Abstract
The evolutionary history of the two structural and functional domains of tRNA is controversial but harbors the secrets of early translation and the genetic code. To explore the origin and evolution of tRNA, we reconstructed phylogenetic trees directly from molecular structure. Forty-two structural characters describing the geometry of 571 tRNAs and three statistical parameters describing thermodynamic and mechanical features of molecules quantitatively were used to derive phylogenetic trees of molecules and molecular substructures. Trees of molecules failed to group tRNA according to amino acid specificity and did not reveal the tripartite nature of life, probably due to loss of phylogenetic signal or because tRNA diversification predated organismal diversification. Trees of substructures derived from both structural and statistical characters support the origin of tRNA in the acceptor arm and the hypothesis that the top half domain composed of acceptor and pseudouridine (TΨC) arms is more ancient than the bottom half domain composed of dihydrouridine (DHU) and anticodon arms. This constitutes the cornerstone of the genomic tag hypothesis that postulates tRNAs were ancient telomeres in the RNA world. The trees of substructures suggest a model for the evolution of the major functional and structural components of tRNA. In this model, short RNA hairpins with stems homologous to the acceptor arm of present day tRNAs were extended with regions homologous to TΨC and anticodon arms. The DHU arm was then incorporated into the resulting three-stemmed structure to form a proto-cloverleaf structure. The variable region was the last structural addition to the molecular repertoire of evolving tRNA substructures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The two structural and functional domains of a typical tRNA, the “top half” containing the acceptor (Acc) and the pseudouridine (TΨC) arms and the “bottom half” containing the dihydrouridine (DHU) and anticodon (AC) arms, may have had different evolutionary histories. In addition, the evolutionary relevance of the variable (Var) region that spans the AC and TΨC arms is unknown. Competing and divergent hypotheses exist regarding which domain is more ancestral (primitive). The primordial AC arm adaptor that embodies the classic genetic code may be more ancient than structures in the top half, because amino acids may have served as cofactors establishing a stereochemical relation with the anticodon or codon in the primordial stage of life (Szathmáry 1999). In contrast, the first tRNA structures may have been “genomic tags” that marked the 3′ ends of ancient RNA genomes for replication by RNA enzymes in the RNA world (Weiner and Maizels 1987). These tags in their simplest form could have been hairpin structures involving a coaxial stack of the Acc and TΨC arms (Maizels and Weiner 1994). The genome tag hypothesis therefore considers the top half of modern tRNA as being the ancient structural and functional domain, while the bottom half arose later to provide additional specificity necessary for codon recognition in mRNA. This idea is consistent with a proposal that the Acc arm precedes evolutionarily the AC arm because AC recognition by aminoacyl-tRNA synthetases (aaRSs) is occasional and idiosyncratic and involves highly variable protein domains (Schimmel et al. 1993). In fact, the Acc arm and the top half harbor roles in almost all macromolecular interactions that involve tRNA (Maizels and Weiner 1994; Schimmel et al. 1993; Schimmel and Ribas de Pouplana 1995), being recognized by aaRSs, RNase P, elongation factor Tu, and rRNA. This is in contrast with the bottom half. In particular, the interaction of aaRSs with the Acc arm drives the highly specific aminoacylation of isoacceptor tRNA with cognate amino acids and defines an operational code probably older than the classic code (Schimmel et al. 1993). The operational code depends fundamentally on the identity of specific nucleotides in the Acc stem and mimics the classic code embedded in the AC arm (Rodin et al. 1993, 1996; Rodin and Rodin 2006a). This establishes a dual complementarity in which pairs of consensus tRNAs with complementary anticodons exhibit bases at the second position of the acceptor stem that are also complementary, and suggests a common origin of Acc and AC arms, a link between operational and classic codes, and a primordial double strand (coding) function in a strand-symmetric RNA world (Rodin and Rodin 2006b). Under this scenario, the Acc arm and the operational code could have established earlier than the AC arm and the classic code, while both codes were being shaped during co-option of the first putative ribozymic aaRSs. However, the early history of the genetic code could have been independent from the evolution of the structural tRNA scaffold. Moreover, the operational and classic codes do not make sense without each other (Rodin and Rodin 2006b), and consequently, a possible “AC first” origin of the genetic code may still be valid (Szathmáry 1999).
We here explore the origin and evolution of tRNA using a novel cladistic approach (Caetano-Anollés 2002a, b) that embeds structure directly into phylogenetic analysis (Pollock 2003). Structural attributes that describe the geometry or the molecular mechanics of RNA molecules were defined as phylogenetic “characters” (and the numerical values they display, as “character states”) and these were used to generate phylogenetic trees, graph representations of evolutionary history that in our case describe the evolutionary relationship among RNA molecules. These characters carry similar phylogenetic signatures and produce phylogenies that are congruent, are similarly rooted, and uncover clear phylogenetic patterns at levels above primary sequence (Caetano-Anollés 2002b, 2005). The general approach has been used to reconstruct a tree of life from rRNA (Caetano-Anollés 2002a), trace the evolution of RNA structures in ribosomes (Caetano-Anollés 2002b), establish deep phylogenetic relationships in Poaceae (Caetano-Anollés 2005), and study the evolution of retrotransposable elements (Sun et al. 2007). Because biological evolution is supported not only by concepts of patterns (e.g., historical reconstruction, tree of life) but also by concepts of processes (e.g., natural selection, molecular drivers of change), phylogenies have the potential to trace the transformation of heritable features that are characteristic of molecular structures and treat these features as molecular phenotypes. We here study these two intimately linked aspects of evolution (patterns and processes), focusing on the tRNA molecule. In doing so, we introduce a new phylogenetic method that produces not only trees of molecules (and, hence, organisms harboring those molecules) but also trees of molecular substructures (Fig. 1). This involves defining new kinds of taxa (substructures) and characters (substructures in molecules), and a criterion of primary homology pertaining substructural repertoires within molecular lineages. We then use these phylogenies to uncover phylogenetic signals embedded in the structure of tRNA and make inferences about the evolutionary origin and history of this ancient molecule.
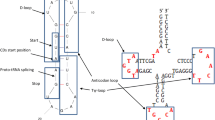
Construction of phylogenetic trees of molecules and substructures. The structure of an RNA molecule (illustrated with tRNAs showing major structural and functional domains and labeled M1, M2, M3, and M4) can be decomposed into substructures (labeled S1, S2, S3, and S4), such as coaxial stem tracts and unpaired regions, that can be studied using features (characters) that describe their geometry (e.g., length of stems [S], hairpin loops [H], bulges and interior loops [B] or unpaired regions [U], or the number of unique molecular features such as modified bases [M] or weak G:U base pairs [W]) or branching, stability and uniqueness (e.g., using morphospace parameters Q, P, and S; see Materials and Methods). These ‘shape’ and ‘statistical’ characters are coded and assigned “character states” (in alphanumeric format) according to an evolutionary model that polarizes character transformation toward an increase in molecular order (character argumentation). Coded characters are arranged in data matrices and subjected to cladistic analyses, generating phylogenies of molecules and substructures. Rooted trees can be used to generate evolutionary heat maps of secondary structure that color two-dimensional structural models of RNA with molecular ancestries
Materials and Methods
Data
The entire set of 571 tRNA sequences with cloverleaf secondary structures was retrieved from Part 2 (Compilation of tRNA Sequences) of the Bayreuth tRNA Database (http://www.uni-bayreuth.de/departments/biochemie/trna; September 2004 edition; see Supplementary Table S1). The structures were derived by comparative analysis using an alignment that is most compatible with the tRNA phylogeny and the three-dimensional (3D) models of tRNA structure that are known (Sprinzl and Vassilenko 2005; Steinberg et al. 1993). While sequences in this database generally predate the availability of complete genome sequences, Part 2 of the database provides a curated set of 571 sequences obtained at the RNA level, each of which contains information on base modifications. This information cannot be found in other databases and represents an important determinant of tRNA structure. There were no technical or methodological assumptions used to select the sequences; the entire set was chosen without any pruning exercise, as it represents the most complete dataset at RNA level currently available.
Computational Approach
The origin and diversification of tRNA molecules and their substructural components were studied directly at the secondary structure level following the three steps outlined in Fig. 1.
-
1.
Character coding. A set of character attributes was used to describe molecular structures that were inferred from nucleic acid sequence by comparative sequence analysis, comparison with crystallographic models, and other criteria. Two different kinds of attributes were used to characterize tRNA structure numerically, “shape” and “statistical” characters. Shape characters describe the geometry of the molecules by measuring, for example, the length in nucleotides of each spatial component of secondary structure (e.g., double helical stems, hairpin loops, bulges). In this study we did not focus on unusual base-pairings or noncovalent interactions important for the tertiary structure of the tRNA molecule, since there are not enough crystallographic models available for comparative analysis of these interactions. Statistical characters describe the branching, stability, and plasticity (uniqueness) of the molecules. These characters take advantage of molecular mechanic aspects of tRNA, such as molecular ensembles that measure conflicting molecular interactions in tRNA folding. To make characters useful, they were appropriately coded so that they could provide maximum phylogenetic signal.
-
2.
Character argumentation. The structural characters described here transform from one character state to another through time and along the branches of the trees and do so following a model of character transformation (a “transformation series”) of linearly ordered and reversible pathways of character state change. We impose, however, an evolutionary direction to transformation by identifying the ancestral state in the pathway. This ‘polarization’ of character state transformation is driven by an evolutionary search of structural order that is supported by statistical mechanics, thermodynamic, and phylogenetic arguments.
-
3.
Phylogenetic tree reconstruction. In this last step, hypotheses about character states and models of character evolution were transformed into hypotheses about evolutionary relationship of groups of molecules (trees of molecules) or groups of substructures (trees of substructures) using methods that minimize evolutionary change, such as maximum parsimony (MP). Data matrices that show how molecules or substructures display character states are used to reconstruct rooted trees in exhaustive, branch-and-bound or heuristic searches of tree space that seek to identify the tree(s) that exhibits the least amount of change.
We illustrate our cladistic analysis as it pertains to shape characters in more detail with an “example run,” a concrete example that shows how we generate a rooted phylogenetic tree describing the evolution of stem substructures from six randomly chosen tRNA molecules (Fig. 2). We first retrieve primary RNA sequence alignments and secondary structures from tRNAs available in the Bayreuth database (Figs. 2A and B). We then score the lengths of paired and unpaired substructures, traveling from the 5′ to the 3′ end of the molecules (Fig. 2B). We then focus on coaxial stems for Acc, DHU, AC, Var, and TΨC regions. For example, character coding for tRNALeu of Phage 4 results in stem lengths of 7, 3, 4, 4, and 5 base pairs, respectively (Fig. 2C). These five digits constitute the first column in a data matrix in nexus format that is used as input text file for equally weighted unconstrained MP analysis (Fig. 2D). This first column represents a phylogenetic character and the numbers character states. The same is done for the other five tRNAs. We then use the input file to generate a rooted phylogenetic tree of stem substructures (Fig. 2E). The nexus file defines the character states of a hypothetical ancestor (under the “ancstates” command) that delimits polarity in character state change. This hypothetical ancestor (anc) has the maximal character state for each character within the tRNA set analyzed and is included in the search for optimal rooted trees with the sole purpose of polarizing character state transformation. No external hypotheses in the form of outgroups are needed to root the trees. These external hypotheses are replaced by the more axiomatic assumption of polarization that is embedded in the model of linearly ordered fully reversible multistate character transformation (see below). We followed similar steps to reconstruct phylogenetic trees of stems and other substructures using complete and partitioned data matrices and both shape and statistical characters.
An “example run” describing the construction of phylogenetic trees of stem substructures. A Aligned primary sequences of five tRNALeu and one tRNASer (Nicotiana rustica). Accession numbers are given under the species name, in parentheses. These sequences were randomly selected from a set of 571 tRNAs describing sequences obtained at the RNA level and the modified bases and gaps are indicated with symbols according to nomenclature in the Bayreuth tRNA Database. Anticodons are underlined in the sequences, and starts and ends of tRNA arms indicated with brackets. Within each tRNA arm, paired bases are marked using “>” and “<” under the sequences. B Complete data matrix derived from the annotated sequence alignment describing the length of specific substructures. The nature of these substructures and corresponding characters is described in supplementary Table S2. C Secondary structure of tRNALeu of phage T4 showing the lengths in base pairs of the five tRNA arms of the molecule. Base pairings are indicated with “=” symbols, and lengths of stems indicated with numbers. D The input file in nexus format used for phylogenetic analysis in PAUP* (Swofford 2002). This text file contains the data matrix, with columns defining characters and rows defining taxa and phylogenetic assumptions. The nexus file defines hypothetical ancestral states with the “ancstates” command under the “begin paup” block. E Rooted phylogenetic tree describing the evolution of stem tRNA substructures. The input file was executed in PAUP* using equally weighted maximum parsimony as the optimality criteria and including the hypothetical ancestor in the search. Symbols used for gaps and modified bases: - , gaps;  , 1-methyladenosine; *, 2-methylthio-N6-isopentenyladenosine; O, 1-methylinosine;
, 1-methyladenosine; *, 2-methylthio-N6-isopentenyladenosine; O, 1-methylinosine;  , N6-(cis-hydroxyisopentenyl)adenosine; B, 2’-O-methylcytidine; ?, 5-methylcytidine;
, N6-(cis-hydroxyisopentenyl)adenosine; B, 2’-O-methylcytidine; ?, 5-methylcytidine;  , 3-methylcytidine; ;, unknown modified guanosine; K, 1-methylguanosine; L, N2-methylguanosine; #, 2’-O-methylguanosine; R, N2,N2-dimethylguanosine; 7, 7-methylguanosine; (, archaeosine; N, unknown modified uridine; 4, 4-thiouridine; !, 5-carboxymethylaminomethyluridine; D, dihydrouridine; P, pseudouridine; ], 1-methylpseudouridine; T, ribosylthymine
, 3-methylcytidine; ;, unknown modified guanosine; K, 1-methylguanosine; L, N2-methylguanosine; #, 2’-O-methylguanosine; R, N2,N2-dimethylguanosine; 7, 7-methylguanosine; (, archaeosine; N, unknown modified uridine; 4, 4-thiouridine; !, 5-carboxymethylaminomethyluridine; D, dihydrouridine; P, pseudouridine; ], 1-methylpseudouridine; T, ribosylthymine
Phylogenetic Characters, Character Coding, and Taxon Selection
Forty-two structural characters were scored (Supplementary Table S2). Character homology was determined by the relative position of substructures in the cloverleaf and coded character states were based on the length (number of bases or base pairs) and number of these substructures. Modified bases were treated as deviations from the cloverleaf model by the Bayreuth database and were not allowed to establish canonical Watson-Crick pairs. Character states were defined in alphanumerical format with numbers from 0 to 9 and letters from A to F. Missing substructures were given the minimum state (0). Each helical stem region was scored as two complementary sequences (5′ and 3′ sides). Fifty-five partitioned data matrices were constructed based on tRNA specificity, taxonomy, or cellular source (Supplementary Table S3). Six additional partitioned data matrices were generated using characters collected from the substructures which function to either stabilize (stems, G:U pairings, and modified bases) or destabilize (bulges, hairpin loops, and other single-stranded regions) tRNAs.
Character and taxa selection may impact phylogenetic reconstruction in different ways. Characters and taxa defining trees of molecules or trees of substructures correspond to substructures in RNA molecules, which are identified to be homologous by topographic correspondence (Caetano-Anollés 2002a). Molecular topography involves mapping substructural features in space in the context of the entire molecule, coding these features, and testing whether encoded features represent true homologies acquired from a common ancestor. In the presence of a model inferred by positional covariance in sequences from a representative group of molecules (in our case confirmed by crystallographic model standards), decisions to code structural homologies are simpler than those used when aligning sequences. In fact, with tRNA this can be done manually. Because most tRNA stems can be described as helical or approximately helical, tRNA structure can be considered an arrangement of helical stems in 3D space. At a secondary structure level, this arrangement can be described well by the six types of tRNA characters that were scored and are listed in Supplementary Table S2 (stems, bulges, weak G:U base pairs, modified bases, hairpin loops, and other unpaired regions). Of these possible characters, stems, weak G:U base pairs, and hairpin loops offer few if any alternatives for coding schemes. Given a secondary structure, hairpin loops will be defined by terminal stems and stems and base pairing within stems will not be affected by the orientation (5′ or 3′ sides) of these substructures as they are scored and coded using the alphanumeric format. In contrast, bulges, modified bases, and other unpaired regions lend themselves to alternative coding schemes that could affect phylogenetic reconstruction. For example, modified bases are distributed on both the 5′ and the 3′ sides along the Acc arm. We could score the modified bases either as one character (i.e., number of modified based on the entire Acc arm; scheme used in this study) or two characters (i.e., number of modified bases on the 5′ side or number of modified based on the 3′ side), and trees of molecules derived from these two different coding strategies could differ. However, in the analysis of other RNA molecules (e.g., Y1 RNA, SRP RNA, and rRNA), we found that different coding schemes involving stems, bulges, and other unpaired regions did not significantly alter the topology of reconstructed trees (Caetano-Anollés 2002a; unpublished data). For example, the 3′ or 5′ sides of bulges and internal loops were coded separately or combined, coaxial stems were split into segments when bulges and internal loops were present and when homology relationships could be identified unambiguously, and unpaired regions of multiloops were coded separately or combined. In all these cases the topology of the trees was generally maintained. We do not expect tRNA molecules would behave any differently.
When considering substructures as taxa, alternative coding schemes of substructures result in alternative hypotheses. In fact, the main conclusions of our studies are drawn from a coding scheme that splits the tRNA molecule into the recognized five major arms of the two structural and functional domains of the molecule. We could have chosen to atomize structure into smaller components, splitting arms into smaller segments and ultimately into the individual nucleotides. However, identifying true homologies by topographic correspondence will be more and more difficult as atomization levels increase, requiring at some point the use of alignment techniques to establish homology relationships. We have therefore chosen to define substructures at a level that diminishes possible conflict in homology determination.
A molecular morphospace defined quantitatively by three statistical parameters, the Shannon entropy of the base-pairing probability matrix (Q), the base-pairing propensity (P), and the mean length of helical stem structures (S) of RNA sequences, was used to depict the degree of conformational order of the molecular components as a point in a 3D order-disorder space (Schultes et al. 1999). In order to describe the statistical properties of the 571 tRNAs, the structures were dissected into five components, the Acc, TΨC, AC, DHU, and Var arms. The unpaired bases along the multiloop were treated as belonging to a hypothetical Acc loop. Q, P, and S were calculated for each of these five structural components and combined to generate a complete data matrix. The tRNAs were folded using RNAfold (http://www.tbi.univie.ac.at/∼ivo/RNA/). Q, P, and S were calculated using the program stoat (V. Knudsen, unpublished) and coded as numbers from 0 to 9 and letters from A to U. The dataset was then partitioned into four subsets categorized by molecules belonging to each of the three superkingdoms of life or viruses/bacteriophages. We also considered possible uncertainties in the secondary structures of mitochondrial and chloroplast tRNAs and either included or excluded these tRNAs from our datasets.
Character Argumentation
Structural features were treated as linearly ordered multistate characters that were polarized by invoking an evolutionary tendency toward molecular order. The choice of linearly ordered characters is appropriate for geometrical and statistical features, as RNA structures change in discrete manner by addition or removal of nucleotides. This causes gradual extension or contraction of geometrical features or changes in gap-recoded continuous-valued statistical parameters. Insertion and deletion (generally rare mutation events when compared to substitutions) are also possible but they are more costly; we consider the cost of insertions and deletions to be proportional to their length. Polarization identifies the ancestral states in the character transformation series and this results in reversible characters that are directional and show asymmetry between gains and losses. The validity of character argumentation and the use of MP has been discussed in detail elsewhere (Caetano-Anollés 2001, 2002a, b, 2005). Operationally, polarization was determined by fixing the direction of character state change using a transformation sequence that distinguishes ancestral states as those thermodynamically more stable. Maximum character states were defined as the ancestral states for stems, modified bases, and G:U base pairs (i.e., structures stabilizing the tRNAs). Minimum states (0) were treated as the ancestral states for bulges, hairpin loops, and other unpaired regions (i.e., structures destabilizing the tRNAs).
Phylogenetic Analysis
All data matrices were analyzed using equally weighted MP as the optimality criterion in PAUP* (Swofford 2002). Note that a more realistic weighting scheme should consider, for example, the evolutionary rates of change in structural features. However, this requires the measurement of evolutionary parameters along individual branches of the tree and the development of an appropriate quantitative model. In the absence of this information, it is most parsimonious and preferable to give equal weight to the relative contribution of each character. The use of MP (the preference of solutions that require the least amount of change) is particularly appropriate and can outperform maximum likelihood (ML) approaches in certain circumstances (Steel and Penny 2000). MP is precisely ML when character changes occur with equal probability but rates vary freely between characters in each branch. This model is useful when there is limited knowledge about underlying mechanisms linking characters to each other (Steel and Penny 2000). Furthermore, the use of large multistep character state spaces decreases the likelihood of revisiting a same character state on the underlying tree, making MP statistically consistent. Depending on the number of taxa in each matrix, MP tree reconstructions were sought using either exhaustive, branch-and-bound, or heuristic search strategies. When the heuristic search strategy was used, 1000 heuristic searches were initiated using random addition starting taxa, with tree bisection reconnection (TBR) branch swapping and the multrees option selected. One shortest tree was saved from each search. Hypothetical ancestors were included in the searches for the most parsimonious trees using the ancstates command. Bootstrap support (BS) values (Felsenstein 1985) were calculated from 105 replicate analyses using “fast” stepwise addition of taxa in PAUP*. Decay analyses (Bremer 1988) were performed using reverse constraints in PAUP*. The g1 statistic of skewed tree length distribution calculated from 104 random parsimony trees was used to assess the amount of nonrandom structure in the data (Hillis and Huelsenbeck 1992). Character reconstruction was implemented in macclade (Maddison and Maddison 2003).
Evolutionary relationships derived from trees of substructures were traced in generic two-dimensional models of tRNA secondary structure that we here call evolutionary heat maps of ancestry. Because reconstructed trees were intrinsically rooted, we established the relative age (ancestry) of each substructure by measuring a distance in nodes from the hypothetical ancestor on a relative 0-1 scale. To do this, we counted the number of nodes in every lineage from the root to the terminals of the tree and divided this number by the maximum number of nodes in a lineage (Caetano-Anollés 2002b). Ancestry values were divided in classes, giving them individual hues in a color scale that was then used to color substructures in a generic cloverleaf secondary structure model of tRNAs.
Results
Phylogenetic Relationships of tRNAs
Phylogenetic analyses of 56 tRNA structural datasets showed that the monophyly of tRNAs belonging to the three superkingdoms or expressing different amino acid specificities was not revealed in most of the parsimonious trees (see Supplementary Tables S1–S3). Figure 3 illustrates general results with representative phylogenies of 17 tRNAAla and 13 Drosophila melanogaster tRNAs derived from structural characters (see Supplementary Tables S4 and S5). In the Ala tree, Archaea and Bacteria each resolved as a monophyletic group, while the Eukarya were paraphyletic. The Drosophila tree revealed the monophyly of tRNASer but failed to identify the monophyletic groupings of either tRNAVal or tRNALys.
Reconstructing phylogenetic trees from tRNA structure. A Strict consensus of four minimal-length trees (109 steps; consistency index [CI] = 0.656, retention index [RI] = 0.752, rescaled consistency index [RC] = 0.531; g1 = −0.751) describing the evolution of tRNAAla. Anticodons are given after the species names. B Strict consensus of three minimal-length trees (166 steps; CI = 0.585, RI = 0.653, RC = 0.413; g1 = −0.827) describing the evolution of tRNAs from Drosophila melanogaster. Anticodons are given after the amino acid names. Trees were derived from equally weighted MP analyses using branch-and-bound searches. Bootstrap values >50% are shown for individual nodes together with decay support indices (in parentheses)
Phylogenetic Relationships of tRNA Substructures
Phylogenetic trees of substructures reconstructed from geometrical characters describing the complete dataset of 571 tRNAs revealed patterns that were traced in evolutionary heat maps of ancestry (Fig. 4). In the tree of stem substructures, the Acc stem was the most basal branch, followed in order by stems of the TΨC, AC, and DHU and Var arms. Since RNA structures are defined by a frustrated conformational interplay of stems and loops, this tree of helical stems defines the fundamental scaffold of structural evolution of the entire molecule. Analyses of hairpin loops placed the hairpin loop of the Var arm at the base of the tree, followed in order by hairpin loops of the TΨC, AC, and DHU arms. Consequently, structural diversification of hairpin loops started to occur once the entire cloverleaf structure had developed. This is appropriately visualized in the corresponding heat maps. Analyses of bulges and internal loops revealed uninformative evolutionary patterns. In contrast, analyses of unpaired segments describing free ends and regions of the multiloop showed that the most ancient unpaired segment was the 5′ free end of the molecule. Unpaired regions spanning TΨC and Acc, DHU and AC, AC and Var, and Acc and DHU, followed in that order. The most derived unpaired substructures were the region spanning Var and TΨC, and the 3′ free end of the molecule that includes the CCA terminus. Finally, analyses of modified bases and weak G:U base pairs showed that base modifications were ancestral, generally more ancient than the use of noncanonical G:U base pairings to stabilize the molecule. Interestingly, the most ancestral base modifications occurred in the multiloop region of the modern tRNA.
Phylogenetic trees of molecular substructures reconstructed from characters describing the geometry of structure in 571 tRNAs. Trees of substructures describe the evolution of stems (S) (4467 steps; CI = 0.961, RI = 0.929; RC = 0.893; g1 = −1.342), hairpin loops (H) (5495 steps; CI = 0.983, RI = 0.976, RC = 0.959; g1 = −1.431), bulges and internal loops (B) (196 steps; CI = 0.933, RI = 0.552, RC = 0.515; g1 = −1.424), unpaired segments describing external segments (free ends) and multiloop regions (U) (2620 steps; CI = 0.889, RI = 0.909, RC = 0.809; g1 = −0.898), and modified bases (M) and noncannonical G:U base pairs (W) in substructures (5915 steps; CI = 0.772, RI = 0.756, RC = 0.584; g1 = −0.641). Only one minimal-length tree was retained in each case using branch-and-bound searches derived from equally weighted MP analyses. Bootstrap values >50% are shown for individual nodes together with decay support indices (in parentheses). Evolutionary heat maps of secondary structure describe inferences of structural evolution derived directly from the trees. The relative scale describes the number of nodes from the hypothetical ancestor at the base of the tree and is unique for trees of S, H, and B substructures, the tree of U substructures, and the tree of M and W substructures, respectively. Question marks indicate unknown evolutionary patterns
Phylogenetic analyses of stem substructures derived from three partitioned datasets of 275 Bacteria (including mitochondria and chloroplast), 220 Eukarya, and 17 virus and bacteriophage tRNAs, respectively, revealed the same topology as that derived from the complete dataset (Fig. 5). Exclusion of mitochondrial and chloroplast tRNA did not alter phylogenetic reconstructions (data not shown). However, the tree of stem substructures derived from the partitioned matrix of 59 archaeal tRNAs showed that the AC stem evolutionarily predated the TΨC stem (Fig. 5).
Phylogenetic trees of molecular substructures showing congruence between trees reconstructed from geometrical and statistical characters for the total dataset of 571 tRNAs and partitioned analyses. Single minimal-length trees of substructures (left column) describing the evolution of stems were recovered from the total dataset (same as the tree in Fig. 3) or from partitioned analyses (133−2121 steps; CI = 0.958–0.980, RI = 0.928–0.966; RC = 0.889–0.947; g1 = −1.164 to −1.392) after branch-and-bound searches. Similarly, single minimal-length trees of substructures (right column) describing the evolution of arms and derived from analyses of statistical features in structure were recovered from the total dataset (43,281 steps; CI = 0.850, RI = 0.654, RC = 0.557; g1 = −1.033) or partitioned analyses (1279–21,049 steps; CI = 0.843–0.903, RI = 0.622–0.791; RC = 0.529–0.715; g1 = −0.659 to −1.173) after branch-and-bound searches. The bacterial partition included mitochondria and chloroplast tRNAs. The viral partition included bacteriophage molecules. Bootstrap values >50% are shown for individual nodes together with decay support indices (in parentheses)
Phylogenetic trees of substructures were also reconstructed from three statistical characters describing features of molecular mechanics (Fig. 5). Trees derived from the complete and partitioned datasets described above revealed the same evolutionary patterns as those derived from geometrical characters.
Discussion
A Phylogenetic Method That Generates Histories of Molecular Evolution Uncovers the Ancestral Nature of the Acc Arm
In this study, we generate histories of structural evolution in RNA molecules. The method we introduce produces phylogenetic trees of molecular substructures. These trees are atypical in that they do not describe the evolution of organismal taxa or molecules embedded in them. Instead they describe the evolution of molecular repertoires of RNA components that are present in the organismal world. Very much as with trees of molecules (Caetano-Anollés 2002a), these trees are derived directly from RNA structure and are intrinsically rooted. Consequently, the ancestral-derived relationships of substructures embedded in the phylogenies can be used to establish which of them are the most ancient (defining a structural origin) and how individual substructures were added (sequentially or in groups) into an evolving RNA molecule (defining a molecular chronology). In a detailed study of tRNA, we used this method to show that the molecule originated in the Acc arm and that tRNA evolved by gradual addition of coaxial stem tracts (Figs. 4 and 5). This result illuminates on the origin and evolution of a molecule that is ancient, ubiquitous, highly diverse (∼50 molecular species in many organisms clustering into amino acid specific groups [e.g. Goodenbour and Pan 2006; Marck et al. 2006]), and fundamental for understanding origins of the genetic code, the protein biosynthetic machinery and life.
Because our phylogenetic method is novel, it is important to discuss some of its major properties.
-
1.
Trees are intrinsically rooted. Phylogenetic reconstruction produces trees that are rooted according to models of character transformation that delimit how individual characters transform from one character state to another along the branches of the trees. In contrast with standard phylogenetic methods, our models incorporate a central hypothesis or axiom that states RNA molecules tend to increase conformational order in evolution (Caetano-Anollés 2001, 2002a). Trees of molecules and substructures are therefore rooted without the need and associated uncertainties of local external hypotheses of relationship, generally taking the form of “outgroup” taxa. As with any phylogenetic method, our analysis rests on the validity of the phylogenetic models that are used. Most importantly, the validity of conclusions about molecular origins depends on the axiomatic component that defines polarization of character transformation. This polarization hypothesis depicts generalized trends in RNA structure that are falsifiable and result in molecules that are less plastic (more unique) but more modular. As such, these trends have been so far supported by a considerable body of evidence: (i) the study of extant and randomized sequences, showing evolution enhances conformational order and diminishes conflicting molecular interactions (known as “frustration”) over that intrinsically acquired by self-organization (Caetano-Anollés 2005; Gultyaev et al. 2002; Higgs 1993; Schultes et al. 1999; Steffens and Digby 1999; Stegger et al. 1984); (ii) experimental verification of a molecular tendency toward order and stability using thermodynamic principles generalized to account for nonequilibrium conditions (Gladyshev and Ershov 1982); (iii) a large body of theoretical evidence that maps the structural repertoire of evolving sequences from energetic and kinetic perspectives (Ancel and Fontana 2000; Higgs 2000; Fontana 2002), with some important predictions confirmed experimentally (Schultes and Bartel 2000) (see discussion below); (iv) phylogenetic congruence between phylogenies generated using geometrical and statistical characters (Caetano-Anollés 2005; this work); and (v) phylogenetic congruence in the reconstruction of rooted trees generated from sequence, structure, and genomic rearrangements at different taxonomical levels (Billoud et al. 2000; Caetano-Anollés 2001, 2002a, b, 2005; Collins et al. 2000; Swain and Taylor 2003). Furthermore, trees rooted with a model that polarizes character transformation in the opposite direction are less parsimonious and are incompatible with accepted organismal classification (e.g., Caetano-Anollés 2002a, 2005). Similarly, congruent reconstructions from RNA structure and orthology and large-scale recombination events in grass genomes support assumptions of polarization in character argumentation (Caetano-Anollés 2005). Note, however, that while the proposed generalized trend in structure appears valid by the criteria outlined above, we do not know what is the nature and stability of selective preferences or constraints acting on primordial tRNA during the early stages of evolution of these molecules. In this regard, increases in redundancy could have driven selection of beneficial traits in molecules during early life, for example, under a phenotypic model in which increasingly adaptive phenotypes evolve (Zhu and Freeland 2006).
-
2.
In the trees, taxa are RNA substructural components. Substructures can be of many kinds, such as structural domains (e.g., arms in tRNA), stems-loops, stems, hairpin loops, internal loops, bulges, unpaired segments in multiloops or external segments, base pairs, and modified bases. To ensure homology, each kind of tRNA substructure was used separately in tree reconstruction (see below). Substructures represent necessarily a finite set defined by the repertoire present in sampled RNAs. This feature is quite unique since traditional phylogenetic and phylogenomic approaches generally deal with taxa that cannot be considered finite. For example, when building organismal phylogenies using morphological or molecular characters, taxa represent species drawn from millions of species that inhabit earth (Bull et al. 1992). The same is true for gene or genome sequences in phylogenomic analyses. In contrast, atoms in molecules are by definition finite and the diversity of an RNA encountered in nature is also finite.
-
3.
The internal nodes of the trees of substructures define relative chronologies of structural diversification. In other words, an internal node delimits the birth of substructures in the context of other substructures in the molecular repertoire. Branches in the trees describe network-like genealogies in which substructural evolution provides the foundation for evolutionary change. Consequently, branch lengths measure character state change at the structural level. An interesting note relates to the putative existence of hard polytomies depicting simultaneous divergence of substructures. This would be the case if duplications or other rearrangements lead to homologous segments in different parts of the molecule. Consequently, absence of hard polytomies in a tree of substructures provides evidence of gradual buildup of substructures in the course of evolution.
-
4.
Trees are built using characters that represent features of structure characteristic of a molecular RNA lineage. These molecular lineages are generally associated with a family of molecules within an organism, defining, for example, a species or individual amino acid and codon specificities in tRNA. Consequently, molecular lineages and associated characters should be relatively independent to each other, satisfying the requirements of character independence in phylogenetic analyses. It is noteworthy that when considering nucleic acid or protein sequences, nucleotide or amino acid sites are related by a process linked to evolution of structure and function that is curbed by epigenetic effects, such as the interaction within and between genes (Felsenstein 1988). Similar arguments could be drawn for morphological characters or features describing higher levels of structural organization. Character independence is therefore difficult to achieve in traditional phylogenetic analyses. In contrast, characters used to build trees of substructures represent features in molecular lineages that are, for the most part, evolving independently from each other.
-
5.
The criterion of primary homology rests on the feature of structure being studied and its associated evolutionary model, and how this feature relates to the substructural taxa analyzed. Features can describe the geometry (e.g., shape characters) or the branching, stability, and plasticity (e.g., statistical characters) of homologous substructural components. Homologous substructures represent those that are of the same kind (e.g., domains, stems, base pairs) and respond to the same evolutionary model defining the character transformation sequences. For example, we reconstruct trees of coaxial stems corresponding to arms in tRNA, separate from trees of hairpin loops or trees of tRNA arms using statistical characters. This is because character change leading to coaxial stem taxa depends on models of character state that are quite different from those governing unpaired segments or Shannon entropy. While taxa (substructures) are implicitly related by a phylogenetic tree describing the evolution of a molecular repertoire and characters (molecules) can be considered relatively free of covariation patterns, and consequently of tendencies of overconfidence in phylogenies, the validity of the reconstruction exercise rests on an adequate sampling of the molecular repertoire. The analyses of 571 tRNAs representing the structures of the entire set of known sequences acquired at the RNA level guarantee an extensive sampling of molecular variants in our study. Note that poor sampling could result in missing substructures and deficient models of molecular evolution.
-
6.
Substructural taxa larger than a nucleotide are necessarily the subject of information compression, generally dependent on how substructures are defined. This could result in information loss and differential weighting of taxa. Their impact, however, decreases if information that is lost constitutes noise and if models are derived from substructures of one kind. A typical information compression example that has a natural rationale (the genetic code) is that of amino acid versus nucleotide sequences. The compression of RNA sequences into structures also has a natural rationale, the mapping of genotype (sequence) into phenotype (structure) delimited by the unique chemistry and folding of RNA biopolymers (Ancel and Fontana 2000; Fontana 2002; Higgs 2000). This mapping has three important properties: (i) the sequence-to-structure map is degenerate, i.e., there are orders of magnitude more sequences than structures; (ii) few common but many rare structures materialize in structure space; and (iii) extensive neutral networks that percolate sequence space define common structures and structural neighborhoods. Because the distribution of sequences that fold into the same structure within neutral networks in RNA is approximately random, the mapping has “space covering” properties. This means that all structures can materialize within relatively few mutational changes in sequence space. This property has been confirmed experimentally using RNA functional switches (Schultes and Bartel 2000). The complexities of sequence-to-structure mapping in tRNA are driven by decreases in frustration and increases in thermodynamic stability of the folding ensemble that ultimately define the cloverleaf structure of the molecule. Based on these considerations, information compression in tRNA is natural and should not bias significantly the phylogenetic reconstruction exercise. Instead, we expect that it will uncover deeper phylogenetic signals that are embedded at higher levels of biological organization and are difficult to retrieve in an analysis of primary sequence.
-
7.
The phylogenetic tree of substructures provides by definition a model of structural evolution. When using shape characters, this model describes an evolutionary process in which RNAs evolve from an originating substructure by addition of nucleotides and base pairs to substructural components. This occurs in defined order. Consequently, the topologies of trees of substructures can be used to build models of RNA evolution (see below). Generally, the phylogenetic relationships of stems determine the evolution of the overall shape of the RNA molecule that is typical of the molecular repertoire studied, and should be considered first. Trees of unpaired structural components define patterns of diversification that do not result in molecular multifurcation and, therefore, provide evolutionary patterns of decoration of the evolving molecule. Substructures describing the nature of base pairs in stems generate trees that depict preference for usage of nucleotides in base pairing interactions. Numbers of paired and unpaired regions describe interruptions in coaxial stacking of helical segments and relative frequency of bulges and internal loops in these segments and generate trees that describe evolution of these geometrical features in RNAs.
The Ancient Origin of the Acc Arm and the Top Half of tRNA
Phylogenetic trees of substructures derived using both geometrical and statistical characters for the entire or partitioned datasets showed that the Acc arm was the most basal substructure, generally followed in order by the TΨC, AC, and DHU and Var arms (Figs. 4 and 5). Overall, the Acc and TΨC arms of the top half domain were more ancestral than the AC and DHU arms of the bottom half domain. These observations support strongly the idea that the tRNAs originated in the Acc arm (Schimmel et al. 1993; Schimmel and Ribas de Pouplana 1995) and agree with the proposal that the bottom half domain evolved later than the top half domain (Maizels and Weiner 1994). In fact, our results are in line with the observations that an RNA minihelix containing the Acc and TΨC arms can be generally charged with the cognate amino acid by modern aaRSs (Martinis and Schimmel 1995). This suggests that primitive tRNAs probably lacked DHU and AC arms. Experimental evidence supports the idea that the nonconserved domains of aaRSs have been added to the conserved structures to facilitate interactions with distal parts of tRNA, such as the AC stem (Schimmel et al. 1993). In this regard, the minihelix domain formed by the stacking of the Acc and TΨC stems probably arose independently and preceded the AC-containing domain (Schimmel and Ribas de Pouplana 1995). Our phylogenetic reconstructions and resulting model of structural evolution (see below) are based on history embedded in the structure of the tRNA itself. Consequently, the indication that indeed the top half predated evolutionarily the bottom half supports strongly a coevolutionary relationship between aaRSs and the two functional domains of tRNA.
The ancestral placement of the Acc arm does not result from its being evolutionarily conserved. Supplementary Fig. S1 shows how character changes in a tree of substructures describing the evolution of stems in tRNA arms were more numerous (562) along the basal branch leading to the Acc arm than those on the other branches of the tree (82–475). This suggests that functional constraints, which can establish at different hierarchical levels of structure with different effects on structural evolution, must be operating mostly at levels other than secondary structure in tRNA. The ancestral placement of the Acc arm does not result from its being longer either. The position of a taxon on a rooted tree rests on shared and derived characters and how these change, but not on the ancestral nature of taxon-associated character states. Therefore, the basal position of the Acc reflects the interplay of length (shape characters) or stability and frustration (statistical characters) of this tRNA substructure in relation to other substructures, showing that they represent important and congruent factors shaping the structure of tRNA. We do expect for an early tRNA structure to become more and more structurally ‘canalized’ (i.e., substructures and the processes that generate them tend to change in ways that facilitate formation of specialized substructures [Ancel and Fontana 2000]) as it establishes new interactions with other functional RNA and proteins, adding new functional constraints to the evolving molecules. These constraints could define not only the structure of modern tRNA but also how that structure is ultimately realized.
Our results focus on tRNA structure and are mute in relation to the actual origin of the genetic code. However, they obviously refute the hypothesis that the AC arm was the origin of tRNA (Szathmáry 1999), the legitimacy of which had already been questioned (Di Giulio 2000). They are also incompatible with evolutionary models in which the tRNA cloverleaf originated directly from a single duplication or ligation of hairpin-like minihelix structures (Di Giulio 1992; Dick and Schamel 1995; Eigen and Winkler-Oswatitsch 1981; Hopfield 1978; Nagaswamy and Fox 2003; Tanaka and Kikuchi 2001). While a recent genomic analysis of the hyperthermophilic archaeal parasite Nanoarchaeum equitans may support indirectly the duplication model (Randau et al. 2005), these models do not address the evolutionary history of the two domains of tRNA. Instead, they partition tRNA into 5′- and 3′-halves, splitting helical regions in Acc and AC arms, while maintaining DHU and TΨC arms intact. If mature tRNAs were formed by joining two hairpins, then DHU and TΨC arms would be evolutionarily equivalent. However, this relationship is not evident in our phylogenetic reconstructions.
The presence of codon-anticodon pairs in the Acc stem of modern tRNA revealed by exhaustive analyses of sequences links the classic and operational codes and suggests that proto-tRNAs were embedded in ancient double-stranded coding genomes (Rodin et al. 1993, 1996; Rodin and Rodin 2006a). This model requires that tRNA molecules evolve from short precursors by duplication or were originally palindromic but, in contrast with proposals that involve duplication of entire 5′- or 3′-halves, is compatible with our results. Partial or total duplications of ancient substructures derived from the Acc stem and homologous to components of the top half could explain the dual complementarity and common origin of Acc and AC stems and still produce structures that were ancestral to the DHU arm.
A Model of tRNA Evolution
With two congruent lines of phylogenetic evidence, one based on shape characters that describe the geometry of molecules and the other based on statistical characters describing molecular mechanics, we here propose an evolutionary model that explains the origin and evolution of tRNA (Fig. 6). This model considers that the modern cloverleaf tRNA structure resulted from gradual addition of structural components to the growing molecule, either by insertion of single or multiple nucleotides or by partial or total duplications. The molecule originated from a short RNA hairpin containing a simple stem-loop structure (Bloch et al. 1985; Dick and Schamel 1995; Eigen and Winkler-Oswatitsch 1981; Hopfield 1978; Tanaka and Kikuchi 2001; Widmann et al. 2005; Woese 1969) homologous to the Acc arm of present-day tRNAs. Trees of substructures derived from the partitioned matrix of archaeal tRNAs (Fig. 5) showed that the AC arm predated the TΨC arm. This suggests that the proto-Acc arm extended its helical structure by stacking or adding an additional arm that was either homologous to the TΨC arm in ancestors of Bacteria and Eukarya or homologous to the AC arm in ancestors of Archaea. These two possible evolutionary routes (Fig. 6) led to two kinds of structures representing Acc-TΨC or Acc-AC helical regions, probably occurring during the onset of organismal diversification. It is noteworthy that the existence of these two evolutionary routes is compatible with whole-genome analysis of protein complements and domain combinations that suggest an early split of the archaeal lineage from a protein architecture-rich communal world by reductive genomic tendencies that were protracted and ultimately led to superkingdom Archaea and then to Bacteria and Eukarya (Wang and Caetano-Anollés 2006; Wang et al. 2007). However, while trees of substructures are compatible with an early rooting of the universal tree in Archaea, trees of molecules showed that tRNAs expressing different amino acid specificities or belonging to an individual superkingdoms were generally not monophyletic (Fig. 3). We hypothesize that this may be due either to loss of phylogenetic signal in tRNA structure (Di Giulio 1999) or to tRNA diversification’s predating true organismal diversification (Widmann et al. 2005). The latter scenario appears more probable, especially if we consider that patterns of representation of protein fold superfamilies in genomes over evolutionary history suggest that reductive tendencies in Archaea occurred before superkingdoms were truly specified (Wang et al. 2007).
A model of tRNA evolution. The model is derived directly from trees of substructures in Figs. 4 and 5 and shows formation of substructures homologous to present-day Acc, TΨC, AC, DHU, and Var arms during the course of evolution. Substructures may have had different functions from those of extant tRNAs. Alternative evolutionary routes A and B may be linked to ancestors of Archaea and Bacteria-Eukarya, respectively
The structures of the two-stem intermediates in the two ancient tRNA lineages were further extended by incorporation of a stem-loop region homologous to present-day AC and TΨC stems, respectively, forming a three-stem proto-tRNA structure containing the Acc, TΨC, and AC regions. The final addition of the DHU stem resulted in the more complicated two-domain structure of present-day tRNAs, containing an ancestral top half domain and a derived bottom half domain. It was only then that the Var region developed between TΨC and AC regions. Interestingly, phylogenetic reconstructions also suggest that structural diversification involving hairpin loops occurred only after the cloverleaf structure was fully realized in evolution (Fig. 4). Similarly, modified bases appeared early in regions homologous to the multiloop structure and then after the AC stem proto-structure was incorporated into the growing tRNA molecule. The use of weak G:U base pairs was derived. Interestingly, diversification of unpaired regions somehow followed the growth of stems in the evolving molecule, with the 5′-terminal free end being the most ancestral and the 3′-terminal free end, including the CCA terminus, the most derived. Remarkably, our suggestion that the 3′-terminal sequence was added only after the entire cloverleaf structure was formed matches inferences derived from statistical analyses of tRNA sequences (Tanaka and Kikuchi 2001). It is noteworthy that CCA-adding enzymes take tRNA transcripts and add these three specific nucleotides to their 3′ ends. Our observation that the 3′ unpaired region is derived suggests that activity and interaction with CCA-adding enzymes were fully established late in evolution, once the cloverleaf structure had been already formed.
Computational analyses of random RNA sequences of 50–90 nucleotides in length suggest that V- and Y-shaped secondary structures may be ancestors of tRNA (Marlière 1983). Coincidentally, these shapes match the evolutionary intermediates of our model. The nucleotide lengths of these structures agree with the suggestion that all tRNAs of prebiotic times assumed the hairpin structure when their length encompassed 73 nucleotides (Rodin et al. 1993). Our model also agrees with the recent proposal that minihelices incorporated two lateral arms later in evolution to form three-stem structures that are similar to the evolutionary intermediates of our model (Muller 2005). Finally, we recently studied the structures of tRNA-derived short interspersed elements (SINEs) and their evolution in plants and eukaryotes (Sun et al. 2007). Trees of substructures showed that SINEs had an ancient origin in the tRNA-derived stem-loop structure and suggested that proto-SINEs were rich in maximal hydrogen-bonding G:C base pairs, two aspects that are fundamental to support the idea that modern viruses are “molecular fossils” of ancient strategies for genomic replication (Weiner and Maizels 1999). Besides showing common evolutionary trends important for tRNA-derived SINE RNA structures, these observations lend further support to the genomic tag hypothesis (Maizels and Weiner 1994; Weiner and Maizels 1987).
Conclusions
Our results support the ancestral nature of the top half domain of tRNA and an origin in the Acc stem, the cornerstone of the genomic tag hypothesis (Maizels and Weiner 1994; Weiner and Maizels 1987). Congruence between phylogenetic trees of substructures derived from structural and statistical characters provides strong support to a model of evolution that explains the structure of tRNA as a progressive series of additions of structural components to the growing molecule. This evolutionary scenario seems to match bioinformatic analyses and experimental biochemical and genetic evidence related to the interaction of tRNA with itself or other molecules. Finally, the phylogenetic methods introduced here have the potential to unravel evolutionary patterns and processes related to the genetic code, chronologies of amino acid charging and usage, and, ultimately, phenomena related to the origins of life. These methods depend not only on the validity of models of character change (and associated character polarization assumptions) but also on the accuracy of structural data utilized and are therefore as good as our inferences and models of structure, whether these are generated from comparative analyses or from crystallographic models (e.g., Selmer et al. 2006; Shi and Moore 2000; Yusupov et al. 2001). These methods at present focus mainly on the structural scaffold of secondary structure and, as such, describe only a minimal set of key structural features in the tRNA molecules. They do not incorporate information on compensatory changes in sequence that maintain base pairing or on tertiary interactions defining 3D structure. However, tRNA structure evolves in 3D space and is evolutionarily constrained by the chemistries and inter- and intramolecular interactions that delimit tRNA function. In the absence of a structural genomic effort focusing on RNA, we lack crystallographic entries describing 3D models of tRNA sufficient for a comparative analysis. Consequently, the evolutionary contributions of 3D structural features of the tRNA molecules other than those delimiting secondary structure (such as long-range contacts between arms) cannot be studied at this time with our cladistic method.
References
Ancel LW, Fontana W (2000) Plasticity, evolvability, and modularity in RNA. J Exp Zool (Mol Dev Evol) 288:242–283
Billoud B, Guerrucci MA, Masselot M, Deutsch JS (2000) Cirripede phylogeny using a novel approach: molecular morphometrics. Mol Biol Evol 17:1435–1445
Bloch DP, McArthur B, Mirrop S (1985) tRNA-rRNA sequence homologies: evidence for an ancient modular format shared by tRNAs and rRNAs. Biosystems 17:209–225
Bremer K (1988) The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42:795–803
Bull AT, Goodfellow M, Slater JH (1992) Biodiversity as a source of innovation in biotechnology. Annu Rev Microbiol 46:219–252
Caetano-Anollés G (2001) Novel strategies to study the role of mutation and nucleic acid structure in evolution. Plant Cell Tissue Org Cult 67:115–132
Caetano-Anollés G (2002a) Evolved RNA secondary structure and the rooting of the universal tree of life. J Mol Evol 54:333–345
Caetano-Anollés G (2002b) Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res 30:2575–2587
Caetano-Anollés G (2005) Grass evolution inferred from chromosomal rearrangements and geometrical and statistical features in RNA structure. J Mol Evol 60:635–652
Collins LJ, Moulton V, Penny D (2000) Use of RNA secondary structure for studying the evolution of RNase P and RNase MRP. J Mol Evol 51:194–2004
Dick TP, Schamel WWA (1995) Molecular evolution of transfer RNA from two precursor hairpins: implications for the origin of protein synthesis. J Mol Evol 41:1–9
Di Giulio M (1992) On the origin of the transfer RNA molecule. J Theor Biol 159:199–214
Di Giulio M (1999) The non-monophyletic origin of the tRNA molecule. J Theor Biol 197:403–414
Di Giulio M (2000) The RNA world, the genetic code and the tRNA molecule. Trends Genet 16:17–18
Eigen M, Winkler-Oswatitsch R (1981) Transfer-RNA, an early gene? Naturwissenschaften 68:282–292
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (1988) Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet 22:521–565
Fontana W (2002) Modelling ‘evo-devo’ with RNA. BioEssays 24:1164–1177
Gladyshev GP, Ershov YA (1982) Principles of the thermodynamics of biological systems. J Theor Biol 94:301–343
Goodenbour JM, Pan T (2006) Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 34:6137–6146
Gultyaev PA, van Batenburg FHD, Pleij CWA (2002) Selective pressures on RNA hairpins in vivo and in vitro. J Mol Evol 54: 1–8
Higgs PG (1993) RNA secondary structure: a comparison of real and random sequences. J Phys I France 3:43–59
Higgs PG (1995) Thermodynamic properties of transfer RNA: a computational study. J Chem Soc Faraday Trans 91:2531–2540
Higgs PG (2000) RNA secondary structure: physical and computational aspects. Quarterly Rev Biophys 33:199–253
Hillis DM, Huelsenbeck JP (1992) Signal, noise, and reliability in molecular phylogenetic analyses. J Hered 83:189–195
Hopfield JJ (1978) Origin of the genetic code: a testable hypothesis based on tRNA structure, sequence, and kinetic proofreading. Proc Natl Acad Sci USA 75:4334–4338
Maddison WP, Maddison DR (2003) MacClade 4: analysis of phylogeny and character evolution, version 4.06. Sinauer Associates, Sunderland, MA
Maizels N, Weiner AM (1994) Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc Natl Acad Sci USA 91:6729–6734
Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H (2006) The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res 34:1816–1835
Marlière P (1983) Computer building and folding of fictitious transfer-RNA sequences. Biochimie 65:267–273
Martinis SA, Schimmel P (1995) Small RNA oligonucleotide substrate for specific aminoacylations. In: Söll D, RajBandary V (eds) tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC, pp 349–370
Muller AWJ (2005) Thermosynthesis as energy source for the RNA world: a model for the bioenergetics of the origin of life. Biosystems 82:93–102
Nagaswamy U, Fox G (2003) RNA ligation and the origin of tRNA. Origins Life Evol B 33:199–209
Pollock D (2003) The Zuckerkandl Prize: structure and evolution. J Mol Evol 56:375–376
Randau L, Münch R, Hohn MJ, Jahn D, Söll D (2005) Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433:537–541
Rodin S, Ohno S, Rodin A (1993) Transfer RNAs with complementary anticodons: Could they reflect early evolution of discriminative genetic code adaptors? Proc Natl Acad Sci USA 90:4723–4727
Rodin S, Rodin A, Ohno S (1996) The presence of codon-anticodon pairs in the acceptor stem of tRNAs. Proc Natl Acad Sci USA 93:4537–4542
Rodin SN, Rodin AS (2006a) Origin of the genetic code: first aminoacyl-tRNA systhetases could replace isofunctional ribozymes when only the second base of codons was established. DNA Cell Biol 25:365–375
Rodin SN, Rodin AS (2006b) Partitioning of aminoacyl-tRNA synthetases in two classes could have been coded in a strand-symmetric RNA world. DNA Cell Biol 25:617–626
Schimmel P, Giegé R, Moras D, Yokoyama S (1993) An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA 90:8763–8768
Schimmel P, Ribas de Pouplana L (1995) Transfer RNA: from minihelix to genetic code. Cell 81:983–986
Schultes EA, Bartel DP (2000) One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science 289:448–452
Schultes EA, Hraber PT, LaBean TH (1999) Estimating the contributions of selection and self-organization in RNA secondary structure. J Mol Evol 49:76–83
Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942
Shi H, Moore PB (2000) The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6:1091–1105
Sprinzl M, Vassilenko KS (2005) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33:D139–D140
Steel M, Penny D (2000) Parsimony, likelihood, and the role of models in molecular phylogenetics. Mol Biol Evol 17:839–850
Steffens W, Digby D (1999) mRNA have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res 27:1578–1584
Stegger G, Hofman H, Fortsch J, Gross HJ, Randles JW, Sanger HL, Riesner D (1984) Conformational transitions in viroids and virusoids: comparison of results from energy minimization algorithm and from experimental data. J Biomol Struct Dynam 2:543–571
Steinberg S, Misch A, Sprinzl M (1993) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 21:3011–3015
Sun F-J, Fleurdépine S, Bousquet-Antonelli C, Caetano-Anollés G, Deragon J-M (2007) Common evolutionary trends for tRNA-derived SINE RNA structures. Trends Genet 23:26–33
Swain TD, Taylor DJ (2003) Structural rRNA characters support monophyly of raptorial limbs and paraphyly of limb specialization in water fleas. Proc R Soc London B 270:887–896
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland, MA
Szathmáry E (1999) The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet 15:223–229
Tanaka T, Kikuchi Y (2001) Origin of the cloverleaf shape of transfer RNA - the double-hairpin model: implication for the role of tRNA intron and the long extra loop. Viva Origino 29:134–142
Wang M, Caetano-Anollés G (2006) Global phylogeny determined by the combination of protein domains in proteomes. Mol Biol Evol 23:2444–2454
Wang M, Yafremava LS, Caetano-Anollés D, Mittenthal JE, Caetano-Anollés G (2007) Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res 17:1572–1585
Weiner AM, Maizels N (1987) tRNA-like structures tag the 3′ ends of genomic RNA moleculesfor replication: implications for the origin of protein synthesis. Proc Natl Acad Sci USA 84:7383–7387
Weiner AM, Maizels N (1999) The genomic tag hypothesis: modern viruses as molecular fossils of ancient strategies for genomic replication, and clues regarding the origin of protein synthesis. Biol Bull 196:327–330
Widmann J, Di Giulio M, Yarus M, Knight R (2005) tRNA creation by hairpin duplication. J Mol Evol 61:524–535
Woese CR (1969) The biological significance of the genetic code. Prog Mol Subcell Biol 1:5–46
Yusupov MM, Yusupov GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883–895
Zhu W, Freeland S (2006) The standard genetic code enhances adaptive evolution of proteins. J Theor Biol 239:63–70
Acknowledgments
We thank Hee Shin Kim, Ajith Harish, Minglei Wang, and Jay Mittenthal for helpful discussions, Joe Felsenstein for comments on phylogenetic reconstruction, Paul Schimmel for suggestions, Sergei Rodin for many critical comments an suggestions that improved the manuscript, and Vegeir Knudsen for writing stoat, a program that codes statistical characters. Both authors designed and performed research, analyzed data, and wrote the paper. This work was supported by National Science Foundation Grant MCB-0343126 (to G.C.A.) and the Critical Research Initiative of the University of Illinois. Any opinions, findings, and conclusions and recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Sun, FJ., Caetano-Anollés, G. The Origin and Evolution of tRNA Inferred from Phylogenetic Analysis of Structure. J Mol Evol 66, 21–35 (2008). https://doi.org/10.1007/s00239-007-9050-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-007-9050-8