Abstract
The connection between microbial community structure and spatial variation and pollution in river waters has been widely investigated. However, water and sediments together have rarely been explored. In this study, Illumina high-throughput sequencing was performed to analyze microbes in 24 water and sediment samples from natural to anthropogenic sources and from headstream to downstream areas. These data were used to assess variability in microbial community structure and diversity along in the Fenghe River, China. The relationship between bacterial diversity and environmental parameters was statistically analyzed. An average of 1682 operational taxonomic units was obtained. Microbial diversity increased from the headstream to downstream and tended to be greater in sediment compared with water. The water samples near the headstream endured relatively low Shannon and Chao1 indices. These diversity indices and the number of observed species in the water and sediment samples increase downstream. The parameters also differ in the two river tributaries. Community structures shift based on the extent of nitrogen pollution variation in the sediment and water samples. The four most dominant genera in the water community were Escherichia, Acinetobacter, Comamonadaceae, and Pseudomonas. In the sediments, the most dominant genera were Stramenopiles, Flavobacterium, Pseudomonas, and Comamonadaceae. The number of ammonia-oxidizing archaea in the headstream water slightly differed from that in the sediment but varied considerably in the downstream sediments. Statistical analysis showed that community variation is correlated with changes in ammonia nitrogen, total nitrogen, and nitrate nitrogen. This study identified different microbial community structures in river water and sediments. Overall this study emphasized the need to elucidate spatial variations in bacterial diversity in water and sediments associated with physicochemical gradients and to show the effects of such variation on waterborne microbial community structures.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
The Fenghe River in Shaanxi Province is a tributary of the Wei-he River (Xu 1990), which originates from the southwest of the Qing-ling Mountains. Along the river, land use varies because of human activities: the headstream of the Fenghe River is minimally exploited, but the downstream of this river is largely influenced by human activities; consequently, these two areas are considerably different in terms of water quality and microbial community structure (Xing et al. 2015).
Because the microbial community reflects and responds to pollution conditions (Buttigieg and Ramette 2014), diversity and community structure vary closely and specifically with water pollution. Natural bacterial communities are highly diverse and dynamic, but community composition follows a predictable temporal pattern (Crump et al. 2009). Microbial communities are characterized on the basis of the phylogenetic type of uncultured community members through sequencing (Ward et al. 1990). The microbial profile of rivers can be elucidated by way of 16S rDNA cloning and through second-generation high-throughput sequencing (Kerstens et al. 2009). This technique is also used to assess bacterial community structure and diversity. Sequencing has also emerged as an efficient tool to examine bacterial communities in various matrices (Staley et al. 2014) including soils, marine waters, and freshwater environments (Ligi et al. 2014). Second-generation high-throughput sequencing is a high-resolution technology used to investigate the RNA population in eukaryotes (Gedalanga et al. 2013). Compared with existing microarray-based techniques, second-generation high-throughput sequencing provides a number of major advantages including enhanced sensitivity, increased dynamic range (Staley et al. 2013), decreased background noise, and improved data mapping precision for genome sequences (Sogin et al. 2006). 16S rDNA sequence data incorporate sequences >1 kb and can be retrieved from published rumen library and public databases. Data are then combined and analyzed to assess the diversity of the rumen microbial ecosystem as indicated by pooled data (Caporaso et al. 2011, 2012; Edwards et al. 2004; Wang et al. 2011).
Interactions between river water and sediments have been investigated with tracer technology (Garneau et al. 2015); nevertheless, the use of microsensors and fine-scale sampling techniques has considerably enhanced spatial resolution, through which water and solid sediment constituents are measured. In our study, these interactions can be confirmed through high-throughput sequencing and statistical analyses.
The effects of water pollution, microbial community structure, and spatial and environmental variations on water in the Fenghe River in Shaanxi Province (China) were analyzed (Leps and Šmilauer 2003). Multidimensional data on community composition, individual population properties, and environmental attributes should be examined, and their multidimensionality should be considered; therefore, CANOCO was employed to correlate environmental data with microbial diversity information (ter Braak 1988). Multivariate statistical analysis was performed to analyze data sets in accordance with the procedure described in the 3.11 release of the CANOCO program (ter Braak 1988).
Materials and Methods
Sample Collection and Analysis
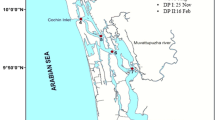
Twenty-four samples were obtained from 12 sampling sites along the Fenghe River in 2014 (Fig. 1). The sites was distributed from the source to the river end of the two tributaries. Water and sediment samples were acquired from each site; water samples were collected from a depth of 0.5 m in the river water, and short sedimentary cores were obtained from the river in November by using a gravity corer with a Plexiglas core tube (150-cm length and 59-mm inner diameter). Afterward, the cores were kept in a vertical position and carefully sectioned in the field. Immediate subsampling can prevent the disturbance of surface sediments during the transportation of the whole-core sediments. As such, all of the sediments were cut into lengths of 1 cm. The subsamples were kept in tightly sealed plastic containers and stored at 4 °C before analysis. The water samples were collected in sterile 20-L carboys, transported to the laboratory, and processed immediately or stored at 15 °C overnight for processing the following day.
Nitrate/nitrogen content was determined through phenol-disulfonic acid spectrophotometry analysis; total phosphorus (TP) was determined using alkaline potassium persulfate digestion-ultraviolet (UV) spectrophotometric method. These methods were performed as defined in “Determination methods for examination of water and wastewater” (Fourth edition) (State Environmental Protection Administration 2002). Ammonia nitrogen (NH4-N) was analyzed with Napierian reagent colorimetric method; nitrite nitrogen (NO2-N) was analyzed with N-(1-naphthyl)-ethylene diamine spectrophotometry; and total nitrogen (TN) was analyzed with alkaline potassium persulfate digestion-UV spectrophotometry (State Environmental Protection Administration 2002).
High-Throughput Sequencing
DNA Extraction and MiSeq Sequencing of 16S rDNA Gene Amplicons
DNA was extracted using an Ezup genomic DNA extraction kit for soil (Sangon Biotech, China, catalog no. SK8264). DNA concentration and quality were determined using a NanoDrop spectrophotometer. The extracted DNA was then diluted to 10 ng/μL and stored at 40 °C for downstream use.
Universal primers 515F (5′GTGCCAGCMGCCGCGGTAA3′) and 909R (5′CCCCGYCAATTCMTTTRAGT3′) with 12 unique barcodes were employed to amplify the V4 hypervariable region of the 16S rDNA gene by sequencing with a Miseq sequencer. Each polymerase chain reaction (PCR) mixture (25 μL) contained 1× PCR buffer, 1.5 mM MgCl2, and 0.4 μm each of deoxynucleoside triphosphates, 1.0 primers, 0.5 U Ex Taq, and 10 ng of soil genomic DNA. The PCR amplification program included an initial denaturation at 94 °C for 3 min followed by 30 cycles of 94 °C for 40 s, 56 °C for 60 s, and 72 °C for 60 s and a final extension at 72 °C for 10 min. Two PCRs were run for each sample, and the two reactions were combined after PCR amplification was performed. The PCR products were subjected to electrophoresis in 1.0 % agarose gel. The band with the correct size was excised and purified using a SanPrep DNA gel extraction kit and quantified with Nanodrop. All of the samples were pooled collectively in equal molar amounts. Sequencing samples were prepared using a TruSeq DNA kit in accordance with the manufacturer’s instructions. The purified library was diluted, denatured, rediluted, and mixed with PhiX (equal to 30 % of the final DNA amount) as described in the protocol of Illumina library preparation. The resulting mixtures were then applied to an Illumina Miseq system and sequenced with a reagent kit v2 2 × 250 bp as described in the manufacturer’s manual.
Sequencing Data Analysis
Sequencing data were processed using QIIME Pipeline Version 1.7.0 (http://qiime.org/). The sequence reads were trimmed and assigned to each sample according to their barcodes. High-quality sequences with an average base quality score >30 and without an ambiguous base “N” were used for downstream analysis. The sequences were then clustered into operational taxonomic units (OTUs) at a 97 % identity threshold. The aligned internal transcribed spacer gene sequences were used to verify the chimera with Uchime algorithm (Edgar et al. 2011). All of the samples were randomly resampled to 4374 reads. Furthermore, calculations were performed on the basis of alpha-diversity (phylogenetic distance whole tree, Chao1 estimator of richness, observed species, and Shannon’s diversity index) and beta-diversity (principal coordinate analysis [PCoA] and UniFrac) analyses. Taxonomy was assigned by using the Ribosomal Database Project classifier (Wang et al. 2007).
Results
Basic Characteristics of Water and Sediment Samples
Elevations in the 12 sampling sites decreased from the upstream to downstream. The location of the “water source” (1846 m) is higher than those of the other areas; as a result, the river water flows by gravity. The temperatures of the W-1, W-2, W-3, W-4, and W-6 samples are lower than those of the other sites. The suspension concentrations and conductivities of these five samples show similar tendencies with temperature (Table 1).
The measured concentration profiles of the 24 individual surface water and sediment species were satisfactorily reproduced. The parameters in both figures vary with changes in the sampling sites. In Fig. 2a, the levels of ammonium nitrogen in samples W-1, W-2, W-3, W-4, W-5, and W-6 are apparently lower than those from the other sites. Nitrate-nitrogen (NO3-N) and TN vary correspondingly in all of the samples. These parameters are lower in W-1, W-2, W-3, W-4, and W-6 than those in the other samples. The trend in nitrogen parameters is similar. Conversely, the pattern of TP is different. W-2 and W-9 exhibit greater TP values than do the other samples. Chemical oxygen demand (COD) also varies from the first to the last samples. W-1, W-2, W-3, W-4, and W-5 samples yield lower CODs than do the other samples. Among the samples, W-11 exhibits the highest biochemical oxygen demand (BOD). Dissolved oxygen (DO) slightly varies in all of the samples as shown in Fig. 2b.
Abundance and Diversity of Microorganisms
With the increasing number of different letters, proportional abundance has been extensively investigated. Accurate prediction of the next letter in the string has been a challenge. Shannon entropy quantifies the uncertainty associated with the prediction. For the 11 upper water samples (sample of W-5 lack), Shannon indices range from 8.70 to 9.89 (mean 9.39). This finding shows that the metabolic diversity of the three types of water samples exhibits similar profiles. In particular, the Shannon indices in W-1, W-2, W-3, W-5, and W-6 sites are the lowest among the samples. These sampling sites are located near the Fenghe River source and are of high altitudes. Sites W-7, W-8, W-9, W-10, W-11, and W-12 show greater Shannon indices because these sites are located in lower altitudes with more human activities in the surroundings compared with the former sites. For the Chao1 index and observed OTU, W-1 exhibits the lowest Chao1 (2844.11) and observed OTUs (1367.9), which clearly separates W-1 from the other water samples. Sites W-4, W-3, and W-6 show relatively lower Chao1 values and OTUs than the other sites. These results are consistent with the findings on Shannon indices.
For the 12 sediment samples listed in Table 2, the Shannon indices range from 9.2 to 10.2 (mean 9.80). Similar to the information obtained from the water samples, these indices show that the metabolic diversity of the 12 types of sediment samples exhibit similar profiles. Samples S-1, S-2, and S-3 are obtained from one upper tributary of the Fenghe River with Shannon indices that continuously increase from 9.41 to 9.85, whereas samples S-5 and S-6 exhibit the lowest values. The Shannon indices of samples S-7, S-8, S-9, S-10, S-11, and S-12 are regularly greater than those of the six former samples. The Chao1 indices of samples S-1, S-2, and S-6 are the lowest. The Chao1 indices and the observed OTUs are consistent with the Shannon indices. Variation tendencies between the two kinds of samples are comparable. Similar to the trend of Chao1 and Shannon indices, diversity indices and observed species increase in the water and sediment samples taken from the headstream to the downstream and differ between the two river tributaries.
Analysis of Microorganism Abundance and Environmental Parameters
The rarefaction curve enables the assessment of differences in species richness among areas with different species-accumulation rates. In Fig. 3, the number of observed species increases as the number of sequences per sample increases. All of the curves in the figure flatten after the sequences per sample reach a relatively large number. The number of observed species for each sample is also indicated by the curves. Meanwhile, the curves from the sediment samples are located above the curves from the water samples, thus showing that the number of observed species is greater in the sediment than in the water samples.
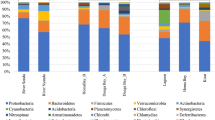
Community structure can be determined from the results of second-generation sequencing (Ligi et al. 2014). The column charts of the structure are shown in Fig. 4, in which genera comprising 4 % of the total sequenced genera are regarded as dominant. As shown in Fig. 4, community structure shifts from the headstream to the downstream. Stramenopiles, Flavobacterium, Pseudomonas, and Comamonadaceae are the four most dominant genera. Stramenopiles (taxonomy ID 33634) are a major line of eukaryotes (Fogg 1996) that currently contains >25,000 known species. Most are algae, ranging from the giant multicellular kelp to the unicellular diatoms, which are the primary components of plankton. Other notable members of the Stramenopiles comprise the generally parasitic oomycetes, including Phytophthora of Irish potato famine infamy and Pythium, which causes seed rot and damping. Flaviobacterium is a genus of motile and nonmotile rod-shaped Gram-negative bacteria with 130 recognized species and three newly proposed species (F. gondwanense, F. salegens, and F. scophthalmum) (Mudarris et al. 1994). Flavobacteria are found in soil and freshwater in various environments. Several species are known to cause diseases in freshwater fish. Pseudomonas is a genus of Gram-negative and aerobic Gammaproteobacteria belonging to the family Pseudomonadaceae and contains 191 validly described species (Euzeby 1997). Comamonadaceae is a family of Betaproteobacteria (Euzeby 1997). Similar to Proteobacteria, Comamonadaceae is Gram negative. These bacteria are aerobic, and most of the species are motile by way of flagella.
Pseudomonas and Comamonadaceae are distributed uniformly in all of the sediment samples, whereas Stramenopiles and Flavobacterium change correspondingly with sampling sites. Flavobacterium in samples S-1 through S-3 are present in large percentages ranging from 10.28 to 21.12 % but are less (<3.52 %) than in the other samples. Stramenopiles constitutes the highest percentage (28 %) in sample S-6 and accounts for 24.07, 12.71, 13.03, and 4.89 % in samples S-5, S-7, S-8, and S-9, respectively. Based on the sample sites and pollution differences shown in Fig. 2, the community structure in the headstream (i.e., samples S-1 through S-3) also differs considerably from that in the downstream (i.e., samples S-10 through S-12). The community structure of the two tributaries varies to a great extent because of nitrogen pollution changes as indicated by the column differences among samples S-6 through S-9 and samples S-1 through S-3 (Fig. 4). Stramenopiles and Flavobacterium clearly respond to regional changes as shown by the representative microbes for the different tributary regions of the Fenghe River.
Variations in stream water geochemistry are correlated with biofilm community composition. In Fig. 5, Escherichia, Acinetobacter, Comamonadaceae, and Pseudomonas are the four most dominant genera detected in the stream water habitats. Escherichia is a genus of Gram-negative, non-spore-forming, facultatively anaerobic, rod-shaped bacteria from the family Enterobacteriaceae (Salmond and Whittenbury 1985). These species are inhabitants of the gastrointestinal tracts of warm-blooded animals, and Escherichia species provide a portion of the microbially derived vitamin K for their host. A number of Escherichia species are pathogenic. Acinetobacter is a genus of Gram-negative bacteria belonging to the wider class of Gammaproteobacteria. Acinetobacter species are oxidase-negative and nonmotile and occur in pairs under magnification. They are important soil organisms because they contribute to the mineralization of aromatic compounds. Comamonadaceae is Gram negative, aerobic, and dominantly motile by way of flagella.
This result differs from the dominant genera obtained from the sediment sample (Fig. 4). The percentages of Escherichia and Acinetobacter are greater in samples W-1 through W-4 and W-6 than those in the other samples, which corresponds to spatial variation and represents the typical domestic species of the river water. Comamonadaceae and Pseudomonas are almost uniformly distributed in the samples and are thus regarded as representative species. The natural community structure gradually changes to accommodate variations in pollution parameters (Newete et al. 2014). This finding reveals that from the headstream to the downstream of the Fenghe River, the quantities of Escherichia and Acinetobacter decrease with the increasing effects of pollution; this phenomenon can be confirmed by the increasing nitrogen content in this part of the river. In general, differences in the community structures of water from the two river tributaries are inconspicuous; conversely, the microbial structure significantly varies on the basis of headstream–downstream analysis. In contrast, analysis of the sediment sample reveals apparent differences between the two tributaries, and these differences could have resulted from the effects of nitrogen pollution. The water microbial composition is influenced by different water types and the surrounding environment, but the bottom sediment slightly affects the water microbial structure.
Ammonia oxidation is the initial and rate-limiting step of nitrification. Ammonia oxidation rates and the abundance of ammonia-oxidizing bacteria have been measured simultaneously, but ammonia oxidation rates have not been fully quantified in conjunction with ammonia-oxidizing archaea (AOA). AOA belongs to archaea and has been identified and widely investigated; several significant questions related to AOA have been raised to promote further studies (You et al. 2009). Studies on archaea often involve sequencing of the amoA gene (Konneke et al. 2005), and the high-throughput sequencing method used in studies provides ample results for archaea communities (You et al. 2009). According to the sequencing results, the most dominant genera in the samples include Candidatus, Nitrososphaerace, Nitrosopumilus, and pGrfC26 (Fig. 6); these four genera belong to the phylum Crenarchaeota, which comprises the widely existent AOA and plays important roles in nitrogen cycling (Dang et al. 2008). Nitrosopumilus is an extremely common archaeon living in seawater and is the first member of group 1a Crenarchaeota to be isolated in pure culture. Gene sequences suggest that the group 1a Crenarchaeota are ubiquitous in the oligotrophic ocean surface and can be found in most noncoastal marine waters around the planet. Interestingly, the relative abundance of archaeal 16S rDNA gene at the genus level in the water and sediment samples differs substantially; the percentage of archaea in sediment samples S-4, S-8, S-9, S-10, and S-11 are >1 %, whereas archaea of the other samples are <1 %. Sediment sample S-11 reveals the highest abundance of pGrfC26, which is an archaeal genus under Crenarchaeota. Nitrososphaerace, which belongs to Crenarchaeota, predominates other species in sediment samples S-8 and S-9, and exhibits good ammonia-oxidizing ability. In general, the variations in the water samples are not as large as those in the sediment samples, which may be attributed to the physical differences between water and sediment.
In the present work, the relative abundance of the archaeal 16S rDNA gene at the genus level differs greatly between the water and sediment samples. In particular, microbial variation within the water samples are not as large as that in the sediment samples. The headstream water and sediment minimally differ in terms of archaeal composition, which in turn differs greatly in the downstream sediment. For the downstream samples, archaeal content in the sediment increases; and samples S-8 through S-11 contain >3.5 % archaea. This observation may have resulted from the variations in the degree of nitrogen pollution downstream as confirmed by the increasing nitrogen content in the water in this part of the river. NH4-N increases after complex biochemical reactions; as a result, the number of AOA increases.
Statistical Analysis
The dissimilarities in microbial composition were analyzed through PCoA based on cosine distance measures (Daniels et al. 2014) and OTU composition. The results show the deviations among the different samples with two axes showing 26.8 and 18.5 % of the total variation in bacterial data (Fig. 8). The figure shows two apparent patterns. River water samples (from W-1 to W-7) accumulate on the upper left side, whereas river sediment samples are slightly scattered on the lower right area, which implies considerable differences between the two kinds of samples. The sediment samples S-1 through S-4 originate from one tributary of the headstream and are positioned away from samples S-6 through S-9, which are derived from the other headstream tributary. The positions of downstream samples S-10 through S-12 in Fig. 7 are distinct from the headstream samples. By comparing the sampling map in Fig. 1 with that in Fig. 7, we find that the geological location of the sampling sites coincides with the coordinate positions. The PCoA also indicates that water samples obtained from the headstream are well separated compared with those from the downstream, but no evidence shows the difference between the two tributaries. In summary, although water and sediment combines and exchanges to some extent, water and sediment are found to be influenced by different parameters. In other words, this research shows that the water and sediments in the Fenghe River do not significantly interact at the microbial level.
Partial mantel test was performed (Table 3) using the whole OTU data and environmental characteristics to analyze the local environmental factors controlling the microbial communities in Fenghe River water. The results show that TN and NH4 are significantly correlated with the community (Mantel r = 0.44917, p < 0.01; Mantel r = 0.69145, p < 0.01), and NO3 is closely related to the community (Mantel r = 0.33727, p < 0.05), indicating that parameters of nitrogen influence aquatic microbes. COD, BOD, and other environmental parameters are not significantly correlated with water microbial community with a Pearson correlation coefficient >0.05. The mantel test results are coherent with the results of the community structures in Figs. 4 and 5.
The large number of sequence data allows the determination of a wide variety of bacterial lineages in sediments and water for reliable statistical analyses. Regularized discriminant analysis (RDA) has been proven as an effective classifier for problems where traditional classifiers fail because of the lack of sufficient training samples, such as in high-dimensional settings. In this study, species data come from the OTU data of the sequencing results, whereas the environmental parameter data come from laboratory tests. The parameters are further separated into site-specific parameters (e.g., elevation, latitude, longitude, river width) and pollution parameters (e.g., NO3-N, NO2-N, and DO). By controlling the parameters of RDA (Aeberhard et al. 1993), we obtained ordination diagrams (axis 1 × axis 2) containing environmental variables and variable samples. The environmental variable arrows point to the expected direction of the steepest increase in the environmental variable values. Each arrow also shows the marginal effect of the particular environmental variable corresponding to the sample scores in the ordination diagram (Braun et al. 1995). In Fig. 8a, the arrows of pH, COD, and BOD form acute angles; this finding indicates that these parameters exhibit a close relationship. In Fig. 8b, the arrows of latitude, river width, and water depth also form acute angles, thus showing their close relationship. For the sample points, the distance between the symbols in the diagram approximates the dissimilarity of their species composition as measured by their Euclidean distance. Considering that the positions of the headstream water samples are close to each other and the downstream samples are combined, we concluded that the community structures of the headstream samples in both figures and those in the downstream samples are relatively similar.
RDA analysis of the interactions and relationships between environmental parameters and microbial communities in the water samples. a DO, TP, pH, TN, temperature, NO3-N, NO2-N, NH4-N, COD, and BOD of the water samples. b Latitude, longitude, elevation, velocity, ORP, width, and depth of the water samples. ORP oxidation–reduction potential
Given the environmental variables and community structure data, we obtained a discriminant connection between sediments and water. For both sample and environmental variables, the sample symbols can be projected perpendicularly to the line overlying the arrow of a particular environmental variable (Solie 1980). The sample points projecting onto the coordinate origin are predicted to correspond to the samples with an average value of environmental variables. DO is concluded as being the only environmental variable with values exceeding the average of those from samples W-1 through W-4, and W-6 (Fig. 8a). In Fig. 8b, W-8, W-8, W-10, and W-11 contain high nitrogen levels. This finding shows the effect of the natural environmental variation on microbial community structure and diversity along the Fenghe River. The variation in nitrogen content is the main factor that influences water and sediment samples.
Discussion
In our study, the sequence of 16S ribosomal DNA was used to characterize bacterial community composition in water and sediment samples from the Fenghe River. Microbial community and diversity in the 12 sampling sites shift from the headstream to the downstream in response to nitrogen pollution and site-specific parameters. The variation tendencies of the two kinds of samples are similar: Microbial diversity increases from the headstream to the downstream, whereas diversity in the sediment is greater than that in the water. Water samples from Guanpingsi, Fengyuk, Xiangyu, Lijiayan, and Wulimiao, which are near the headstream and located at low latitudes, are less affected by human activities with relatively small Shannon and Chao1 indices and OTUs. The diversity indices (Chao1 and Shannon indices), as well as the number of observed species in the water and sediment, increase from the headstream to the downstream and differ between the two river tributaries.
Although some insights suggest interactions between water and sediments, the interaction is not evident in these sequencing results. The four most dominant genera differ between the water and sediment samples. Escherichia, Acinetobacter, Comamonadaceae, and Pseudomonas dominate the water samples, whereas Stramenopiles, Flavobacterium, Pseudomonas, and Comamonadaceae are dominant in the sediment. Although the community structure between the water and the sediment varies considerably, Pseudomonas and Comamonadaceae are universal and are abundantly found in both samples; this may have been caused by the strains of these bacteria, which produce antagonistic substances that restrain the growth of other bacteria and potentially confer them their competitive advantages (Aguirre-von-Wobeser et al. 2015). Stramenopiles and Flavobacterium in the sediments clearly respond to regional changes and may be used as representative microbes for different regions of the Fenghe River. Escherichia and Acinetobacter are the main typical microbes of the original water. The microbial community structure changes from the headstream areas to the downstream areas, which is consistent with the diversity index data. However, differences are found between the headstream and the downstream the water samples, and differences in microbial community structure are found among the two tributaries (Fig. 7). For archaea, the most dominant genera in the water and sediment samples are Candidatus, Nitrososphaera, Nitrosopumilus, and pGrfC26. The number of archaea, especially AOA, increases in the downstream sediment. This observation is attributed to the aggravation of NH4-N in the corresponding area: As the ammonium levels in the environment increase, the quantity of ammonia-transforming archaea, such as AOA, also increases.
The physicochemical properties of water and sediments vary; thus, the microbial community structure changes with environmental variations (Staley et al. 2015). Consequently, the difference between the two river conditions results in obvious discrepancies between the sediment and water samples. Despite the lack of vested data on sediment condition parameters, the figure of PCoA indicates that samples S-1 through S-3 exhibit different physical and chemical properties; this difference can be anticipated as listed in Table 1. The partial mantel test and RDA analysis of the environmental parameters and microbial structure of the samples further confirm the specific influence of TN, ammonia, and NO3-N pollution on the communities in the samples. The samples with high nitrogen pollution are positioned close to one another in the RDA plot, and their community structures respond effectively to nitrogen variables in Fig. 8a. Velocity and elevation exhibit environmental variations; as such, microbial community structure and diversity vary from headstream areas to downstream areas. Variation in nitrogen content, not elevation, is the main factor that influences water and sediment samples.
Conclusions
Using sequencing technology, we provided qualitative information on the composition of bacteria and archaea in the Fenghe River as well as microbial community data in water and sediment samples from the headstream area to the anthropogenic areas. We used the 16S rDNA gene sequences of the river samples obtained according to a taxa cutoff set at 97 % similarity to assess the biodiversity of bacterial communities. By minimizing variations in the site-specific and pollution parameters, we performed analyses at taxonomical levels using some statistical methods. The results showed differences in bacterial assemblages between the water and the sediment samples, and the Fenghe River indices indicated the existence of diverse microbial communities in these regions. Specifically, the sediment samples exhibited greater bacterial diversity and diversity indices (Chao1 and Shannon indices). The number of observed species in the water and sediment samples increased from the headstream areas to the downstream areas, and this finding differs between the two sampling river tributaries. The community structure shifted from headstream to downstream. Stramenopiles, Flavobacterium, Pseudomonas, and Comamonadaceae are the four most dominant genera in sediment. Conversely, Escherichia, Acinetobacter, Comamonadaceae, and Pseudomonas are the four most dominant genera detected in the water habitats. Stramenopiles and Flavobacterium in the sediments clearly respond to regional changes and can be used as he representative microbes of different regions in the Fenghe River. Moreover, Escherichia and Acinetobacter may be the typical microbes of the original water. The microbial community structure not only changes from headstream to downstream areas but also between the two distributaries. For archaea, the most dominant genera in the water and sediment samples are Candidatus, Nitrososphaera, Nitrosopumilus, and pGrfC26; archaeal variations in the water samples are not comparable with those in the sediment. W-1 through W-6 are from highly elevated locations, which affect the microbial community structure and diversity along the Fenghe River. In addition, variations of nitrogen content are possibly the main environmental factor that influences the water and sediment. Finally, we found that the water and sediment do not significantly interact with each other at the microbial level. The microbial structure in the water differed between headstream and downstream samples and thus is influenced by different water-pollution conditions. In the sediment, differences between the two tributaries are evident. However, interactions between river water and sediment should be further investigated at the microbial level. The combination of environmental parameter analyses and second-generation 16S sequencing may provide a good approach.
References
Aeberhard S, Coomans D, Devel O (1993) Improvements to the classification performance of Rda. J Chemometr 7(2):99–115
Aguirre-von-Wobeser E, Eguiarte LE, Souza V, Soberon-Chavez G (2015) Theoretical analysis of the cost of antagonistic activity for aquatic bacteria in oligotrophic environments. Front Microbiol 6:490
Braun BS, Freiden R, Lessnick SL, May WA, Denny C (1995) Identification of target genes for the Ewing’s Sarcoma EWS/FLI fusion protein by representational difference analysis (RDA). Mol Cell Biol 15(8):4623–4630
Buttigieg PL, Ramette A (2014) A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol 90(3):543–550
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al (2011) Global patterns of 16S rDNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516–4522
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al (2012) Ultra-high-throughput microbial community analysis on the IlluminaHiSeq and MiSeqplatforms. ISME J 6:1621–1624
Crump BC, Peterson BJ, Raymond PA et al (2009) Circumpolar synchrony in big river bacterioplankton. Proc Natl Acad Sci USA 106(50):21208–21212
Dang HY, Zhang XX, Sun J, Li TG, Zhang ZN, Yang GP (2008) Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology 154:2084–2095
Daniels L, Budding AE, de Korte N et al (2014) Fecal microbiome analysis as a diagnostic test for diverticulitis. Eur J Clin Microbiol 33(11):1927–1936
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Edwards JE, McEwan NR, Travis AJ, Wallace RJ (2004) 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86(3):263–281
Euzeby JP (1997) List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47(2):590–592
Fogg GE (1996) Algae: An introduction to phycology. In: van den Hoek C, Mann DC, Jahns HJ (eds). Nature 381(6584):660–660
Garneau C, Sauvage S, Probst A, Sánchez-Pérez JM (2015) Modelling of trace metal transfer in a large river under different hydrological conditions (the Garonne River in southwest France). Ecol Model 306:195–204
Gedalanga P, Kotay SM, Sales CM, Butler CS, Goel R, Mahendra S (2013) Novel applications of molecular biological and microscopic tools in environmental engineering. Water Environ Res 85(10):917–950
Kerstens HHD, Crooijmans RPMA, Veenendaal A et al (2009) Large scale single nucleotide polymorphism discovery in unsequenced genomes using second generation high throughput sequencing technology: applied to turkey. BMC Genom 10:479
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437(7058):543–546
Leps J, Šmilauer P (2003) Multivariate analysis of ecological data using CANOCO. Cambridge University Press, New York
Ligi T, Oopkaup K, Truu M et al (2014) Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput 16S rRNA amplicon sequencing. Ecol Eng 72:56–66
Mudarris M, Austin B, Segers P, Vancanneyt M, Hoste B, Bernardet JF (1994) Flavobacterium scophthalmum sp. nov., a pathogen of turbot (Scophthalmus maximus L.). Int J Syst Bacteriol 44(3):447–453
Newete SW, Erasmus BFN, Weiersbye IM, Byrne MJ (2014) The effect of water pollution on biological control of water hyacinth. Biol Control 79:101–109
Salmond GPC, Whittenbury R (1985) Biology of microorganisms, 4th edition—Brock TD, Smith DW, Madigan MT. Nature 314(6006):49–49
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM et al (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci 103:12115–12120
Solie LP (1980) The development of high-performance Rda devices. IEEE Sonic Ultrason Trans 27(3):164–165
Staley C, Unno T, Gould TJ et al (2013) Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115(5):1147–1158
Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ (2014) Bacterial community structure is indicative of chemical inputs in the Upper Mississippi River. Front Microbiol 5:524
Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ (2015) Species sorting and seasonal dynamics primarily shape bacterial communities in the Upper Mississippi River. Sci Total Environ 505:435–445
State Environmental Protection Administration (2002) Determination methods for examination of water and wastewater. China Environmental Science Press, Beijing
Ter Braak CJF (1988) CANOCO—an extension of DECORANA to analyze species-environment relationships. Vegetatio 75(3):159–160
Wang L, Xu PH, Song T, Li HE (2011) Variation tendency of water quality and its causing effect of the Fenghe River. J Northwest Univ (Nat Sci Ed) 3:29
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rDNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Ward DM, Weller R, Bateson MM (1990) 16s rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345(6270):63–65
Xing Y, Si YX, Hong C, Li Y (2015) Multiple factors affect diversity and abundance of ammonia-oxidizing microorganisms in iron mine soil. Arch Environ Contam Toxicol 69(1):20–31
Xu JX (1990) Complex response in adjustment of the Weihe River channel to the construction of the Sanmenxia reservoir. Z Geomorphol 34(2):233–245
You J, Das A, Dolan EM, Hu Z (2009) Ammonia-oxidizing archaea involved in nitrogen removal. Water Res 43(7):1801–1809
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grants Nos. 51178048 and 51378064) and BNU Discretionary Foundation (Grant No. 2014KJJCB22).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, S., Sun, Y., Zhao, X. et al. Sequencing Insights into Microbial Communities in the Water and Sediments of Fenghe River, China. Arch Environ Contam Toxicol 71, 122–132 (2016). https://doi.org/10.1007/s00244-016-0277-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00244-016-0277-5