Abstract
To study the expression profiles of lncRNA and mRNA in the calcium oxalate monohydrate-attached HK-2 cells, and investigate the association between critical lncRNA expression level and renal injury. The HK-2 cells were treated with crystal suspension of calcium oxalate. The effects of calcium oxalate crystals on the growth of HK-2 cells were determined by MTT assay. Total RNA was extracted and the lncRNA and mRNA expression profiles were analyzed by high-throughput transcriptase sequencing platform HiSeq 2500. The profile of identified lncRNAs and mRNAs were verified by real-time PCR and their potential function was analyzed by Gene Ontology database and KEGG signal pathway analysis. Calcium oxalate crystals adhered to the surface of HK-2 cells in few minutes and showed obvious cytotoxicity. RNA seq results showed that there were 25 differentially expressed lncRNAs in HK-2 cells treated with calcium oxalate crystals, of which 9 were up-regulated and 16 were down-regulated. The difference was verified by real-time PCR which showed statistically significant (P < 0.05). Calcium oxalate crystals have a significant effect on lncRNA and mRNA expression in human renal epithelial cells, which may play critical roles in kidney stone-mediated renal injury.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Kidney stone disease, also known as nephrolithiasis or urolithiasis, is one of the oldest diseases known to medicine. Its estimated that 1–15% people suffered kidney stone at some point in their life, and the prevalence and incidence of kidney stone is reported to be increasing all over the world [1, 2]. Kidney stone as a consequence of renal epithelial cell exposure to oxalate and calcium oxalate crystals, can cause blockage of the ureter, blood in the urine, vomiting or painful urination, resulting in damage of the kidneys. Due to the high prevalence and predominance in working-age adults, the kidney stone disease has a substantial impact on the individual and society.
Most common types of human kidney stones (approximately 75%) are calcium oxalate and calcium phosphate, which are calcareous (calcium containing) stones, radio-opaque and non-calcareous stones [3, 4]. There are multiple risk factors (include high urine calcium levels, certain foods, calcium supplements, obesity, some medications, hyperparathyroidism, gout and not drinking enough fluids) reported for stone formation in kidney, and most of them due to combination of genetics and environmental factors [5]. Kidney stone formation is a complex multistep process that includes supersaturation, crystal nucleation, growth, and aggregation [4, 6], and this process partially due to metabolic disorders [7]. However, some evidences indicate that kidney stone formation is a result of nanobacterial disease akin to Helicobacter pylori infection and peptic ulcer disease [8,9,10,11]. In addition, some researchers have long-standing interests in the potential role of macromolecules based on the observation that all human kidney stones consist of a stone matrix [12, 13], which account for 2–3% of their total dry weight and consists of macromolecules generally present in the urine actively participates in the assembly of kidney stones [14].
As details of human stone formation have accumulated, however, the mechanisms of kidney stone formation and growth are not fully understood. In this study, we generated a calcium oxalate monohydrate (COM)-HK-2 cell model, we measured the cytotoxicity of COM to the renal tubular epithelial cells and analyzed its mRNA and lncRNA expression profiles by RNA Seq approach. Our findings provide a new direction on mechanism study of kidney stone formation, which may improve the understanding of kidney stone for urologists, nephrologists and primary care providers.
Materials and methods
Cell culture
The HK-2 cell line obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen Corporation, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 100 U/mL Penicillin G, 100 µg/mL Streptomycin (Sigma Corporation of America, Ronkonkoma, NY, USA).
COM crystal preparation and cell exposure
Commercial calcium oxalate monohydrate (COM) crystals were purchased from SIGMA-ALDRICH (C0350000, Sigma Aldrich, St. Louis, MO, USA), and suspended in DMEM medium at a concentration of 100 µg/mL. Confluent cells treated with 1 ml/ cm2 COM crystals suspension for 24 h at 37 °C were used for total RNA extraction and RNA-sequencing analyses.
MTT assay
Cell proliferation was assessed by MTT assay according to the manufacturer’s protocol (Roche Diagnostics GmbH, Mannheim, Germany). The formed formazan crystals were dissolved by adding 100 µl/well acidic SDS buffer [10% SDS, 0.16% (6 mol/l) HCl and 5% isobutyl alcohol] and incubating overnight in a CO2-free incubator at 37 °C. The optical density (OD) 570 absorption was measured in a microplate reader (Perkin Elmer Victor3 1420 Multilabel Plate Counter, PerkinElmer, Inc., Waltham, MA, USA). Experiments were repeated three times, and data were represented as the mean of five-replicate wells ± standard error.
RNA extraction and sequencing
Total cell RNA was extracted from each individual sample using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc. USA). Ribosomal RNA(rRNA) was removed by RiboGone™ rRNA Removal Kit (TaKaRa, Japan), and rRNA free residue was cleaned up by ethanol precipitation. Purity and quantity of RNA were measured using Nanodrop equipment and SDS gel electrophoresis. 3 µg RNA per sample was used for libraries preparation by NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations. The sequencing libraries were prepared by random fragmentation of cDNA sample, followed by 5′ and 3′ adapter ligation. Adapter-ligated fragments were then PCR amplified and gel purified. For cluster generation, the library was loaded into a flow cell where fragments were captured on a lawn of surface-bound oligos complementary to the library adapters. Each fragment was then amplified into distinct, clonal clusters through bridge amplification. When cluster generation was complete, the libraries were ready for sequencing [15, 16] (performed by Macrogen, lnc., Seoul, Korea, http://www.macrogen.com/) on an Illumina Hiseq 2500 platform. Individual gene expression level was interpreted by fragments per kilobase of transcript per million mapped (FPKM) value. FDR and log2FC was used for differential mRNA transcripts and lncRNAs screening with a screening condition of FDR < 0.05 and |log2FC| > 1.
Real-time PCR
Real-time PCR was performed to validate the expression of identified lncRNA using LightCycler 480II Real-Time PCR System and SYBR® Green PCR Master Mix (TaKaRa, Japan). Each real-time PCR reaction (in 25 µL) involved 12.5 µL 2 × SYBR Green Realtime PCR Master Mix, 1 µL of each primer, 2 µL cDNA and 8.5 µL H2O. The cycling conditions included an initial single cycle (95 °C for 3 min), and followed by 40 cycles (95 °C for 15 s; 57 °C for 15 s; 72 °C for 20 s). All amplifications were followed by dissociation curve analysis of the amplified products. Specific primers were designed using the NCBI, specificities were confirmed with BLAST and gene expression levels were normalized with RPS20 to attain the relative expression by using \({{\text{2}}^{( - \Delta \Delta {C_{\text{t}}})}}\) value methods. The statistical difference in gene expression was analyzed as published previously [17,18,19]. The correlation between the results of RNA-seq and real-time PCR was calculated using correlation test.
KEEG analysis
KEEG (Kyoto Encyclopedia of Genes and Genomes) is a database of signaling pathway that describes gene regulatory networks. In this study, Rich Factor and Q value were used to perform the figures. Rich factor refers to the ratio of the number of genes located in the pathway entry to the total number of genes located in the pathway entry in the gene, and the higher the Rich Factor, the higher the degree of enrichment. Q value is the P value after multiple hypothesis verification, in the range 0–1, P < 0.05, indicating significant enrichment.
Data analysis
SPSS17.0 software was used for statistical analysis, quantitative data were analyzed by one-way ANOVA, and the variance was analyzed by rank sum test. The difference between the samples was analyzed by LSD method. The results were expressed as mean ± standard deviation. P < 0.05 for the difference was defined as statistically significant.
Results
COM treatment significantly inhibit the growth of HK-2 cells
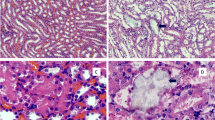
Human proximal tubule epithelial cells HK-2 was treated with calcium oxalate crystal suspension at concentration of 100 µg/mL. Results showed that calcium oxalate crystals attached to the surface of HK-2 cell in a few minutes (Fig. 1a), and obviously exhibited cytotoxicity to the HK-2 cells, which could significantly inhibit cell growth of HK-2 cells (Fig. 1b) and cause cell morphology changes.
Calcium oxalate monohydrate could significantly inhibit HK-2 cell proliferation. a Representative images showed the adhesion of COM crystals to HK-2 cells (indicated by red arrows). b Cell proliferation assay by MTT. HK-2 cells were seeded equally at the density of 5 × 103 per well in 96-well plates and cultured with 100 µg/mL COM suspension or DMEM medium in the incubator. After 1–4 days treatment, viable cells were determined by incubating with 0.5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide 100 µL per well in phenol-red free culture medium for 4 h in the incubator, the formed formazan dyes were dissolved by acid SDS buffer and the OD570 absorption was measured in a micro plate reader. Result showed that HK-2 cells treated with COM crystals suspension proliferated at slower rates than the controls. *P < 0.05; **P < 0.01
Alteration of mRNA expression profiles in COM-treated HK-2 cells
We surveyed the mRNA expression profiling of COM-treated HK-2 cells compared to un-treated control by high-throughput transcriptase sequencing platform HiSeq 2500, and found that there were totally 25 mRNA expression significantly changed in HK-2 cells after the COM treatment, including 11 up-regulated and 14 down-regulated mRNA (Fig. 2). However, only 9 of them were protein-coding genes and the other 16 of these genes were pseudogenes (Tables 1, 2). To validate the RNA-seq results, we performed real-time PCR to verify the expression level of identified mRNA in the COM-treated HK-2 cells. The results showed that expression patterns of these mRNA were consistent with the RNA-Seq findings.
Differential expressed mRNA cluster analysis. a Cluster analysis. A hierarchical heat map showing the transformed expression values for differential expressed mRNA transcripts. Red shows higher expression and blue shows lower expression. b Differential expressed genes statistics. FDR and log2FC was used for differential mRNA transcripts screening with a screening condition of FDR < 0.05 and |log2FC| > 1. Result showed that 14 mRNA transcripts were down-regulated and 11 mRNA transcripts were up-regulated in COM crystal treated HK-2 cells
Alteration of lncRNA expression profiles in COM-treated HK-2 cells
We analyzed the lncRNA expressions in COM-treated HK-2 cells, and found that there were 25 lncRNAs exhibited different expression pattern in COM-treated HK-2 cells, among which 9 were up-regulated and 16 were down-regulated (Fig. 3; Tables 3, 4) as compared to un-treated control group.
Differential expressed lncRNA analysis. a Scatter plot showed the differential and non-differential expression lncRNAs. b Differential expressed lncRNA statistics. FDR and log2FC was used for differential lncRNAs screening with a screening condition of FDR < 0.05 and |log2FC| > 1. Result showed that 9 lncRNAs were up-regulated and 16 lncRNAs were down-regulated in COM crystal suspension treated HK-2 cells
To validate the data obtained from RNA Seq, we used real-time PCR to verify the expression pattern of identified mRNA and lncRNA in COM-treated HK-2 cells. Results showed that the identified mRNA and lncRNA were exhibited significant differences between the COM-treated group and controls.
KEEG analysis of deregulated lncRNAs and mRNAs
To further predict the functional role of the candidate lncRNAs and mRNAs, we performed Gene Ontology (GO) analysis and KEEG analysis to study the significant signaling pathways in COM-treated HK-2 cells. Interestingly, most of the derogated genes involved in COM-treated HK-2 cells were infectious diseases. GO terms with highest number of differentially expressed genes (DGEs) were related to “infectious disease”, “immune system” and “signal transduction” (Fig. 5a). Furthermore, significantly enrichment GO terms (P value < 0.05) were mainly including “Pertussis”, “Hepatitis B” and “Toll-like receptor signaling pathway” (Fig. 5b). These results suggested important roles these genes involved in human health and disease regulations, and some of them might related to kidney stone formation.
Discussion
The most favorable theory for human stone disease is that oxalate-induced injury to renal tubular epithelial cells promotes adherence of COM crystals accumulation and result in kidney stone formation [20, 21]. Accumulating evidences demonstrated that calcium oxalate stimulation result in series consequences, including gene expression profiling changes and cell dysfunction [22, 23]. However, the mechanism of this multi-factorial disorder disease is still not fully understood.
As modern techniques developing, new opportunities are arising. A large number of high throughput platforms have been performed to analyze the miRNA expressions in kidney stone formation [24, 25]. However, previous studies mainly focus on DNA, mRNA and protein levels using microarray or proteomics [22, 24, 26]. Here, we generated a COM-HK-2 cell model and analyzed the mRNA and lncRNA expression profiles by high-throughput transcriptase sequencing platform HiSeq 2500. RNA-seq has accelerated the discovery and characterization of lncRNAs, a new class of biologically significant RNA transcripts. To our knowledge, lncRNAs in COM stimulated proximal renal tubular cells is little known at present.
In this study, we identified several mRNAs and lncRNAs which were involved in signal transduction, signaling molecules and interaction, and infectious diseases. The results were validated by real-time PCR. 14 mRNAs (including TICAM2, DNAJC25-GNG10, VPS4A, ARHGEF18 and BORCS7) exhibited downregulation and 11 mRNAs (including FOSB, IL6, ZNF709 and POLR212) showed up-regulation in COM crystal treated HK-2 cells (Figs. 2, 3, 4). According to the KEGG pathway annotation, most of these genes were related to infectious diseases, immune system and signal transduction (Fig. 5). For example, TICAM2 is related to primary bacterial infectious disease and mycobacterium tuberculosis 1, and involved in innate immune system and regulated by c-FLIP [27]; VPS4A plays a critical role in necroptosis and vesicle-mediated transport [28]; FOSB encodes a leucine zipper protein which can dimerize with proteins of JUN family, related to Toll-like receptor signaling pathway and ERK signing, suggesting that COM crystals may activate immune system and ERK signaling through upregulation of FOSB expression [29], suggesting that these genes might perform functions on COM-induced immune response and signaling transduction. The mechanisms and signaling pathways involved in the kidney stone formation are still largely unclear, our studies provide an insight on the signaling pathways regulated in COM adhesion progression.
Validation of the differential expressed lncRNAs by quantitative real-time PCR. RNA samples from COM-treated HK-2 cells and control used for RNA-sequencing were validated by quantitative real-time PCR. a lncRNAs up-regulated in COM-treated HK-2 cells. b lncRNAs down-regulated in COM-treated HK-2 cells. Results showed that all the differential expressed lncRNAs exhibited consistent expression pattern by RNA Sequencing and quantitative real-time PCR analysis. *P < 0.05; **P < 0.01
KEGG annotation and pathway enrichment for neighbor gene functions of predicated lncRNAs and mRNA. a Pathways of identified differentially expressed genes involved in metabolism and human diseases were analyzed. The number ahead the column means the number of differentially expressed genes in this term. b Top 20 pathway enrichment. P value indicated the significance of the enrichment of individual pathway. P < 0.05 for the difference was defined as statistically significant. Results showed that “Pertussis” and “Toll-like receptor signaling pathway” were the most significant pathways during the COM treatment in HK-2 cells. GO terms with highest number of differentially expressed genes (DGEs) were related to “infectious disease”, “immune system” and “signal transduction”. These results suggested important roles these genes involved in human health and disease regulations, and some of which might relate to kidney stone formation
Moreover, we also demonstrated that 9 lncRNAs showed up-regulation and 16 lncRNAs down-regulated in the COM crystal-treated HK-2 cells. LINC00339 (also known as HSPC157) exhibited the highest up-regulation level, which located on chromosome 1 and associated with endometriosis of ovary, potentially had a strong effect on WNT4 and CDC42 gene expression [30, 31]. NIFK-AS1, which is the antisense RNA of NIFK gene, showed the maximum downregulation level in COM crystal-treated HK-2 cells. The NIFK gene, which encodes a protein may play role in mitosis and cell-cycle progression, can promote cancer progression via CK1α/β-catenin in metastasis and Ki-67 dependent cell proliferation [32]. Multiple evidences suggest that the expression of lncRNAs can regulate and have high correlation with the expression of neighboring mRNAs [33, 34]. These data indicating that COM crystals decrease cell proliferation of HK-2 cells (Fig. 1) might through NIFK, WNT4 and CDC42 regulation by lncRNAs. However, these predicted functions of identified lncRNAs require experimental verification.
In conclusion, we first generated the expression profile of mRNA and lncRNA in COM crystal-treated HK-2 cells based on transcriptome RNA-seq approach. 25 mRNAs and lncRNAs were identified and validated by real-time PCR. Bioinformatic analysis demonstrated that some of the identified genes are involved in infectious system and signal transduction processes, suggesting which may be associated with kidney stone formation. As the mechanism of kidney stone formation has not yet been understood, our study should provide valuable resource for further studies.
References
Romero V, Akpinar H, Assimos DG (2010) Kidney stones: a global picture of prevalence, incidence, and associated risk factors. Rev Urol 12(2–3):e86-96
Morgan MS, Pearle MS (2016) Medical management of renal stones. BMJ 352:i52. https://doi.org/10.1136/bmj.i52
Parmar MS (2004) Kidney stones. BMJ 328(7453):1420–1424. https://doi.org/10.1136/bmj.328.7453.1420
Aggarwal KP, Narula S, Kakkar M, Tandon C (2013) Nephrolithiasis: molecular mechanism of renal stone formation and the critical role played by modulators. Biomed Res Int 2013:292953. https://doi.org/10.1155/2013/292953
Grases F, Costa-Bauza A (2016) Simplified methods for the evaluation of the risk of forming renal stones and the follow-up of stone-forming propensity during the preventive treatment of stone-formation. Urolithiasis 44(1):77–82. https://doi.org/10.1007/s00240-015-0838-5
Hamamoto S, Taguchi K, Fujii Y (2011) Molecular mechanism of renal stone formation. Clin Calcium 21(10):1481–1487
Domingos F, Serra A (2011) Nephrolithiasis is associated with an increased prevalence of cardiovascular disease. Nephrol Dial Transplant 26(3):864–868. https://doi.org/10.1093/ndt/gfq501
Ciftcioglu N, Bjorklund M, Kuorikoski K, Bergstrom K, Kajander EO (1999) Nanobacteria: an infectious cause for kidney stone formation. Kidney Int 56(5):1893–1898. https://doi.org/10.1046/j.1523-1755.1999.00755.x
Liu GL, Zheng XY, Xie LP (2010) Could the urolithiasis be treated by anti-nanobacterial therapy? Med Hypotheses 74(4):755. https://doi.org/10.1016/j.mehy.2009.11.025
Agababov RM, Abashina TN, Suzina NE, Vainshtein MB, Schwartsburd PM (2007) Link between the early calcium deposition in placenta and nanobacterial-like infection. J Biosci 32(6):1163–1168
Silay MS, Miroglu C (2007) The risk of urolithiasis recurrence may be reduced with anti-nanobacterial therapy. Med Hypotheses 68(6):1348–1350. https://doi.org/10.1016/j.mehy.2006.10.027
Khan SR, Joshi S, Wang W, Peck AB (2014) Regulation of macromolecular modulators of urinary stone formation by reactive oxygen species: transcriptional study in an animal model of hyperoxaluria. Am J Physiol Renal Physiol 306(11):F1285-1295. https://doi.org/10.1152/ajprenal.00057.2014
Khan SR, Kok DJ (2004) Modulators of urinary stone formation. Front Biosci 9:1450–1482
Boyce WH (1968) Organic matrix of human urinary concretions. Am J Med 45(5):673–683
Maina S, Edwards OR, Barbetti MJ, de Almeida L, Ximenes A, Jones RA (2016) Deep sequencing reveals the complete genome sequence of sweet potato virus G from East Timor. Genome Announc 4(5):e00957–16. https://doi.org/10.1128/genomeA.00957-16
Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, Kim YT, Kim JI, Kang JH, Seo JS (2012) A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 22(3):436–445. https://doi.org/10.1101/gr.133645.111
Xu L, Wang Z, Li XF, He X, Guan LL, Tuo JL, Wang Y, Luo Y, Zhong HL, Qiu SP, Cao KY (2013) Screening and identification of significant genes related to tumor metastasis and PSMA in prostate cancer using microarray analysis. Oncol Rep 30(4):1920–1928. https://doi.org/10.3892/or.2013.2656
Xu L, Wang Z, He SY, Zhang SF, Luo HJ, Zhou K, Li XF, Qiu SP, Cao KY (2016) Bax-interacting factor-1 inhibits cell proliferation and promotes apoptosis in prostate cancer cells. Oncol Rep 36(6):3513–3521. https://doi.org/10.3892/or.2016.5172
Wang Z, Wang Y, Zhang J, Hu Q, Zhi F, Zhang S, Mao D, Zhang Y, Liang H (2017) Significance of the TMPRSS2:ERG gene fusion in prostate cancer. Mol Med Rep 16(4):5450–5458. https://doi.org/10.3892/mmr.2017.7281
Al-Atar U, Bokov AA, Marshall D, Teichman JMH, Gates BD, Ye ZG, Branda NR (2010) Mechanism of calcium oxalate monohydrate kidney stones formation: layered spherulitic growth. Chem Mater 22(4):1318–1329. https://doi.org/10.1021/cm901751g
Evan AP, Worcester EM, Coe FL, Williams J, Lingeman JE (2015) Mechanisms of human kidney stone formation. Urolithiasis 43(1):S19-S32. https://doi.org/10.1007/s00240-014-0701-0
Liu Z, Jiang H, Yang J, Wang T, Ding Y, Liu J, Wang S, Ye Z (2016) Analysis of altered microRNA expression profiles in the kidney tissues of ethylene glycol-induced hyperoxaluric rats. Mol Med Rep 14(5):4650–4658. https://doi.org/10.3892/mmr.2016.5833
Koul HK, Menon M, Chaturvedi LS, Koul S, Sekhon A, Bhandari A, Huang M (2002) COM crystals activate the p38 mitogen-activated protein kinase signal transduction pathway in renal epithelial cells. J Biol Chem 277(39):36845–36852. https://doi.org/10.1074/jbc.M200832200
Wang B, Wu B, Liu J, Yao W, Xia D, Li L, Chen Z, Ye Z, Yu X (2014) Analysis of altered microRNA expression profiles in proximal renal tubular cells in response to calcium oxalate monohydrate crystal adhesion: implications for kidney stone disease. PLoS One 9(7):e101306. https://doi.org/10.1371/journal.pone.0101306
Koul S, Khandrika L, Meacham RB, Koul HK (2012) Genome wide analysis of differentially expressed genes in HK-2 cells, a line of human kidney epithelial cells in response to oxalate. PLoS One 7(9):e43886. https://doi.org/10.1371/journal.pone.0043886
Chen S, Gao X, Sun Y, Xu C, Wang L, Zhou T (2010) Analysis of HK-2 cells exposed to oxalate and calcium oxalate crystals: proteomic insights into the molecular mechanisms of renal injury and stone formation. Urol Res 38(1):7–15. https://doi.org/10.1007/s00240-009-0226-0
Funami K, Matsumoto M, Oshiumi H, Inagaki F, Seya T (2017) Functional interfaces between TICAM-2/TRAM and TICAM-1/TRIF in TLR4 signaling. Biochem Soc Trans 45(4):929–935. https://doi.org/10.1042/BST20160259
Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, Konig J, Larios J, Poser I, Muller-Reichert T, Scheuring S, Roux A, Gerlich DW (2017) Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 19(7):787–798. https://doi.org/10.1038/ncb3559
Gajewski PA, Turecki G, Robison AJ (2016) Differential expression of FosB proteins and potential target genes in select brain regions of addiction and depression patients. PLoS One 11(8):e0160355. https://doi.org/10.1371/journal.pone.0160355
Albertsen HM, Chettier R, Farrington P, Ward K (2013) Genome-wide association study link novel loci to endometriosis. PLoS One 8(3):e58257. https://doi.org/10.1371/journal.pone.0058257
Powell JE, Fung JN, Shakhbazov K, Sapkota Y, Cloonan N, Hemani G, Hillman KM, Kaufmann S, Luong HT, Bowdler L, Painter JN, Holdsworth-Carson SJ, Visscher PM, Dinger ME, Healey M, Nyholt DR, French JD, Edwards SL, Rogers PA, Montgomery GW (2016) Endometriosis risk alleles at 1p36.12 act through inverse regulation of CDC42 and LINC00339. Hum Mol Genet 25(22):5046–5058. https://doi.org/10.1093/hmg/ddw320
Lin TC, Su CY, Wu PY, Lai TC, Pan WA, Jan YH, Chang YC, Yeh CT, Chen CL, Ger LP, Chang HT, Yang CJ, Huang MS, Liu YP, Lin YF, Shyy JY, Tsai MD, Hsiao M (2016) The nucleolar protein NIFK promotes cancer progression via CK1alpha/beta-catenin in metastasis and Ki-67-dependent cell proliferation. Elife. https://doi.org/10.7554/eLife.11288
Wang Y, Xue S, Liu X, Liu H, Hu T, Qiu X, Zhang J, Lei M (2016) Analyses of long non-coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci Rep 6:20238. https://doi.org/10.1038/srep20238
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25(18):1915–1927. https://doi.org/10.1101/gad.17446611
Acknowledgements
The authors thank Dr. Deng Qiong for the suggestions of revising the manuscript; Dr. Zhi Fan and Dr. Sun Rui for their statistical analysis provided.
Funding
This study was financially supported by Shenzhen Science and Technology Program (Nos. JCYJ20150402144905865 and JCYJ20170307141840188); Longhua Science and Technology Innovation Fund (Nos. 20150925A0410013 and 20170919A1030346).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Z., Zhang, Jw., Zhang, Y. et al. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing in calcium oxalate monohydrate-stimulated renal tubular epithelial cells. Urolithiasis 47, 225–234 (2019). https://doi.org/10.1007/s00240-018-1065-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00240-018-1065-7