphological plasticity, molecular diversity, and species composition inferred using two isolation media. Inoculum 54:11 [abstract]
Arnold AE, Maynard Z, Gilbert G (2001) Fungal endophytes in dicotyledonous neotropical trees: patterns of abundance and diversity. Mycol Res 105:1502–1507
Arnold AE, Maynard Z, Gilbert G, Coley PD, Kursar TA (2000) Are tropical fungal endophytes hyperdiverse? Ecol Lett 3:267–274
Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre AE (2003) Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci USA 100:15649–15654
Baayen RP, Bonants PJM, Verkley G, Carroll GC, van der Aa HA, de Weerdt M, van Brouwershaven IR, Schutte GC, Maccheroni W, Jr. Glienke de Blanco C, Azevedo JL (2002) Nonpathogenic isolates of the citrus black spot fungus, Guignardia citricarpa, Identified as a cosmopolitan endophyte of woody plants, G. mangiferae (Phyllosticta capitalensis). Phytopathology 92:464–477
Bayman P, Angulo-Sandova P, Báez-Ortiz Z, Lodge DJ (1998) Distribution and dispersal of Xylaria endophytes in two tree species in Puerto Rico. Mycol Res 102:944–948
Bayman P, Baker JL, Mahoney NE (2002a) Aspergillus on tree nuts: incidence and associations. Mycopathologia 155:161–169
Bayman P, Cotty PJ (1991) Improved media for selecting nitrate-nonutilizing mutants in Aspergillus flavus. Mycologia 83:311–316
Bayman P, González EJ, Fumero JJ, Tremblay RL (2002b) Are fungi necessary? How fungicides affect growth and survival of the orchid Lepanthes rupestris in the field. J Ecol 90:1002–1008
Bayman P, Lebrón LL, Tremblay RL, Lodge DJ (1997) Fungal endophytes in roots and leaves of Lepanthes (Orchidaceae), New Phytol 135:143–149
Berthaud J, Charrier A (1988) Genetic resources of Coffea. In: Clarke RJ, Marcrae R, (Eds). Coffee, vol 4: Agronomy, Elsevier Applied Science, London, pp 1–42
Cabral D, Stone JK, Carroll GC (1993) The internal mycobiota of Juncus spp.: microscopic and cultural observations of infection patterns. Mycol Res. 97:367–376
Carroll GC (1995) Forest endophytes: pattern and process. Can J Bot 73:S1316–S1324
Colwell, RK (2000) Statistical estimates of species richness and shared species from samples. http://viceroy.eeb.uconn.edu/ estimates
Colwell RK, Coddington JA (1994) Estimating terrestrial biodiversity through extrapolation. Philos T Soc B 345:101–118
Cotty PJ, Bayman P, Egel D, Elias K (1994) Agriculture, Aspergillus and aflatoxins. In: Powell KA, Renwick A, Peberdy J E (Eds). The Genus Aspergillus, Plenum Press, New York, pp 1–27
Davis EC, Franklin JB, Shaw AJ, Vilgalys R (2003) Endophytic Xylaria (Xylariaceae) among liverworts and angiosperms: phylogenetics, distribution, and symbiosis. Am J Bot 90:1661–1667
Dix NJ, Webster, (1995) Fungal Ecology. Chapman & Hall, London
Dodd JC, Estrada A, Jeger MJ (1992) Epidemiology of Colletotrichum gloeosporioides in the tropics. In: Bailey JA, Jeger MJ (Eds). Colletotrichum: Biology, Pathology and Control. CAB International, Wallingford, UK pp 308–325
Freeman S, Rodriguez RJ (1993) Genetic conversion of a fungal plant pathogen to a non-pathogenic, endophytic mutualist. Science 260:75–78
Gamboa MA, Bayman P (2001) Communities of endophytic fungi in leaves of a tropical timber tree (Guarea guidonia: Meliaceae). Biotropica 33:352–360
Gamboa MA, Laureano S, Bayman P (2002) Does size matter? Estimating endophytic fungal diversity in leaf fragments. Mycopathologia 156:41–45
Guo LD, Hyde KD, Liew ECY (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences, New Phytol 147:617–630
Hamilton G (1999) Insider trading. New Scientist 6/26/99:42–46
Hawksworth DL, Rossman AY (1997) Where are all the undescribed fungi? Phytopathol 87:888–891
Hindorf H (1973) Colletotrichum population on Coffea arabica L. in Kenya II. Qualitative and quantitative differences in the Colletotrichum population. Phytopathol Z 77:216–234
Lebrón L, Lodgej DJ, Laureano S, Bayman P (2001) Where is the gate to the party? Inoculum 52:46 [Abstract]
Lee JS, Ko KS, Jung HS (2000) Phylogenetic analysis of Xylaria based on nuclear ribosomal ITS 1-5.8S-ITS2 sequences. FEMS Microbiol Lett 187:89–93
Lee SB, Taylor JW 1990. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JS, White TJ (Eds). PCR Protocols: A Guide to Methods and Applications, Academic Press, New York, pp 282–287
Little AH, Wadsworth FH (1964) Common Trees of Puerto Rico and the Virgin Islands, vol. I. USDA Agric Handb 249, Washington, DC
Liston A, Alvarez-Buylla E (1995) Internal transcribed spacer sequences of conifers: “There is a fungus among us.” Inoculum 46:26 [Abstract]
Lodge DJ, Fisher PJ, Sutton BC (1996) Endophytic fungi of Manilkara bidentata leaves in Puerto Rico. Mycologia 88:733–738
Lodge DJ (1997) Factors related to diversity of decomposer fungi in tropical forests. Biodiv Conserv 6:681–688
Monaghan RL, Polishook JD, Pecore VJ, Bills GF, Nallin-Omstead Streicher SL (1995) Discovery of novel secondary metabolites from fungi—is it really a random walk through a random forest? Can J Bot 73:S925–S931
Okane I, Nakagiri A, Ito T (2001) Identity of Guignardia sp. inhabiting ericaceous plants. Can J Bot 79:101–109
Ortiz-Gaicía S, Gernandt DS, Stone JK, Johnston PR, Chapela IH, Salas-Lizana R, AlvarezBuylla ER (2003) Phylogenetics of Lophodermium from pine. Mycologia 95: 846–859
Osono T, Takeda H (1999) Decomposing ability of interior and surface fungal colonizers of beech leaves with reference to lignin decomposition. Euro J Soil Biol 35:51–56
Petrini O, Petrini LE, Rodrigues KF (1995) Xylariaceous endophytes: an exercise in biodiversity. Fitopatol Bras 20:531–539
Ploetz RC, Zentmyer GA, Nishijima WT, Rohrbach KG, Ohr HD (1994) Compendium of Tropical Fruit Diseases. APS Press, American Phytopathological Society, St. Paul, MN
Polishook JD, Ondeyka JG, Dombrowski AW, Peláez F, Platas G, TeranAM (2001) Biogeography and relatedness of Nodulisporium strains producing nodulisporic acid. Mycologia 93:1125–1127
Punithalingam, E (1980) Plant diseases attributed to Botryodiplodia theobromae Pat. Bibliotheca Mycologia 71: 1–123
Ramos-Mariano RL, Fernandes de Lira RV, Barbosa ds Silveira E, Menezes M (1997) Levantamento de fungos endofíticos e epifíticos em folhas de coqueiro no nordeste do Brasil. I. Freqüêcia da populaçãao fúngica e efeito de hospedeira. Agrotrópica 9:127–134
Ribeiro WRC, Bolkan HA (1981) Micoflora de frutos de tomate comercializados no Distrito Federal. Fitopatol Bras 6:367–375
Rodrigues KF (1994) The foliar fungal endophytes of the Amazonian palm Euterpe oleracea. Mycologia 86:376–385
Rogers JD (2000) Thoughts and musings on tropical Xylariaceae. Mycol Res 104:1412–1420
Smith H Wingfield MJ, Crous PW, Coutinho TA (1996) Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa .S Afr J Bot 62:86–88
Stone JK (1988) Fine structure of latent infections by Rhabdoclineparkeri on Douglas-fir, with observations on uninfected epidermal cells. Can J Bot 66: 45–54
Strobel GA, Long DM (1996) Endophytic microbes embody pharmaceutical potential. ASM News 64:263–268
Sturz AV, Christie BR, Nowak J (2000) Bacterial endophytes: potential role in developing sustainable systems of crop protection. Crit Rev Plant Prot 19:1–30
Thurston HD, (1998) Tropical Plant Diseases, 2nd ed. APS Press, American Phytopathological Society, St. Paul, MN
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JS, White TJ (Eds). Protocols: A Guide to Methods and Applications, Academic Press, New York, pp 315–322
Zhaing W, Wendel JF, Clark LG (1997) Bamboozled again! Inadvertent isolation of fungal rDNA sequences from bamboos (Poaceae: Bambusoideae). Mol Phylogenet Evol 8:205–2l7
Zhou S, Stanosz GR (2001) Relationships among Botryosphaeria species and associated anamorphic fungi inferred from analyses of ITS and 5.8s rDNA sequences. Mycologia 93:516–527
Acknowledgments
This project was made possible by support from NASA-IRA, NIH-RCMI, and by FIPI (UPR’s Fondos Institucionales para la Investigación). We thank Miguel Angel Gamboa for advice.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Santamaría, J., Bayman, P. Fungal Epiphytes and Endophytes of Coffee Leaves (Coffea arabica). Microb Ecol 50, 1–8 (2005). https://doi.org/10.1007/s00248-004-0002-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-004-0002-1