Abstract
Differential emergence and diversity of bacterial communities from activated sludge in response to varied cultural conditions using 2,4-dichlorophenoxyacetic acid (2,4-D) were investigated by coupling molecular analyses based on 16S rDNA with functional genes. We employed three different cultural conditions: (1) a culture sequentially fed a high concentration (300 mg/L) of 2,4-D (HS); (2) a culture continuously fed a low concentration (10 mg/L) of 2,4-D (LC); and (3) a serial batch culture in which 1% (v/v) of culture was transferred to a fresh medium containing a high concentration (300 mg/L) of 2,4-D (HB). The HS and LC bioreactors were operated for 3 months and HB was repeatedly transferred for 1 month. The 2,4-D was stably degraded under all the cultural conditions tested. PCR amplification and cloning-based analysis of functional genes using community DNAs from the cultures revealed five different oxygenase genes that may be involved in the initial step of 2,4-D degradation. All five gene-types were present in HS, while one of the five genes, type V (tftA) was not detected in LC. Quantitative PCR analysis showed that in HS, Ralstonia eutropha JMP 134 type-tfdA4 (type I) was the most abundant in copy number (2.0 ± 0.1 × 107 copies/μg DNA) followed by RASC type-tfdA (type II) (1.8 ± 1.0 × 106 copies/μg DNA), putative cadA-like gene (type IV) (2.6 ± 0.8 × 105 copies/μg DNA), cadA gene (type III) (1.3 ± 1.0 × 104 copies/μg DNA), and tftA gene (type V) (3.5 ± 1.1 × 103 copies/μg DNA). Similar results were obtained in LC. In contrast, HB contained only type I and type III genes, and the type I gene was five orders of magnitude greater in copy number than the type III gene. Denaturing gel gradient electrophoresis (DGGE) analysis of PCR, amplified 16S rDNA fragments of bacterial communities in the three different cultures showed low similarity coefficient values (≤0.35) when compared to the original activated sludge, suggesting that 2,4-D amendment caused a drastic change in the bacterial community. Particularly, HB showed only six bands (16–18 bands in the other cultures) and very low similarity coefficient values when compared to the other communities (0.10 to HS, 0.17 to LC, and 0.0 to original sludge). These results indicated that serial batch culturing (HB) resulted in a phylogenetically limited number of 2,4-D degrading bacteria carrying limited catabolic genes whereas more diverse 2,4-D degraders and catabolic genes were present in HS and LC. Therefore, the approach used for monitoring should be taken into account when one evaluates the population dynamics of contaminant-degrading bacteria at bioremediation sites.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) has frequently been used as a model chemical to investigate the evolution and diversity of catabolic genes involved in the degradation of anthropogenic contaminants in the environment [3, 14, 16, 17, 18, 19, 20, 21, 22, 25, 27, 33]. Although the chemical structure of 2,4-D is relatively complex, it is readily degraded and used as a carbon source by various environmental microorganisms [8, 12, 28, 29, 32]. A number of 2,4-D degrading bacteria have been isolated and their catabolic genes have been characterized [2, 9, 10, 13, 22, 24, 30]. Currently there are four genes that have been found that can carry out the first step in the degradation pathway. Two are α-ketoglutarate dependent 2,4-D dioxygenases, tfdA from Ralstonia eutropha JMP134 [8, 14, 19, 25, 33] and tfdA from Burkholderia sp. strain RASC [30]. There is 78.5% nucleotide sequence identity between these two genes. Very recently, cadAB genes have been found in Bradyrhizobium sp. strain HW13 [22], which is a slow-growing bacterium in the α-subdivision of Proteobacteria isolated from a pristine environment. This finding was a significant breakthrough in 2,4-D degradation studies, since TfdA has been thought to be primarily responsible for 2,4-D metabolism in the environment. The cadAB genes have no sequence similarity with the tfdA gene but has 46% and 44% deduced ammo acid sequence identity to the tftAB genes, respectively, from a 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) degrading bacterium Burkholderia cepacia AC1100 [5, 6]. Both the cadAB and tftAB genes differ from tfdA because they code for two-component ring-hydroxylating oxygenases. The CadAB transformation of 2,4-D to 2,4-dichlorophenol differs from TftAB because a third subunit CadC, a P-450 ferredoxin, is also required, TftAB can transform 2,4-D to 2,4-dichlorophenol, but B. cepacia AC1100 apparently lacks other genes required to completely mineralize 2,4-D [15].
Many of the 2,4-D-degrading bacteria isolated from heterogeneous environments such as agricultural soils, sediments, waste treatment facilities, and pristine soils have been obtained by using batch enrichment cultures [8, 17, 19, 21]. The repeated subculturing in 2,4-D-rich medium is highly selective, so it often results in the isolation of copiotrophic fast-growing microbial species. Currently it is not clear if isolates obtained using this approach are representative of the phylogenetic and functional diversity of 2,4-D-degrading bacterial strains originally residing in the environment. To understand the factors important in microbial degradation of anthropogenic compounds in the environment it is essential to have methods to detect and quantify the responsible functional genes.
In this study we used the first gene in the 2,4-D degradation pathway as a model system to test the feasibility of adapting a real-time PCR method, which uses an internal quenching probe, to quantify catabolic genes in environmental samples. Because of the limited number of sequences currently available in the cad/tft family degenerate primers were determined to obtain more sequences related to these genes. Primers specific for five genes required for the first step in 2,4-D metabolism were determined and PCR conditions were optimized using well-characterized 2,4-D degraders. Using activated sludge as the initial inoculum three different enrichment cultures were then used to quantify the number of these catabolic genes present under varied cultivation conditions: (1) a culture sequentially fed a high concentration (300 mg/L) of 2,4-D; (2) a culture continuously fed a low concentration (10 mg/L) of 2,4-D; and (3) a serial batch culture in which 1% (v/v) of culture was repeatedly transferred to a fresh medium containing a high concentration (300 mg/L) of 2,4-D. Measurements of 2,4-D were made throughout the experiment to ensure that it was being degraded. Bacterial communities that developed in the three cultures were examined using denaturing gradient gel electrophoresis (DGGE) analysis of 16S rDNA amplified by PCR. The richness of 2,4-D-degrading isolates obtained by cultivation was also determined and the isolates were compared to the community from which they were isolated by PCR-DGGE. The phylogenetic diversity of 2,4-D-degrading bacteria isolates was also determined for comparison to functional diversity.
Materials and Methods
Enrichment of Activated Sludge with 2,4-D
A sample of activated sludge was taken from an aeration tank in a municipal sewage treatment plant in Ibaraki prefecture, Japan. The initial concentration of activated sludge was ~3000 mg/L (dried weight of sludge collected on a 0.22-μm pore-size filter) at startup of enrichment. The activated sludge sample was enriched under three different cultural conditions to reflect different environmental conditions: (1) HS. A 1.5-L reactor culture fed sequentially with a high concentration (300 mg/L) of 2,4-D for 3 months. When 2,4-D (300 mg/L) was completely degraded, the culture was allowed to settle, 1 L of the supernatant was discarded carefully to avoid losing biomass and replaced with the same volume of fresh 2,4-D medium. (2) LC. A laboratory-activated sludge unit continuously fed a low concentration (10 mg/L) of 2,4-D at a flow rate of 2.2 L per day for 3 months. The unit consisted of an aeration tank (working volume, 1.1 L) and a settling tank (volume, 0.4 L). The hydraulic retention time was 0.5 day. All the biomass from the settling tank was returned to the aeration tank to minimize loss of biomass from the reactor. (3) HB. A conventional serial batch culture in a high concentration (300 mg/L) of 2,4-D. Two milliliters of the 3-month microbial consortium from HS was inoculated into 198 ml of 2,4-D basal medium contained in a 500-mL volume flask. When the added 2,4-D was almost degraded, 2 mL of culture was transferred to fresh medium. This successive subculturing was repeated 11 times over a month.
Basal mineral medium (2,4-D medium) with no additional organic substrates as previously described [21] was used for all cultivations. In all experiments 2,4-D was added from a 15,000 mg/L stock solution made by dissolving the chemical in 0.1 M NaH2PO4 buffer (pH 7.0). All cultures were maintained at 25°C. Air was continuously supplied to HS and LC at a rate of 2 L per min and to HB by shaking at 200 rpm. Concentration of 2,4-D in all three different cultures were periodically (at 2- to 4-day intervals) measured with a high-performance liquid chromatograph (HPLC, System Gold, Beckman Instruments, Fullerton, CA, USA) equipped with a Hypersil ODS column (4.6 × 250 mm; Agilent Technologies, CA, USA), using 50% acetonitrile–50% acidic water (pH 2.6) as the mobile phase.
DNA Extraction
DNA was extracted from the original sludge inoculum and from the microbial communities that developed under the treatments described above. Samples (10 mL) of the three different enrichment cultures were centrifuged at 8000 × g at 4°C for 15 min, and the pellets were stored at −20°C until use. Total community DNAs were extracted from the pellets using the Fast DNA extraction kit (BIO101, Inc., Carlsbad, CA, USA) according to the manufacturer’s specifications. DNA concentrations were measured using the PicoGreen dsDNA quantitation kit (Molecular Probes, Eugene, OR, USA) according to the manufacturer’s specifications. The extracted DNAs were all diluted to the same concentration, 30 ng DNA/μL.
PCR Amplification and Cloning of 2,4-D Catabolic Genes
To determine the presence and to differentiate the genes required for the first step in 2,4-D degradation in environmental samples, gene-specific primers were determined by aligning all sequences known with this function. Initially three primer sets were determined type 1, type II, and Deg cad/tft (Table 1). The type I primer set was specific for tfdA genes representative of R. eutropha JMP134 and type II for the Burkholderia sp. strain RASC group. There was only one sequence available for each of the two other known 2,4-D catabolic genes, cadA [22] and tftA [5, 6] therefore degenerate primers (deg cad/tft) were determined that targeted both genes.
The deg cad/tft primer set was tested on the HS and LC community DNA extracts by conducting PCR, then cloning and sequencing the products. Each PCR mixture contained 30 ng of purified community DNA, 0.4 μM of each primers, 200 μM of each dNTP, 1 × PCR buffer (30 mM Tris/HCl pH 8.05, 100 mM KCl), 1.5 mM MgCl2, 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA, USA) in a final volume of 50 μL. PCR amplification consisted of 45 cycles of 1 min at 95°C, 1 min at 50°C, and 2 min at 72°C. The PCR products were purified with MicroSpin S-400 HR columns (Amersham Biosciences), ligated to pT7 Blue plasmid (pT7 Blue-2 LIC vector kit) (Novagen, CN Biosciences, Darmstadt, Germany) and transformed into Escherichia coli DH5α (Competent High, Toyobo, Osaka, Japan). The nucleotide sequences of 81 clones were determined using an Applied Biosystems model 377 DNA sequencer. Sequences were aligned using the Clustal W program [31]. Analysis revealed three distinct sequences that were designated type III-cadA, type IV-putative cadA-like, and type V-tftA. Within each type the sequences were almost 100% identical therefore primer sets were determined to distinguish each type (Table 1) within a community. Optimal annealing temperatures ranging from 50 to 65°C were tested for PCR amplifications of all five primer sets (types I to V) using the PCR reagent concentrations described above. PCR amplification consisted of initial denaturation at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s annealing temperature (50–65°C range), and 30 s at 72°C, and a final extension at 72°C for 10 min. R. eutropha JMP134 [9], Burkholderia sp. strain RASC [30], and the E. coli recombinants which harbored types III to V genes were used as positive and negative controls. The optimal annealing temperature was determined to be 60°C for all primer sets and was used for subsequent PCR amplifications.
Quantitative PCR
To quantify the five different gene types within a community we adapted a real-time PCR method that uses an internal quenching probe [4, 23]. To increase specificity to the target genes in addition to the primers, probes specific for the five gene types were determined (Table 1). The probes were labeled with BODIPY FL (D-6140, Molecular Probes) at their 5′- or 3′-end via an aminohexylphosphate linker with a six-carbon spacer and were blocked from extension with 3′-phosphate, if necessary. All oligonucleotides were obtained from Espec Oligo Service Corp. (Tsukuba, Japan) and used without further purification. PCR with fluorescence monitoring was performed in 20-μL volumes per reaction in a real-time PCR instrument (LightCycler, Roche Diagnostics, Mannheim, Germany) following the method reported previously [23]. Briefly, each reaction contained 0.5 μM of each type-specific primer (Table 1), 50 nM BODIPY FL-labeled internal probe, 160 μM of each dNTP, 50 mM Tris, pH 8.3 (25°C), 1.5 mM MgCl2, 250 μg/mL bovine serum albumin, 0.5 U of Taq polymerase (Gene Taq, Nippon Gene Co. Ltd., Tokyo, Japan), and 0.32 μL of TaqStart antibody (Clontech Laboratories Inc., Palo Alto, CA, USA). Templates used in amplifications were standards known to contain the target genes, total community DNAs (30 ng/μL) extracted from HS, LC, HB, and the original sludge.
Total DNAs from R. eutropha JMP134 (for type I), Burkholderia sp. RASC (for type II), and the E. coli recombinants which harbored types III to V genes were used to make standards for quantitative PCR. The target genes (Table 1) were amplified by PCR, products were purified using a gel extraction kit (Qiagen Inc., Hilden, Germany) and then nucleotide sequences were determined to confirm the products. Products were quantified with the PicoGreen dsDNA quantitation kit using λ-DNA as the standard (Molecular Probes). The PCR products were serially diluted to make standards ranging from 101 to 109 copies of each target sequence.
Rapid-cycle PCR was performed using an initial denaturation at 95°C for 2 min followed by 45 cycles of 95°C for 15 s, 60°C for 18 s, and extension at 72°C for 10 s. Temperature transition rates were programmed for 20°C /s. Fluorescence was acquired twice in each cycle, at the end of denaturation (Fd) and annealing (Fa). The fluorescence-quenching ratio (Q) at each cycle was estimated by the following equation:
Q = { 1 − (Fa/Fd)/P } × 100
where P is the fluorescence ratio of (Fa/Fd) at the cycle when a significant decrease in the (Fa/Fd) value was observed. The value of P was used for proportional background subtraction [23]. For quantification, crossing points with a certain threshold line were obtained from the fluorescence-quenching ratio (on the logarithmic scale) versus cycle number plots. The values of crossing points were plotted against the log of the initial template copy number to make a standard curve. The copy numbers of the targeted type gene in the community DNAs extracted from the three different cultures (HS, LC, and HB) and the original sludge were determined from the standard curve.
DGGE Analysis of Bacterial Communities
The variable V3 region of 16S rRNA gene was PCR amplified with primers 357F (5′-CCT ACG GGA GGC AGC AG-3′) with a GC-clamped (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G-3′) on the 5′ end and 518R (5′-ATT ACC GCG GCT GCT GG-3′), which amplifies a 193-bp fragment in E. coli [26]. PCR amplification was performed in an Eppendorf Mastercycler gradient (Eppendorf, Hamburg, Germany). Each PCR mixture contained 150 ng of purified community DNA, 0.5 μM each of primers, 200 μM of each dNTP, 1 × PCR buffer (30 mM Tris/HCl pH 8.05, 100 mM KCl), 1.5 mM MgCl2, and 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems) in a final volume of 100 μL. The tubes were first incubated for 10 min at 95°C. Touchdown PCR [11] was then performed by using 20 cycles of denaturation at 95°C for 1 min, annealing at 65°C (then decreasing the temperature by 1°C every second cycle until the touchdown temperature of 56°C was reached) for 1 min, and primer extension at 72°C for 1 min. Ten additional cycles were carried out using an annealing temperature of 55°C followed by a final extension at 72°C for 10 min.
The PCR products were purified with MicroSpin S-400 HR columns (Amersham Biosciences, Piscataway, NJ, USA) and analyzed by DGGE using a DCode™ System (Bio-Rad Laboratories, Hercules, CA, USA). PCR samples were applied directly onto 8% (w/v) polyacrylamide gels in 0.5 × TAE [20 mM Tris-acetate (pH 7.4), 10 mM acetate, 0.5 mM disodium EDTA] with denaturing gradient ranging from 20 to 70% [100% denaturant corresponded to 7 M urea and 40 % (v/v) formamide]. Electrophoresis was performed for 5 hr at 130 V and 25 mA (Bio-Rad Power PAC 1000) and at a constant temperature of 60°C. After electrophoresis, the gel was incubated in ethidium bromide (0.5 mg/L) for 30 min then photographed on a UV transilluminator with a digital camera (EDAS 290 Electrophoresis Documentation and Analysis system, Kodak, Tokyo, Japan).
All samples were run on a single DGGE gel to compare communities from the different enrichments tested. Bands in the DGGE gels were visually identified and distinguished by the distance migrated in the gel. Each band was scored as being present or absent in each of the samples. Dice similarity coefficients [7], Cs, were used to quantify similarity between the communities. The similarity coefficient, Cs, was determined as follows: Cs = 2j/(a + b), where a was the number of DGGE bands in a culture, b was the number of DGGE bands in the other culture, and j was the number of common DGGE bands.
Isolation of 2,4-D-Degrading Bacteria
To compare results obtained from molecular genetic analysis to those obtained by cultivation, bacteria were isolated from HS (day 90), LS (day 90), and HB (day 30) cultures using 0.1 × PTYG agar medium or 2,4-D agar medium containing 10 or 300 mg/L 2,4-D as previously described [21]. After 3–10 days of incubation at 25°C, a total of ~200 colonies were randomly selected and transferred onto 2,4-D (100 mg/L)- BTB (0.001% bromthymol blue used as a pH indicator) plates. Colonies that turned the 2,4-D-BTB plates yellow [21], which indicated acid was being produced from 2,4-D degradation, were picked and further purified on the same medium. Their 2,4-D-degrading ability was then confirmed by HPLC using single colonies grown in fresh 2,4-D (100 mg/L) medium with no additional organic substrates. Each 2,4-D-degrading isolate was compared to the community from which they were isolated by PCR-DGGE as described above.
Phylogenetic Analysis
The entire 16S rRNA gene of each isolate was amplified with fD1 (5′-AGA GTT TGA TCC TGG CTC AG-3′) as forward primer (E. coli positions 8 to 27) and modified rPl (5′-GGT TAC CTT GTT ACG ACT T-3′) as reverse primer (E. coli positions 1510 to 1492) [34]. PCR reagents were same as described above and the amplification consisted of 35 cycles of 1 min at 95°C, 1 min at 50°C, and 2 min at 72°C. The PCR products were purified with MicroSpin S-400 HR columns (Amersham Biosciences) and partial nucleotide sequences (~500 bp sequences) were determined with the forward primer as sequence primer using an Applied Biosystems model 377 DNA sequencer Nucleotide sequences were compared with sequences in the National Center for Biotechnology Information (NCBI) GenBank database using the BLASTn program [1]. The 16S rDNA sequences of the isolates and representative 2,4-D-degrading bacteria were aligned with ARB program (Ludwig W et. al., ARB: a software environment for sequence data, unpublished data [http://www.arb-home.de/]). Subsequently, the alignments were corrected by visual inspection considering the secondary structure of the 16S rRNA. All gaps and unidentified positions were eliminated in calculation, and the phylogenetic analysis of the sequences was performed by applying ARB parsimony method in the software package.
Neucleotide Sequence Determination
The nucleotide sequences obtained in this study have been deposited in the DNA Data Bank of Japan (DDBJ) database under the following accession nos.: strain HBH2, AB088537; strain HBL7, AB088542; strain LCL2, AB088550; strain LCL3, AB088551; strain LCH2, AB088546; strain HBL1, AB088538; strain HBH3, AB088433; strain HBL5, AB088540; strain LCH1, AB088545; strain HBL6, AB08854; strain LCH4, AB088548; strain LCL1, AB088549; strain HBL4, AB088539; strain HSP2, AB088544; strain HSH6, AB088543; strain LCH3, AB088547; strain LCP2, AB088552; type III gene, AB088553; type IV gene, AB088554; type V gene, AB088555.
Results
Degradation of 2,4-D in the Three Different Cultures
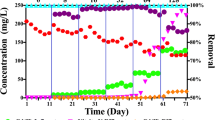
The patterns of 2,4-D degradation in HS, LC, and HB are shown in Fig. 1. HS showed a long lag period and 45 days was required to completely degrade the first amendment of 2,4-D (Fig. 1a). The subsequent twelve 2,4-D amendments did not have the same lag phase and it took only 4 to 7 days for 2,4-D to be degraded. In LC it took 40 days for the effluent 2,4-D to go below 0.1 mg/L, the detection limit of HPLC, and then it remained below detection for the last 3 months of operation (Fig. 1b). Throughout the 11 rounds of subculturing of HB 2,4-D was rapidly degraded within 2 to 4 days without a lag period (Fig. 1c), likely because the inoculum had been preconditioned in the HS bioreactor.
Degradation of 2,4-D in the three different cultures inoculated with activated sludge: (a) a culture sequentially fed a high concentration (300 mg/L) of 2,4-D (HS); (b) a continuous culture amended with a low concentration (10 mg/L) of 2,4-D (LC); and (c) a repeated batch culture with a high concentration (300 mg/L) of 2,4-D (HB) in which 1% (v/v) of the culture was transferred to a fresh medium and subcultured (11 times within a month as indicated in the figure). Symbols, closed circle: 2,4-D concentration in the reactor (a and c) and effluent 2,4-D concentration (b); open circle: influent 2,4-D concentration (b).
PCR Amplification and Classification of 2,4-D Catabolic Genes
Five primer sets were determined in this study to distinguish the different oxygenases known to catalyze the first step of 2,4-D degradation. The specificity of the primers for PCR was determined by amplification of the expected product size (Table 1) from the positive and no products in the negative controls. The PCR primer set designated type I was specific for tfdA from JMP134 and type II for tfdA from RASC (Figs. 2a, b). There was very little information available on isolates carrying the newly discovered cadA and tftA genes; therefore a degenerate primer set was first used to capture genes present in mixed communities that had similar sequences. Three different sequences were found within the 81 clones we screened and designated cad gene type (type III), putative cad-like gene type (type IV), and tft type (type V). The former had 78.0% nucleotide identity (87.7% amino acid sequence similarity) to the cadA gene and only 53.9% amino acid identity to tftA. The type IV sequence had 63.7% nucleotide identity (60.0% amino acid identity) to cadA and 60.4% nucleotide identity (54. 1% amino acid identity) to tftA. The type V sequence had 99.6% nucleotide identity (98.8% amino acid sequence identity) to the tftA gene. The high amino acid sequence similarity of the type III and type V genes to the known genes suggest they have the same function. Type IV has less similarity; therefore to be conservative we have designated it a putative cad-like gene. Optimal annealing temperature for all primer sets was 60°C, which was used for all subsequent analyses. When the five primer sets were tested using total community DNAs from enrichment cultures, HS was found to contain all genes types. Whereas LC had types I through IV, HB had types I and III, and only type V was present in the original sludge used as an inoculum (Fig. 2).
PCR amplification products of the five gene types (panel a: type I; panel b: type II; panel c: type III; panel d: type IV; panel e: type V) from total community DNAs (30 ng) extracted from the original sludge, HS, LC, and HB. Lane M: 100 bp DNA Ladder marker (New England Biolabs, Inc., Beverly, MA, USA); lane 1: positive control; lane 2: the original activated sludge not enriched with 2,4-D; lane 3: HS; lane 4: LC; lane 5: HB; lane 6: negative control (without template); lane 7: negative control (type II gene in panel a, type I gene in panel b, c, d, and e); lane 8: negative control (type III gene in panel a and b, type II gene in panel c, d, and e); lane 9: negative control (type IV gene in panel a, b, and c, type III gene in panel d and e); lane 10: negative control (type V gene in panel a, b, c, and d, type IV gene in panel e). The positive control DNAs used were as follows, panel a: total DNA of Ralstonia eutropha JMP134 for the type I gene; panel b: total DNA of Burkholderia sp. strain RASC for the type II gene; panel c to e: E. coli recombinant which harbors each type gene (c: type III; d: type IV; e: type V).
Quantification of the Five Types of 2,4-D Catabolic Genes
Copy numbers of the five types of catabolic genes in the total community DNAs was quantified using a rapid-cycle PCR method with quenching probes to determine their frequency. Typical quenching curves generated by real-time quantification by rapid-cycle PCR and a standard curve used for quantification are shown in Fig. 3. Quantification indicated that the copy number of gene type I (JMP134 type-tfdA) was one to five orders of magnitude greater than those of the other four types of genes in all cultures (Table 2). All of the genes were present in HS, while only one of the five genes, type V (tftA), was present in the original sludge. However, the copy number of type V in the sludge and HS was equivalent. The highest number of the type I gene was detected in HB, and the only other gene, type III, was five orders of magnitude lower in number.
Real-time quantification of type I gene (JMP134 type-tfdA) by using a specific primer set and a quenching probe labeled with BODIPY FL. PCR amplification was conducted using the total community DNAs (30 ng) extracted from the three cultures and the original activated sludge as templates together with the purified PCR product at 109 to 102 copy numbers per reaction as standard. Amplification was accompanied by decrease in fluorescence intensity in response to PCR product accumulation. The fluorescence-quenching ratio at each cycle was plotted on logarithmic scale (a). The quenching curves shift to earlier cycles as the amount of initial target gene increases from 102 to 109. Standard curve for quantification of type I gene was depicted by plotting the initial template concentration (initial copy number) versus cycle number at cross point where each quenching curve met a certain threshold line (b). Symbols, closed circle: total DNA from HS; open square: total DNA from LC; open circle: total DNA from HB; dashed horizontal line: threshold line (a).
DGGE Analysis of Bacterial Communities
DGGE analysis of PCR amplified 16S rDNA fragments of bacterial communities in the three different 2,4-D enrichment cultures and the original activated sludge showed different community fingerprint patterns (Fig. 4). There were a similar number of bands observed in the original sludge, HS, and LC profiles, 16, 16, and 18 respectively. There were only 6 bands in HB. However, the similarity coefficient values indicated that the complexities of the community fingerprints were not very similar to the original sludge inoculum or to each other. The similarity coefficient value between the original sludge and HS, LC and HB was 0.31, 0.35 and 0.0, respectively, where 1 is an exact match and 0 is no match. The value between HS and LC was 0.29, HS and HB was 0.10, and LC and HB was 0.17.
DGGE profiles (negative image) of PCR amplified 16S rDNA fragments of bacterial community in the original activated sludge that was not amended with 2,4-D (Ori.) and the three different cultures (HS, LC, and HB). DGGE bands of the 16S rDNA fragments of 2,4-D degrading bacterial isolates that correspond to bands in the DGGE community profiles are LCL3 matching band f in LC; HBH2 matching band f in HB; HBL5 matching band d in HB; HBL6 matching band c in HB; HSH6 matching band e in HS; and HSP2 matching band b in HS.
Isolation of 2,4-D Degrading Bacteria
Of the 200 isolates from the three enrichment cultures, 60 initially turned the 2,4-D BTB medium yellow, indicating the substrate was being degraded. However, the phenotype was very unstable in most of the isolates and the trait was lost during purification. Finally 17 isolates (two from HS, eight from LC, and seven from HB) capable of degrading 2,4-D as sole carbon and energy source were obtained for further analysis. Phylogenetic analysis based on partial 16S rDNA sequences (about 500 bp) revealed that the isolates were affiliated with only four genera: Ochrobactrum, Ralstonia, Comamonas, and Pseudomonas, each of which belongs to the α-, β-, or γ-Proteobacteria (Fig. 5). PCR amplification using primers specific for the five types of genes encoding enzymes that catalyze the first step of 2,4-D degradation indicated that all of the isolates harbored only JMP134 type-tfdA (type I) gene (data not shown). All the PCR-DGGE profile of the 2,4-D degrading isolates had corresponding bands in the community fingerprints from which they were isolated. For example, the intense bands in the community profiles labeled b, c, d, e and f matched the bands from strains HSP2, HBL6, HBL5, HSH6, and LCL3 and HBH2, respectively. However, majority of bands in the DGGE profiles did not have a corresponding isolate and in particular the highly intense band labeled “a” found in HS and LC (Fig. 4). This is not surprising because all members of the community may not be able, to completely mineralize 2,4-D and would not have been obtained using the cultivation approach we used. Also only a fraction of isolates initially isolated on 2,4-D media maintained the phenotype and were used for further characterization.
Phylogenetic tree based on the nucleotide sequences of partial 16S rDNA (about 500 bp) of 2,4-D-degrading strains isolated from HS, LC, and HB. The first two letters of the isolate’s strain name indicates the culture from which it was isolated. The third letter indicates the agar medium used for the initial isolation, H: 2,4-D (300 mg/L)-BTB plate; L: 2,4-D (10 mg/L)-BTB plate; P: 0.1 × PTYG plate. Microorganisms previously described as 2,4-D-degraders are indicated in boldface. DDBJ/GenBank, accession numbers: strain HBH2, AB088537; strain HBL7, AB088542; strain LCL2, AB088550; strain LCL3, AB088551; strain LCH2, AB088546; Ralstonia sp. TFD41, AF067833; strain HBL1, AB088538; strain HBH3, AB088433; strain HBL5, AB088540; strain LCH1, AB088545; Ralstonia eutropha JMP134, AF139729; Burkholderia sp. RASC, AF049544; Comamonas terrigena, AJ420326; strain HBL6, AB088541; Rhodoferax fermentans TFD23, AF049536; Variovorax sp. HW1, D89026; strain LCH4, AB088548; strain LCL1, AB088549; strain HBL4, AB088539; Pseudomonas putida, AF307865; strain HSP2, AB088544; Sphingomonas sp. B6-10, AF049534; Bradyrhizobium sp. HW13, D89027; Ochrobactrum anthropi, AJ242580; strain HSH6, AB088543; strain LCH3, AB088547; strain LCP2, AB088552; Aquifex pyrophilus, M83548. The scale bar indicates the number of nucleotide changes per base position analyzed.
Discussion
To better understand the diversity of functional genes in communities a real-time PCR method was adapted to quantify the genes for the first step of the 2,4-D degradation pathway. Application of the method to communities evolved under different cultural conditions indicated that the gene type and abundance were different (Fig. 2 and Table 2). Of the five gene types tested, the type I gene was detected from all of the three cultures at one to five orders of magnitude greater than the other types of genes, whereas it was not detected in the original sludge. Others have speculated that bacterial strains harboring different types of catabolic genes may emerge when different concentrations of resources are fed to a microcosm. Ka et al. [19] isolated 47 dominant 2,4-D degrading bacterial strains from agricultural plots with and without 2,4-D treatment at three different concentrations, and classified them into four groups based on 2,4-D catabolic gene typing. The isolates grouped according to 2,4-D amendment concentrations and treatment periods.
Quantification of the various genes easily demonstrated why the JMP134 type-tfdA gene has been isolated in most studies. Although this gene type was below detection in the original inoculum it became the dominant gene type under all cultural conditions tested. Despite the dominance of this single gene type the PCR-DGGE community profiles indicated that the community members were very different from each other (Fig. 4) with similarity coefficients 0.3. The 2,4-D-degrading isolates obtained in this study aided in data interpretation. A number of the 2,4-D-degrading bacterial isolates corresponded to different intense bands in PCR-DGGE profiles of the community from which they were obtained. Phylogenetic analysis indicated that the isolates belonged to the α-, β-, and γ-Proteobacteria (Fig. 5). However, all these isolates harbored only the type I gene. This illustrates the difference between analyses of community based on their function versus diversity. There is apparently redundancy of functional genes in phylogenetically diverse bacteria.
There were also differences in the numbers and types of the other initial 2,4-D catabolic genes present under the various cultural conditions. In HS, all of the gene types were detected although the tfdA type genes (type I and II) were dominant. A similar pattern was obtained in LC except that type V was missing. The biomass of both cultures had been retained in the reactors without significant loss allowing the survival and/or retention of slower growing organisms. This may be the primary reason why all the gene types could have coexisted with the dominant gene. In particular the type V gene may have been present in HS because of biomass retention and was not involved in 2,4-D degradation. By contrast, only the type I and type III genes were detected in HB. The frequent dilution of HB by transferring the culture would favor faster growing organisms: It would not be surprising if microbes harboring the type I gene are fast growers, since most of the previously described organisms harboring type I grow fast [21]. However, those explanations are highly speculative, and the question could have been addressed more definitively had we been able to isolate 2,4-D degraders harboring the respective genes. However, attempts at isolation of a variety of organisms covering all types of genes were unsuccessful. This likely occurred because an overwhelming number of strains carried the type I gene. Alternatively the strains harboring the other gene types could not completely mineralize 2,4-D or grew on other metabolites present in the bioreactors and would therefore not have been selected.
By using degenerate primers for the amplification of cadA and tftA family genes a third gene sequence was found in 2,4-D-degrading microbial communities. This demonstrates the wealth of information that is still unknown despite the considerable amount of research that has been done in 2,4-D degradation genetics and physiology. The 2,4-D-degrading activity of the three types of genes designated III to V remains to be clarified, but it is very likely that types III and type IV genes are involved in 2,4-D metabolism, since they emerged after 2,4-D amendment in HS and LC (Table 2, Figs. 2c, d). Interestingly, the type V gene was detected from the original activated sludge and HS but not from the other cultures, suggesting that the strains harboring the tftA gene may not be selected when 2,4-D is the sole carbon source. This result is consistent with the previous report that B. cepacia AC1100 harboring tftAB genes did not grow in 2,4-D and the enzyme had greater affinity for 2,4,5-T transformation [15].
The coupling of molecular analysis of microbial community based on PCR-amplified 16S rDNA with quantification of functional genes is a useful approach to understand community structure, diversity, and function. Different cultural conditions were chosen to reflect different scenarios that may occur in the environment to test whether the method would indeed find differences in the presence and numbers of functional genes present in a community. Our results demonstrated that different 2,4-D amendments and cultural conditions greatly influenced the diversity and abundance of catabolic genes for 2,4-D degradation which can be measured using a molecular genetic approach but missed when using a traditional cultivation method. Serial batch cultivation using high concentrations of 2,4-D (HB) resulted in a phylogenetically limited range of 2,4-D-degrading bacteria carrying limited catabolic gene types, whereas bioreactors with retained biomass (LC and HS) led to a wider variety of 2,4-D catabolic genes. Therefore, this should be taken into account if one is using cultivation to evaluate and monitor the population of contaminant-degrading microorganisms at sites where biostimulation is used to clean up a contaminated environment. Future research will include tests using environmental samples.
References
SF Altschul W Gish W Miller EW Myers DJ Lipman (1990) ArticleTitleBasic local alignment search tool. J Mol Biol 215 403–410 Occurrence Handle10.1006/jmbi.1990.9999 Occurrence Handle1:CAS:528:DyaK3MXitVGmsA%3D%3D Occurrence Handle2231712
MA Bhat M Tsuda K Horiike M Nozaki CS Vaidyanathan T Nakazawa (1994) ArticleTitleIdentification and characterization of a new plasmid carrying genes for degradation of 2,4-dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Appl Environ Microbiol 60 307–312 Occurrence Handle1:CAS:528:DyaK2cXitV2ltbk%3D Occurrence Handle7509586
GR Chaudry GH Huang (1988) ArticleTitleIsolation and characterization of a new plasmid from a Flavobacterium sp. which carries the genes from the degradation of 2,4-dichlorophenoxyacetate. J Bacteriol 170 3897–3902 Occurrence Handle2842290
AO Crockett CT Wittwer (2001) ArticleTitleFluorescein-labeled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal Biochem 290 89–97 Occurrence Handle10.1006/abio.2000.4957 Occurrence Handle1:CAS:528:DC%2BD3MXht1artLc%3D Occurrence Handle11180941
CE Danganan RW Ye DL Daubaras L Xun AM Chakarabarty (1994) ArticleTitleNucleotide sequence and functional analysis of the genes encoding 2,4,5-trichlorophenoxyacetic acid oxygenase in Pseudomonas cepacia AC1100. Appl Environ Microbiol 60 4100–4106 Occurrence Handle1:CAS:528:DyaK2MXhvFGmtbs%3D Occurrence Handle7527626
CE Danganan S Shankar RW Ye AM Chakrabarty (1995) ArticleTitleSubstrate diversity and expression of the 2,4,5-trichlorophenoxyacetic acid oxygenase from Burkholderia cepacia AC1100. Appl Environ Microbiol 61 4500–4504 Occurrence Handle1:CAS:528:DyaK2MXpslCmtbw%3D Occurrence Handle8534119
LR Dice (1945) ArticleTitleMeasures of the amount of ecologic association between species. Ecology 26 297–302
RH Don JM Pemberton (1981) ArticleTitleProperties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol 145 681–686 Occurrence Handle1:CAS:528:DyaL3MXhtFOlurs%3D Occurrence Handle6257648
RH Don JM Pemberton (1985) ArticleTitleGenetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J Bacteriol 161 466–468 Occurrence Handle1:CAS:528:DyaL2MXhtVOitbk%3D Occurrence Handle3968031
RH Don AJ Weightman HJ Knackmuss KN Timmis (1985) ArticleTitleTransposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134 (pJP4). J Bacteriol 161 85–90 Occurrence Handle1:CAS:528:DyaL2MXhs1Wntrg%3D Occurrence Handle2981813
RH Don PT Cox BJ Wainwright K Barker JS Mattick (1991) ArticleTitle“touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19 4008 Occurrence Handle1:CAS:528:DyaK3MXmtVShu70%3D Occurrence Handle1861999
WC Evance BSW Smith HN Fernley JI Davies (1971) ArticleTitleBacterial metabolism of 2,4-dichlorophenoxyacetate. Biochem J 122 543–551 Occurrence Handle5123888
F Fukumori RP Hausinger (1993) ArticleTitleAlcaligenes eutrophus IMP 134 “2,4-dichlorophenoxyacetic acid monooxygenase” is an α-ketoglutarate-dependant dioxygenase. J Bacteriol 175 2083–2086 Occurrence Handle1:CAS:528:DyaK3sXitVyitbY%3D Occurrence Handle8458850
RR Fulthorpe C McGowan OV Maltseva WE Holben JM Tiedje (1995) ArticleTitle2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol 61 3274–3281 Occurrence Handle1:CAS:528:DyaK2MXnvVyisLk%3D Occurrence Handle7574638
RA Haugland DJ Schlemn RP Lyons III PR Sferra AM Chakrabarty (1990) ArticleTitleDegradation of the chlorinated phenoxyacetate herbicides 2,4-dichlorophenoxyacetic acid and 2,4,5-trichlorophenoxyacetic acid by pure and mixed bacterial cultures. Appl Environ Microbiol 56 1357–1362 Occurrence Handle1:CAS:528:DyaK3cXkslCmsrw%3D Occurrence Handle2339889
DA Hogan DH Buckley CH Nakatsu TM Schmidt RP Hausinger (1997) ArticleTitleDistribution of the tfdA gene in soil bacteria that do not degrade 2,4-dichlorophenoxyacetic acid (2,4-D). Microb Ecol 34 90–96 Occurrence Handle10.1007/s002489900038 Occurrence Handle1:CAS:528:DyaK2sXltVWksbc%3D Occurrence Handle9230096
WE Holbon BM Schroeter VG Matheson RH Olsen JK Kukor VO Biederbeck AE Smith JM Tiedje (1992) ArticleTitleGene probe analysis of soil microbial populations selected by amendment with 2,4-dechlorophenoxyacetic acid. Appl Environ Microbiol 58 3941–3948 Occurrence Handle1476437
K Itoh R Kanda Y Sumita H Kim Y Kamagata K Suyama H Yamamoto RP Hausinger JM Tiedje (2002) ArticleTitletfdA-like genes in 2,4-D degrading bacteria that belong to Bradyrhizobium–Agromonas–Nitrobacter–Afipia cluster in α-Proteobacteria. Appl Environ Microbiol 68 3449–3454 Occurrence Handle10.1128/AEM.68.7.3449-3454.2002 Occurrence Handle1:CAS:528:DC%2BD38Xlt1SntLg%3D Occurrence Handle12089027
JO Ka WE Holben JM Tiedje (1994) ArticleTitleGenetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soil. Appl Environ Microbiol 60 1106–1115 Occurrence Handle1:CAS:528:DyaK2cXis1Gmsbs%3D Occurrence Handle8017907
JO Ka WE Holben JM Tiedje (1994) ArticleTitleUse of gene probe to aid in recovery and identification of functionally dominant 2,4-dichlorophenoxyacetic acid-degrading populations in soil. Appl Environ Microbiol 60 1116–1120 Occurrence Handle1:CAS:528:DyaK2cXis1Gmsbg%3D Occurrence Handle8017908
Y Kamagata RR Fulthorpe K Tamura H Takami LJ Forney JM Tiedje (1997) ArticleTitlePristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxy acid-degrading bacteria. Appl Environ Microbiol 63 2266–2272 Occurrence Handle1:CAS:528:DyaK2sXjsFCltbg%3D Occurrence Handle9172346
W Kitagawa S Takami K Miyauchi E Masai Y Kamagata JM Tiedje M Fukuda (2002) ArticleTitleNovel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J Bacteriol 184 509–518 Occurrence Handle10.1128/JB.184.2.509-518.2002 Occurrence Handle1:CAS:528:DC%2BD38XjtVyrug%3D%3D Occurrence Handle11751829
S Kurata T Kanagawa K Yamada M Torimura Y Kamagata T Yokomaku K Kurane (2001) ArticleTitleFluorescent quenching-based quantitative detection of specific DNA/RNA using a BODIPY FL-labeled probe or primer. Nucleic Acids Res 29 e34 Occurrence Handle10.1093/nar/29.6.e34 Occurrence Handle1:STN:280:DC%2BD3M3ks1Omuw%3D%3D Occurrence Handle11239011
OV Maltseva C McGowan R Fulthorpe PJ Oriel (1996) ArticleTitleDegradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology 142 1115–1122 Occurrence Handle1:CAS:528:DyaK28XjtFyisr8%3D Occurrence Handle8704953
C McGowan R Fulthorpe A Wright JM Tiedje (1998) ArticleTitleEvidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microbiol 64 4089–4092 Occurrence Handle1:CAS:528:DyaK1cXms1emsrY%3D Occurrence Handle9758850
G Muyzer C Ellen DE Waal AG Uitterlinden (1993) ArticleTitleProfiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction–amplified genes coding for 16S rRNA. Appl Environ Microbiol 59 695–700 Occurrence Handle1:CAS:528:DyaK3sXit1Kktrk%3D Occurrence Handle7683183
CH Nakatsu R Korona RE Lenski FJ De Bruijn TL Marsh L Forney (1998) ArticleTitleParallel and divergent genotypic evolution in experimental population of Ralstonia sp. J Bacteriol 180 4325–4331 Occurrence Handle1:CAS:528:DyaK1cXlvVCmtb0%3D Occurrence Handle9721265
MH Rogoff JJ Reid (1956) ArticleTitleBacterial decomposition of 2,4-dichlorophenoxyacetic acid. J Bacteriol 71 303–307 Occurrence Handle1:CAS:528:DyaG28XltlSgtA%3D%3D Occurrence Handle13306699
GL Silton LT Fan LE Erickson SM Lee (1986) ArticleTitleBiodegradation of 2,4-D and related xenobiotic compounds. Enzyme Microb Technol 8 395–403 Occurrence Handle10.1016/0141-0229(86)90145-6
Y Suwa AD Wright F Fukumori KA Nummy RP Hausinger WE Hollben LJ Forney (1996) ArticleTitleCharacterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl Environ Microbiol 62 2464–2469 Occurrence Handle1:CAS:528:DyaK28XjvFGnu7Y%3D Occurrence Handle8779585
JD Thompson DG Higgins TJ Gibson (1996) ArticleTitleClustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680
JM Tiedje M Alexander (1969) ArticleTitleEnzymatic cleavage of the ether bond of 2,4-dichlorophnoxyacetate. J Agric Food Chem 17 1080–1084 Occurrence Handle10.1021/jf60165a012 Occurrence Handle1:CAS:528:DyaF1MXkvFymsr8%3D
TR Vallaeys R Futhorpe AM Wright G Soulas (1996) ArticleTitleThe metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microbiol Ecol 20 163–172 Occurrence Handle10.1016/0168-6496(96)00027-X Occurrence Handle1:CAS:528:DyaK28XjvFGhtLs%3D
WG Weisbrug SM Barns DA Pelletier DJ Lane (1991) ArticleTitle16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 173 697–703 Occurrence Handle1987160
Acknowledgment
The Japanese Society for the Promotion of Science supported this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, T., Kurata, S., Nakatsu, C. et al. Molecular Analysis of Bacterial Community Based on 16S rDNA and Functional Genes in Activated Sludge Enriched with 2,4-Dichlorophenoxyacetic Acid (2,4-D) under Different Cultural Conditions. Microb Ecol 49, 151–162 (2005). https://doi.org/10.1007/s00248-003-1035-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-003-1035-6