Abstract
Species richness and geographical phenotypic variation in East African lacustrine cichlids are often correlated with ecological specializations and limited dispersal. This study compares mitochondrial and microsatellite genetic diversity and structure among three sympatric rock-dwelling cichlids of Lake Tanganyika, Eretmodus cyanostictus, Tropheus moorii, and Ophthalmotilapia ventralis. The species represent three endemic, phylogenetically distinct tribes (Eretmodini, Tropheini, and Ectodini), and display divergent ecomorphological and behavioral specialization. Sample locations span both continuous, rocky shoreline and a potential dispersal barrier in the form of a muddy bay. High genetic diversity and population differentiation were detected in T. moorii and E. cyanostictus, whereas much lower variation and structure were found in O. ventralis. In particular, while a 7-km-wide muddy bay curtails dispersal in all three species to a similar extent, gene flow along mostly continuous habitat appeared to be controlled by distance in E. cyanostictus, further restricted by site philopatry and/or minor habitat discontinuities in T. moorii, and unrestrained in O. ventralis. In contrast to the general pattern of high gene flow along continuous shorelines in rock-dwelling cichlids of Lake Malawi, our study identifies differences in population structure among stenotopic Lake Tanganyika species. The amount of genetic differentiation among populations was not related to the degree of geographical variation of body color, especially since more phenotypic variation is observed in O. ventralis than in the genetically highly structured E. cyanostictus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The origin of biological diversity is of profound interest to evolutionary biologists, not least because the amount of variation is distributed unevenly across groups of organisms. Cichlid fish of the East African Great Lakes, for example, range among the most diverse species flocks in vertebrates (Fryer 1959; Fryer and Iles 1972; Dominey 1984; Sturmbauer 1998; Kocher 2004; Salzburger and Meyer 2004). The lacustrine radiations of cichlids in Lakes Malawi, Victoria, Tanganyika, and several smaller lakes produced enormous numbers of endemic, ecologically, and morphologically highly differentiated, species within short evolutionary time spans (Salzburger et al. 2002, 2005; Verheyen et al. 2003; Joyce et al. 2005). Estimates of the ages and numbers of cichlid species for the East African Great Lakes range from 9 to 12 million years (my), with ∼250 species, for Lake Tanganyika; 4 to 9 my, with ∼700–800 species, for Lake Malawi; and 0.2 to 0.5 my, with ∼500–700 species, for Lake Victoria (Kornfield and Smith 2000; Snoeks 2001; Turner et al. 2001). The fast speciation rates in lacustrine cichlids have been associated with a variety of mechanisms and conditions, including natural and sexual selection, hybridization, and habitat complexity and instability (Fryer and Iles 1972; Kornfield and Smith 2000; Kocher 2004; Salzburger and Meyer 2004; Schliewen and Klee 2004; Seehausen 2004). There is compelling evidence of sympatric divergence in small West African (Schliewen et al. 1994; Schliewen and Klee 2004) and Nicaraguan crater lake cichlids (Barluenga et al. 2006), and to some extent also in Lake Victoria (Seehausen and van Alphen 1999), but in more mature species flocks such as Lakes Malawi and, especially, Tanganyika, diversification has been attributed largely to allopatric processes (Sturmbauer 1998).
Among the most severe events repeatedly reshaping the East African Great Lakes’ lacustrine environments, fluctuations in the water level associated with paleoclimatic changes and geologic activity were shown to have affected the population structure and speciation patterns in all three Great Lakes simultaneously (Sturmbauer et al. 2001; Verheyen et al. 2003). Rossiter (1995) coined the term “species pump” for the diversifying action of fluctuating lake levels and the associated recurrent cycles of population fusion and fragmentation. Radiations of several cichlid lineages in Lake Tanganyika, for example, coincide with major water level changes (Sturmbauer et al. 2003; Brandstätter et al. 2005; Duftner et al. 2005; Koblmüller et al. 2005), and phylogeographic patterns of several benthic species provide evidence of recurrent population shifts coupled with changes of shoreline structure and water depth (Verheyen et al. 1996; Sturmbauer et al. 1997, 2001, 2005; Rüber et al. 1999; Baric et al. 2003).
The potential for allopatric diversification in East African cichlids is best illustrated by one of the most species-rich clades, the mbuna haplochromines of Lake Malawi, which are comprised of stenotopic inhabitants of rocky coasts. With little to no gene flow across habitat barriers such as sandy and deep-water sections (van Oppen et al. 1997a; Arnegard et al. 1999; Markert et al. 1999; Danley et al. 2000; Rico and Turner 2002), numerous local variants are likely to have evolved allopatrically on a small spatial scale (Fryer and Iles 1972; Ribbink et al. 1983). Genetic differentiation among non-mbuna cichlid specialists of rocky and sandy habitat in Lake Malawi is equally dictated by habitat structure (Pereyra et al. 2004), whereas neither benthic generalists nor pelagic and demersal species developed significant genetic structure across larger geographic distances (Shaw et al. 2000; Taylor and Verheyen 2001; Pereyra et al. 2004).
Although even short habitat discontinuities act as strong migration barriers to some rock-dwellers of Lake Malawi, they seem to disperse freely along continuous rocky shorelines (van Oppen et al. 1997a; Arnegard et al. 1999; Rico and Turner 2002). We do not know of studies on the population structure of Lake Victoria cichlids, but a representative of the “Lake Victoria superflock” from an unstructured satellite lake displayed little if any genetic structure (Abila et al. 2004). In contrast, studies of rock specialists from Lake Tanganyika demonstrated high population differentiation along continuous stretches of shoreline as well as across habitat barriers (Rüber et al. 2001; Taylor et al. 2001; Duftner et al. 2006). The present study compares genetic differentiation and diversity in nuclear and mitochondrial markers in representatives of three endemic tribes of Lake Tanganyika, Eretmodus cyanostictus (Eretmodini), Ophthalmotilapia ventralis (Ectodini), and Tropheus moorii (Tropheini; Poll 1986). The tribes span a large part of the phylogenetic diversity of cichlids in Lake Tanganyika (Fig. 1) (Salzburger et al. 2002). The monogamous, biparental mouthbrooding Eretmodini are one of the eight seeding lineages of the Lake Tanganyika radiation (Salzburger et al. 2002). Reduced swim bladders are believed to constrain dispersal of E. cyanostictus; the species has been shown to be genetically structured in other localities (Taylor et al. 2001; Rüber et al. 2001) and provides a reference to evaluate the level of differentiation in the other species. Ectodini and Tropheini derive from the ancestor of the “redefined H-lineage” (Salzburger et al. 2002), a particularly species-rich lineage of mouthbrooding cichlids exhibiting a wealth of morphological, ecological, and behavioral diversity. Even within tribes, diversity is high: the 35 Ectodini species encompass maternal and biparental mouthbrooders, as well as specialized rock- and sand-dwellers with different feeding modes (Koblmüller et al. 2004). The ectodine representative in this study, O. ventralis, is a rock-dwelling, polygamous, maternal mouthbrooder, whose males defend territories around a nest site (bowers) consisting of a layer of sand on a flat rock surface, and are visited by gravid females for spawning (Konings 1998). The 25 Tropheini species endemic to Lake Tanganyika are member of the “modern haplochromines” and sister-group to the species flocks of Lakes Malawi and Victoria and several other East African riverine and lacustrine cichlids (Salzburger et al. 2005). Within the Tropheini, the genus Tropheus is renowned for pronounced geographical color variation: more than 100 different color morphs have been described (Schupke 2003). Although classified into several nominal species, the members of the genus—with the exception of T. duboisi—are polyphyletic (Sturmbauer and Meyer 1992; Sturmbauer et al. 1997, 2003, 2005; Baric et al. 2003). Males and females of the monogamous, maternal mouthbrooders hold individual territories except for a short period prior to spawning, when females move into their mates’ territories to boost-feed and mature their eggs (Yanagisawa and Nishida 1991; Sturmbauer and Dallinger 1995; Egger et al. 2006).
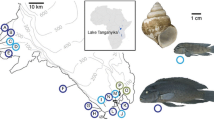
Sampling sites and study species. A Lake Tanganyika, with the locations of the sample sites along the southwestern shoreline. Bathymetric lines are approximated according to Fig. 1 of Gasse et al. (1989). B Cladogram representing the phylogenetic relationships of the main lineages of Lake Tanganyika cichlids (according to Salzburger et al. 2002, 2005). Photographs show the three study species.
The three species occur sympatrically in great abundance on all rocky shore sections of southern Lake Tanganyika. Numbers of adult individuals in two 400-m2-transect squares differing in slope and substrate structure (sizes of pebbles and rocks) averaged 108 for adult T. moorii, 84.5 for adult E. cyanostictus, and 108 for adult and subadult O. ventralis (Sturmbauer et al., unpublished data). E. cyanostictus reaches its highest density in very shallow areas not included in these transects, and it is likely that the total census size for E. cyanostictus is at least as high as that of T. moorii. Studies in northern Lake Tanganyika report average numbers of about 1 Tropheus/m2 at depths of <4 m (Kohda and Yanagisawa 1992) and 0.1 Tropheus/m2 at a depth of 3–6 m (Yanagisawa and Nishida 1991), and densely packed territories ranging from 0.8 m2 (Kawanabe 1981) to 6.1 m2 (Yanagisawa and Nishida 1991; Sturmbauer and Dallinger 1995). Given what is known about the mating system of the three species (Yanagisawa and Nishida 1991; Konings 1998; Morley and Balshine 2003), effective population size is probably highest in the permanently pair-bonding, biparental mouth- brooders E. cyanostictus, somewhat lower in the temporarily pair-bonding, maternal mouthbrooders T. moorii, and lowest in the polygynous O. ventralis.
E. cyanostictus, O. ventralis, and T. moorii were sampled from five locations that span both continuous rocky shoreline and a potential migration barrier in the form of a river mouth forming a muddy bay (Fig. 1). Identical sampling sites for the three species allow us to examine the impact of habitat on the genetic structure of the benthic rock-dwelling populations. If, as in the rock-dwelling haplochromines in Lake Malawi, gene flow is controlled for the most part by habitat discontinuities in the distantly related species of this study as well, we expect to observe congruent population structure with strong differentiation across the barrier but gene flow along the continuous habitat, in all three species. In contrast, differences in population structure among species would demonstrate the importance of species-specific qualities such as dispersal ability, degree of stenotopy, and philopatric behavior.
Materials and Methods
Population samples were taken from Eretmodus cyanostictus (n = 138), Tropheus moorii (n = 179), and Ophthalmotilapia ventralis (n = 135) at five localities along the southwestern Zambian shore of Lake Tanganyika (Fig. 1). Four of the sampling sites, Tongwa (08°40′S, 30°53′E), Katukula (08°43′S, 30°57′E), Funda (08°46′S, 30°59′E), and Katoto (08°48′S, 31°01′E) are spaced 9.5, 6.5, and 5.5 km apart on steep, mostly continuous rocky shoreline, whereas Kasakalawe (08°47′S, 31°04′E) is separated from Katoto by the sandy habitat stretch of Mbete Bay at the estuary of Izi River. The characterization of the habitat is based on surveillance of the shoreline between sampling sites and on observations made during specimen collection and snorkeling at the sampling sites. The shore between Katoto and Tongwa is very steep and we know from our field observations (including dives to depths of 20 m) that the steep shores in this area are rocky until depths of 20 m or more. The species investigated here occur mainly at depths of between 1 and 20 m. However, we cannot exclude minor interruptions of the rocky habitat between Katoto and Tongwa by the inflow of seasonal streams or by very short sandy stretches.
The census of the three species reported in the introduction was conducted in September 2004 in two 20 × 20-m transects, one situated on shallow-sloping, pebbly shore in front of Tanganyika Lodge, Kasakalawe, at a depth of from 1.75 to 2.85 m, and one at a more steeply sloping, more sediment-rich shore in front of Toby Veall’s Lodge in Kalambo (depth, 0.57–3.57 m; 08°36′S, 31°11′E). Fish were counted while snorkeling along 2-m-wide rows parallel to the shoreline. Counts for each of the 10 rows per transect were repeated three times, and the averages of the replicate values per row were summed to yield the total number in the 400-m2 transect.
Total DNA was extracted from ethanol preserved fin clips by proteinase K extraction, followed by sodium chloride extraction and isopropanol precipitation (Bruford et al. 1998). Standard protocols and universal primers (L-Pro [Meyer et al. 1994], primer E “TDK-D” [Lee et al. 1995]) were used to amplify and sequence partial proline tRNA and mitochondrial control region sequences (Baric et al. 2003; Sturmbauer et al. 2005). Alignments included the first section of the mtDNA control region only and comprised 359 bp, with 60 variable and 27 parsimony-informative sites in E. cyanostictus; 361 bp, with 67 variable and 55 parsimony-informative sites in T. moorii; and 358 bp, with 23 variable and 13 parsimony-informative sites in O. ventralis. Sequence data were deposited in GenBank under accession numbers AJ295902–AJ295905, AJ295912, AJ295913, AJ489719, AJ489723, AJ491061, AJ491975–AJ491993, AJ491995–AJ492048, AJ492050–AJ492060, AJ492062–AJ492109, AJ492111–AJ492145, AJ492994, AY660819, AY660820 for T. moorii, EF035309–EF035446 for E. cyanostictus, and EF035174–EF035308 for O. ventralis.
Populations were typed at three microsatellite loci, but as the loci were not equally informative in each species, locus sets were not fully overlapping between species. The following loci were used: E. cyanostictus—Pzeb 3 (van Oppen et al. 1997b), TmoM11 (Zardoya et al. 1996), and UNH002 (Kellogg et al. 1995); O. ventralis—UME 002, UME003 (Parker and Kornfield 1996), and TmoM11; and T. moorii—Pzeb 3, UME002, and TmoM11. The fluorescently labeled PCR products were separated on an ABI 373, and later on an ABI 377, automatic sequencer (Applied Biosystems) and sized by comparison to ROX GS 500 internal size standard (Applied Biosystems) using GENESCAN software (Applied Biosystems). Except for one locus-population combination (TmoM11 in E. cyanotictus, Funda; Table 1), observed heterozygosity complied with Hardy Weinberg expectations (GENEPOP version 3.4; Raymond and Rousset 1995) after correcting for multiple tests (Benjamini and Hochberg 1995). Indexes of marker polymorphism were calculated with FSTAT (Goudet 2001) and are given in Table 1. O. ventralis from Funda were not genotyped for microsatellite alleles, as the total extract was used for mtDNA amplification due to poor DNA quality caused by unintentional use of low-grade ethanol for sample preservation in the field.
Mitochondrial diversity indexes were calculated in DNASP version 4.00 (Rozas et al. 2003). One sample (T. moorii from Katoto) was about twice as large as the other population samples, and the effect of large sample size on diversity indexes was assessed by random resampling of 30 haplotypes (1000 iterations) from the full sample and recalculation of the diversity indexes. The significance of interspecific differences in diversity was addressed by a Mann-Whitney rank sum test (SIGM ASTAT 3.9; SPSS Inc.); differences among populations within species were tested against a null distribution obtained by randomizing haplotypes between populations and recalculation of the indices (Muñoz-Fuentes et al. 2005). Haplotype genealogies were reconstructed using a statistical parsimony approach with a 95% cutoff as implemented in TCS (Clement et al. 2000). Population structure was estimated from mitochondrial data based on haplotype frequencies (F ST) and uncorrected genetic distances between haplotypes (ΦST), and based on microsatellite allele frequencies (F ST), over all populations within species and in pairwise intraspecific population comparisons using ARLEQUIN (Schneider et al. 2000). Type I errors in pairwise population comparisons were controlled using the method of Benjamini and Hochberg (1995). Haplotype sharing between populations was calculated in MICROSAT (as compiled by E. Minch; http://www.hpgl.stanford.edu/projects/microsat/) by coding haplotypes as homozygous diploid loci. Sequence divergence within and between populations was calculated in DNASP based on absolute numbers of differences, and in MEGA version 3.0 (Kumar et al. 2004) applying a Kimura two-parameter model. Analyses of historical population demography involved the calculation of mismatch distributions and estimation of the time parameter τ = 2μt, where μ is the mutation rate per generation per haplotype (Rogers and Harpending 1992), under the finite alleles model (Schneider and Excoffier 1999) as implemented in ARLEQUIN. DNASP was used for calculation and significance tests of Fu’s (1997) F S and Fu and Li’s (1993) D* and F*. In addition, a coalescent-based parameter estimation using the program FLUCTUATE was performed (Kuhner et al. 1998) employing estimates of base frequencies and transition:transversion ratios obtained under the HKY model from PAUP* (Swofford 2001). FLUCTUATE uses a maximum likelihood criterion to estimate parameters of population growth (g in units of μ −1 t −1, where μ is the mutation rate per site per generation) and present-time effective population size (θ = 2N ef μ for mitochondrial genes) applying a model of exponential population growth (g > 0) or decline (g < 0). Twenty short chains, of 200 steps, and five long chains, of 200,000 steps, were run three times for each population to ensure convergence of the estimates.
Results
Genetic Variation
Across populations, genetic variation, represented by the number of different haplotypes, haplotype diversity, and nucleotide diversity (Table 2), was high in E. cyanostictus and T. moorii, but significantly lower in O. ventralis (p = 0.008 for each index). Populations (across species and locations) differed most strongly in nucleotide diversity (coefficient of variation [CV] = 56.7%) and number of different haplotypes (CV = 41.3%), whereas haplotype diversity values were less variable (CV = 8.4%). There was no significant difference in diversity indexes among populations of O. ventralis. E. cyanostictus from Kasakalawe displayed significantly lower haplotype diversity than the population in Katukula and significantly lower nucleotide diversity than each of the populations along the western shore. In T. moorii, several pairs of populations exhibited significant differences in one or more indexes (Fig. 2). Sample sizes were similar for all populations (mean n = 28, SD = 3.3) except for T. moorii from Katoto (n = 60), such that higher variability indexes for this population could be a sampling artifact. A resampling procedure yielded a lower average number of different haplotypes in subsamples, n = 30, but undiminished average gene and nucleotide diversity (Table 2).
Mitochondrial genetic diversity measured as number of haplotypes, haplotype diversity, and nucleotide diversity in E. cyanostictus (black), T. moorii (gray), and O. ventralis (white), per population and in a pooled sample. Error bars represent standard deviations. Numbers indicate populations (according to x-axis labels) with significantly lower or higher values for a given diversity index than observed in the population concerned. *Average number of haplotypes in subsamples of n = 30.
The average number of nucleotide differences in E. cyanostictus was 5.74 nucleotides. Along the continuous shoreline, haplotype sharing amounted to 20%–35% among neighboring populations (Table 3) but decreased with geographic distance (Fig. 3). No haplotypes were shared across Mbete Bay, and all but one haplotype of the Kasakalawe population were resolved as a monophyletic clade (Fig. 4). T. moorii sequences differed on average by 11.53 nucleotides and fell into three clades, which remained unconnected in the statistical parsimony network (Fig. 5). Haplotypes of the Kasakalawe population were found exclusively in clades I and II, both of which also contained haplotypes of the Katoto population. The remaining Katoto samples grouped in clade III together with haplotypes from the other locations along the continuous western shoreline. Only two individuals north of Katoto bore haplotypes belonging to clade II. As in E. cyanostictus, shared haplotypes were most frequent between neighboring locations (Table 3, Fig. 3). A high proportion of missing haplotypes in the network of E. cyanostictus (41.5%) and T. moorii (34.7%, 50%, and 32.2% empty nodes in clades I, II, and III, respectively) may reflect large effective population sizes of these species, consistent with census counts and mating system (see Introduction). Haplotype divergence in O. ventralis was low, with an average difference of 2.66 nucleotides between sequences. The majority of the individuals displayed one of the four most common haplotypes (Fig. 6), and haplotype sharing was high across all populations regardless of geographic distance (Table 3).
Haplotype network of T. moorii. Circle diameters are proportional to the number of individual sequences per haplotype. Small open circles represent unsampled haplotypes. Note that some of the empty nodes are occupied by haplotypes from other locations in the phylogeographic study of the genus Tropheus by Sturmbauer et al. (2005). Average numbers of nucleotide substitutions between clades are 16.6 for clades I and II, 21.6 for clades II and III, and 22.0 for clades I and III.
Population Structure
Mitochondrial population differentiation was highly significant (p < 0.001) in each of the three species (E. cyanostictus, ΦST = 0.260, F ST = 0.041; T. moorii, ΦST = 0.413, F ST = 0.034; O. ventralis, ΦST = 0.215, F ST = 0.066). The ΦST estimates are congruent with the genetic structure discernible in the haplotype genealogies: due to limited haplotype sharing and deep sequence divergence, the highest ΦST value was detected in T. moorii, while the comparatively small ΦST value in O. ventralis reflects low sequence divergence and high haplotype sharing. The occurrence of the highest F ST value in O. ventralis, however, might be an artifact of the interaction between population polymorphism H S and F ST estimates (Hedrick 2005). Higher genetic variability in E. cyanostictus and T. moorii than in O. ventralis causes F ST to underestimate genetic structure in the first two species relative to the latter. A standardized measure of genetic differentiation, F’ ST, is obtained by expressing F ST as a proportion of its theoretical maximum (Hedrick 2005) and yields F’ ST values of 75% for E. cyanostictus, 79% for T. moorii, and 33% for O. ventralis, with the lowest value for O. ventralis in accordance with ΦST estimates and the haplotype genealogies.
Pairwise population comparisons highlighted the strong effect of the habitat barrier between Kasakalawe and the locations along the continuously rocky western shoreline. Kasakalawe populations of each species were clearly differentiated from all other conspecific populations west of Mbete Bay (Table 4). No haplotypes were shared between E. cyanostictus from Kasakalawe and the populations west of the bay, and Kasakalawe haplotypes were almost exclusively grouped in a separate cluster with an average of 1.9% sequence divergence from the western haplotypes (Fig. 4). In T. moorii, the area of Mbete Bay delimits the distribution of two distinct mitochondrial lineages that arose 700,000–945,000 years before present (YBP) (TCS-8-G west and TCS-1-A2 and TCS-1-A4 east of the bay [Sturmbauer et al. 2005]). Hence, the average sequence divergence across the bay is high (6.1%), but in contrast to complete lineage sorting observed in E. cyanostictus, T. moorii from Kasakalawe and Katoto—the populations immediately east and west of Mbete Bay—share several haplotypes (Fig. 5). Nevertheless, population differentiation across the bay was highly significant (Table 4). Highly significant differentiation between populations across Mbete Bay was also observed in O. ventralis, although the three most common haplotypes were shared between populations across the bay (Fig. 6), and average sequence divergence between Kasakalawe and the western populations was low (0.98%).
While population differentiation across Mbete Bay was evident in all three species, the species differed with regard to the degree of genetic differentiation among populations along the continuous shoreline. In E. cyanostictus, differentiation values between neighboring populations were low and sometimes (dependent on the algorithm) nonsignificant, whereas gene flow is apparently reduced between more distant locations. Genetic differentiation in T. moorii was significant in all population pairs, with the exception of a nonsignificant F ST value between the neighboring Funda and Katoto, which was, however, opposed by a high and significant ΦST estimate. O. ventralis displayed no genetic structure along the continuous habitat. With only four samples along the continuous shore, it was not possible to use Mantel matrix randomizations to test the correlation between geographic and genetic distances in E. cyanostictus and T. moorii, but scatter plots of distance measures indicate an influence of geographic distances on mitochondrial gene flow (Fig. 3).
Estimates of pairwise population differentiation from microsatellite data were largely consistent with mitochondrial ΦST values. The highest differentiation estimates in each species were observed between the sample from Kasakalawe and the remaining populations. However, not all of the significant mitochondrial differentiation estimates were reflected by significant nuclear estimates (Table 5). The closest congruence between marker sets was observed in E. cyanostictus, where the highest significance levels were assigned to population comparisons including Kasakalawe and Tongwa, while lower or no structure was found among the remaining pairs. In contrast to the distinct mitochondrial structure in T. moorii along the continuous shoreline, microsatellite differentiation estimates among the Funda, Katukula, and Tongwa populations were low and not significant. Lack of nuclear structure was supported by all three loci for Katukula-Tongwa, and by Pzeb3 and UME002 for Funda-Katukula and Funda-Tongwa, while allele frequencies of TmoM11 were significantly different in these population pairs (Funda-Katukula, F ST = 0.036, p < 0.001; Funda-Tongwa, F ST = 0.220, p < 0.01). In O. ventralis, microsatellite differentiation between Kasakalawe and the remaining populations was also less pronounced than the structure detected in mitochondrial sequences, and the two most distant populations, Kasakalawe and Tongwa, did not differ significantly in their allele frequencies after correction for multiple comparisons. Both nuclear and mitochondrial markers indicated panmixis in O. ventralis along the continuous shoreline.
Historical Population Demography
Star-like genealogies (Figs. 4–6) and unimodal mismatch distributions in several of the analyzed populations suggested expansion of effective sizes at slightly different periods in the (in evolutionary terms) recent past, but results from different tests for population expansion were not always congruent (Fig. 7). A certain amount of gene flow among neighboring populations and past fusion of haplotype lineages was evident in our data, and inconsistencies in the tests may represent their varying sensitivity to divergent immigrant haplotypes mimicking the pattern of long-term constant population size. E. cyanostictus generally complied with a model of population expansion. In all populations, Fu’s F S was negative and significant, whereas the negative values of Fu and Li’s D* and F* were not significant. Growth parameters (g) obtained from FLUCTUATE were positive but not very high, and their 95% confidence intervals did not include zero except for the population from Katoto (Fig. 7). Mismatch distributions also showed deviations from unimodality in Katoto and, particularly, in Tongwa, consistent with somewhat lower g estimates for these populations; again, this may be due to gene flow between populations. The confidence intervals of time estimates for population growth along the western shoreline overlapped largely (95% CI of τ = 2.8–9.7), whereas the expansion of the Kasakalawe population was dated somewhat more recently (95% CI of τ = 1.3–3.9).
Tests for population expansion in E. cyanostictus, T. moorii, and O. ventralis. The undifferentiated populations of O. ventralis from Katoto to Tongwa were combined in one sample. Bar charts represent observed mismatch distributions among haplotype sequences; lines represent expected distributions based on parameter estimates and their 95% confidence limits. The x-axes are drawn to scale to facilitate comparison of mode positions, with the exception of the graphs for T. moorii from Kasakalawe and Katoto. Boxes within charts report parameter estimates and 95% confidence intervals of τ , the significance level of the difference between observed and expected mismatch distributions p (SSD), and F S, D*, and F* statistics with respective probability values. The separate table shows the range of parameter estimates g and θ obtained from three replicate runs of the FLUCTUATE software.
Lineage admixture interfered with tests of population expansion in T. moorii populations in Kasakalawe and Katoto. A bimodal mismatch distribution for the population in Katukula was caused by two divergent haplotypes, which were most likely due to immigration from Katoto (Fig. 5) and were consequently excluded from tests for population growth. With 95% CIs of τ values ranging from 1.7 to 8.1, significantly negative F S values, nonsignificant D* and F*, and FLUCTUATE growth parameters of between 330 and 580 (95% CI > 0), populations in Funda, Katukula, and Tongwa showed signs of expansion at approximately the same period as E. cyanostictus. Inferences of the demographic history of O. ventralis differed from those of the other species. Neither the population in Kasakalawe nor the pooled sample of the undifferentiated populations from Katoto to Tongwa on the western shore displayed unambiguous signatures of recent expansion. Mismatch distributions were flat, but in agreement with expectations for very recent expansion. However, confidence intervals for τ estimates were wide, and included negative growth parameter (g) estimates for Kasakalawe (Fig. 7). Nonsignificant Fu and Fu/Li statistics for Kasakalawe conflicted with population expansion, and although significantly negative values of F S, D*, and F* for the western shore sequences could be interpreted as evidence for background selection (Fu 1997), this is unlikely for mitochondrial sequence data. Combining all O. ventralis samples into a single analysis produced results similar to those of the analysis of western shore populations except for a shift of the estimated expansion period to a 95% CI of τ = 1.28–7.34.
The parameter estimates of present-day effective population size, θ, obtained from FLUCTUATE under a model of exponential population expansion, ranged from 0.08 to 0.10 for all populations of E. cyanostictus; from 0.06 to 0.12 for T. moorii from Funda and Tongwa, and higher (θ = 0.18–0.20) for T. moorii from Katukula; and from 0.03 to 0.04 for O. ventralis in Kasakalawe and 0.08 to 0.09 for O. ventralis along the western coast (Fig. 7). These θ values are likely to overestimate the actual effective population sizes (θ = 0.1 corresponds to N e = 1,100,000–1,500,000 assuming a mutation rate of 3.25% to 4.4% per million years). When population size changes were eliminated from the model, values of θ (estimate of long-term stable effective size) were considerably lower (0.027–0.042 in the populations of E. cyanostictus; 0.029–0.040 in T. moorii except for Katoto, where θ = 0.060 (note that this population is comprised of two mitochondrial lineages); and 0.009 and 0.020 for O. ventralis from Kasakalawe and from the western shoreline, respectively; (data not shown). Although still somewhat high, the more realistic θ estimates under a model of stable population size perhaps refute the model of exponential population growth in the recent past. Under both models, however, the relation of θ values among the three species, with distinctly higher effective population sizes in E. cyanostictus and T. moorii than in O. ventralis, is plausible given their census counts and mating systems.
Discussion
Differences in Mitochondrial Genetic Structure and Diversity Among Species
Tropheus moorii and Eretmodus cyanostictus showed great genetic diversity and population differentiation, whereas much lower variation and structure were found in Ophthalmotilapia ventralis. Species-specific patterns of population structure in the three rock-dwellers are consistent with differences and similarities in their responses to habitat structure and innate dispersal behavior. All three species show a break in genetic continuity at Mbete Bay, which constitutes a 7-km-long interruption of the rocky shoreline. Gene flow across the bay may be curbed not only by the geographical extension of the habitat barrier, but also by differences in water chemistry or temperature due to inflow of the small Izi River into the bay.
The shore between Katoto and Tongwa is steep and lacks structures that would indicate large interruptions of the rocky habitat associated with the steep coastline in this area. Data from our population samples along this section of the lake therefore suggest that gene flow along relatively short stretches (20 km) of mostly continuous habitat is (1) low and controlled by distance in E. cyanostictus, as differentiation increases with geographic distance, but is nonsignificant between some neighboring populations; (2) highly restricted in T. moorii and controlled by distance as well as strong site philopatry and/or minor habitat discontinuities, since differentiation is significant even over the short distances and habitat composition that allow gene flow in E. cyanostictus; and (3) unrestrained in O. ventralis.
Previous phylogeographic analyses suggested that lake level fluctuations and habitat structure contribute to the population structure in rock-dwelling species (Tropheus [Sturmbauer and Meyer 1992; Sturmbauer et al. 1997, 2005; Baric et al. 2003], E. cyanostictus [Verheyen et al. 1996; Sturmbauer et al. 1997; Rüber et al. 1999, 2001]). In particular, a strong phylogeographic pattern disrupted by the occasional presence of closely related and even identical haplotypes on opposite sides of the lake found in Tropheus, Eretmodus, and another highly structured rock-dweller, Variabilichromis moorii, suggests incidental gene flow in association with periods of low lake level (Sturmbauer et al. 1997, 2001; Duftner et al. 2006). Furthermore, this and other recent population genetic analyses of sympatric species indicate that species-specific differences in traits related to dispersal superimpose on extrinsic forces and create variable patterns of population structure and diversity (this study; Duftner et al. 2006; Koblmüller et al. 2006).
The current shoreline of the shallow southern tip of Lake Tanganyika was formed by the latest rise in lake level between 11,000 and 17,000 YBP (Gasse et al. 1989; Sturmbauer et al. 2001). The magnitude of the preceding lowstand is under debate, and estimates range from 150 to 400 m below the present level (Rüber et al. 2001 and references therein). Depending on the scale of the lowstand, the northward shift of the shoreline and the associated displacement of littoral populations was more or less drastic (Fig. 1), but even a 150-m drop would have displaced all populations south of Katukula. With the subsequently rising lake level, littoral species such as E. cyanostictus and T. moorii moved with the coastal habitat, as the lake extended south- and westward and increased the length of shoreline with suitable habitat. Comparable levels of genetic diversity and congruent reconstructions of demographic history and effective population sizes suggest similar responses to habitat changes in the two species, and the observed differences in population structure may mostly originate from species-specific dispersal behavior following recolonization of the novel habitat. Stochastic variation of the lineage sorting process is ruled out as the single source of the observed differences by the overall congruence between pairwise nuclear and mitochondrial differentiation estimates. A reduced swim bladder enables E. cyanostictus to inhabit wave-exposed rocky habitats, but poses a physical constraint on dispersal across open water (Konings 1998). The short range of gene flow between E. cyanostictus populations along continuously rocky shore (Rüber et al. 2001; Taylor et al. 2001; this study) is in line with the species’ poor dispersal capacities, but evidence of undifferentiated populations also suggests that migration is not uncommon within the individuals’ physical limitations. In contrast, the high level of haplotype structuring in T. moorii may be governed mainly by behavioral constraints on (at least) female dispersal, and only secondarily by distance and major habitat barriers. The phylogeographic distribution of mitochondrial control region haplotypes of Tropheus populations around the entire lake has been studied in considerable detail, and haplotype lineages were defined to classify the highly diverse group into 10 clades derived from a common ancestor 700,000–945,000 YBP (Sturmbauer and Meyer 1992; Baric et al. 2003; Sturmbauer et al. 2005). The swampy Mbete Bay delimits the geographical distribution of two of these divergent haplotype lineages: haplotypes from Funda, Katukula, Tongwa, and farther north to the Lufubu River estuary (clade III in Fig. 5) belong to a geographically restricted, derived lineage (TCS-8 “G” of Sturmbauer et al. 2005), whereas haplotypes from Kasakalawe (clades I and II in Fig. 5) are part of a widespread ancestral lineage (TCS-1 “A” of Sturmbauer et al. 2005), and both haplotype lineages co-occur in Katoto. The confined distribution of the two lineages implies that migrants have not been exchanged regularly across Mbete Bay and along the western shoreline; rather, the distribution of haplotype lineages may be due primarily to sporadic migration in conjunction with lake level fluctuations (Baric et al. 2003; Sturmbauer et al. 2005). In addition to the data presented here, the permanent influence of the Mbete Bay area on gene flow and migration in cichlids is evidenced by the reciprocal monophyly of Neolamprologus caudopunctatus (Lamprologini) east and west of Mbete Bay, separated by a split dating to 170,000–260,000 YBP (Koblmüller et al. 2006). As inhabitants of the shallow rocky habitat (<20 m deep), the distribution of O. ventralis must have been influenced by lake level changes as well, but high levels of gene flow provide for genetic continuity among populations in the absence of habitat barriers. Gene flow in O. ventralis may be due to dispersing juveniles. In contrast to E. cyanostictus and T. moorii, where juveniles hide among pebbles near the parental territories (Konings 1998; Morley and Balshine 2002), dispersal of juvenile O. ventralis is mediated by mouth-brooding mothers moving around in schools and, later, by juvenile aggregations in the open inshore water (Konings 1998). Low nucleotide diversity suggests smaller long-term female effective population sizes in O. ventralis than in E. cyanostictus and T. moorii, but DNA sequence-based inference of the demographic history of the populations was inconclusive and did not unambiguously support a model of recent population expansion.
Estimates of Population Structure from Nuclear Markers
The patterns of population structure discussed above were inferred from maternally inherited mitochondrial DNA sequences and represent the evolutionary history of the female part of the population. However, the differences in population structure between species may be further enhanced or alleviated by behavioral differences between the sexes. Based on data from three microsatellite loci, we cannot quantitatively compare mitochondrial and nuclear differentiation estimates, but certain trends emerge nevertheless. Nuclear differentiation was significant across the habitat barrier in all three species, suggesting that discontinuous habitat restricts both male and female dispersal. Among populations along the continuous shoreline, congruent pairwise differentiation estimates were obtained from nuclear and mitochondrial markers in E. cyanostictus, while both marker systems suggested panmixis in O. ventralis. Nuclear differentiation differed from mitochondrial estimates in T. moorii: while mitochondrial differentiation was significant among population pairs along the western shore, the absence of nuclear differentiation among populations from Funda, Tongwa, and Katukula raises the possibility of male-biased gene flow along continuous habitat. A more thorough investigation of nuclear gene flow among neighboring T. moorii populations should be undertaken with a larger nuclear data set and sex-specific sampling.
Genetic Structure and Diversity of E. cyanostictus in Different Regions
In agreement with previous findings (Rüber et al. 2001; Taylor et al. 2001), genetic structure was high in E. cyanostictus. Unlike in these studies, however, a lack of differentiation between neighboring populations along a continuous habitat suggested that gene flow is not infrequent across distances of 5 to 10 km, if dispersal is not constrained by any habitat barriers. Differentiation estimates were higher between populations along a mostly continuous stretch of rocky coast (but interspersed by small sandy beaches) in the Democratic Republic of Congo than among the populations along the Zambian coast (microsatellite F ST—Zambia, 0.098 and 0.074 [Taylor et al. 2001; this study], and DR Congo, 0.127 [Rüber et al. 2001]; mitochondrial FST—Zambia, 0.041 [this study], and DR Congo, 0.263 [Rüber et al. 2001]; mitochondrial ΦST—Zambia, 0.260 [this study], and DR Congo, 0.843 [based on data of Rüber et al. 2001]). Genetic diversity in almost-identical segments of mitochondrial DNA was lower in the Congolese than in the Zambian populations (DR Congo, average π = 0.0076, SD = 0.0024, and average H E = 0.903, SD = 0.053 [calculated from Rüber et al. 2001]; for Zambia, see Table 2). Nucleotide diversity across populations, however, was higher on the Congolese coast (π = 0.040) than in Zambia (π = 0.016). Congolese and Zambian populations are reciprocally monophyletic, and the average number of nucleotide substitutions per site between the two groups of populations is D XY = 0.057. After subtracting intrapopulation polymorphism, the mean net distance of D A = 0.03 between the two groups suggests a split between 340,000 and 460,000 YBP, applying the rate of 6.5%–8.8% divergence per my estimated for the control region segment used in these studies (Sturmbauer et al. 2001). This estimate overlaps with a period of high lake level between ∼670,000 and 390,000 YBP, following the most severe lake lowstand, ∼650 m below the present level, 1.1 my BP [Cohen et al. 1997). With rising water level, the southward extension of the lake increased, and gene flow between populations that had been geographically close during the major lowstand was curbed by distance and habitat barriers. The subsequent lake level declines to between 150 and 300 m below the present level (Cohen et al. 1997) interrupted the structure of populations inhabiting the shallow southern areas of the lake but caused mainly vertical dislocations of populations on steeply sloping shores, where rocky habitat extends into deep water (Rüber et al. 2001). The observed differences in genetic differentiation and diversity estimates between Congolese and Zambian populations are consistent with a model of longstanding allopatry of the Congolese steep shore populations and recurrent population fusion in the shallow Zambian lake section.
Population Structure and Geographic Color Variation
Population fragmentation often coincides with geographic color variation in Lake Malawi mbuna (Arnegard et al. 1999; Smith and Kornfield 2002; Rico and Turner 2002; Rico et al. 2003). Geographical variation in color patterns is also exhibited by many benthic species in Lake Tanganyika (Konings 1998). In T. moorii and O. ventralis, the genetic structure among populations in our study area corresponds with the distribution of geographical color variation. Within the “Bright Blue Ventralis” variant of O. ventralis occurring at our study sites, geographical types are distinguishable by varying black markings on their body, and a discontinuity in the distribution of morphs occurs at Mbete Bay (Kohda et al. 1996). T. moorii also exhibits phenotypic differences across Mbete Bay, but a small proportion of eastern phenotypes was found at one study site west of the bay (Kohda et al. 1996). According to Fig. 1 of Kohda et al. (1996), this site was located somewhat north of our sampling site in Katoto, where authors of the present study (S.B., W.S., and C.S.) found T. moorii to display a color pattern intermediate between the Kasakalawe population and the phenotype of the populations northwest of Katoto. Both Kohda and coworkers’ (1996) and our own account of color patterns in populations immediately northwest of Mbete bay are consistent with the co-occurrence of western and eastern haplotype lineages in this area.
Despite the congruence between color type distribution and genetic similarity among populations observed in two species in the present study, there is no general correlation between the amount of color variation and the amount of neutral genetic population differentiation in the Lake Tanganyika rock-dwellers that have so far been examined in a population genetics framework. Notwithstanding the deep genetic divergence between Congolese and Zambian populations of E. cyanostictus, and the significant differentiation observed on smaller scales, Konings (1998) does not distinguish different phenotypes in the southern distribution area of the species from Congo to Tanzania. In contrast, more pronounced phenotypic variation is displayed by O. ventralis along its distribution encompassing the southern two-thirds of the lake (Konings 1998), although our study indicates less substructure and higher levels of gene flow in this species. The genus Tropheus comprises over 100, often highly distinct, morphs (Schupke 2003), most of which correspond to allopatrically distributed populations; apparently, diversification in color patterns coincides with highly restricted gene flow in this taxon. Another genetically highly structured rock-dweller, Variabilichromis moorii (tribe Lamprologini), however, displays virtually no phenotypic variation across its entire range (Duftner et al. 2006).
Conclusions
Genetic diversity and population differentiation were high in T. moorii and E. cyanostictus, whereas much lower variation and structure were found in O. ventralis. While dispersal is curtailed by a muddy bay in all three species, gene flow along continuous habitat appeared to be controlled by distance in E. cyanostictus, further restricted by behavioral philopatry in T. moorii, and unrestrained in O. ventralis. The diversity in population structure observed among rock specialists of Lake Tanganyika contrasts with the more uniform findings emerging from studies of haplochromine Lake Malawi rock-dwellers, where gene flow was generally limited by habitat barriers but not by distance alone. The enormous diversification and species richness of rock-dwelling Lake Malawi haplochromines have been related to observations of distinct population structuring (Pereyra et al. 2004; Genner and Turner 2005). Phenotypic diversification and population substructuring are not generally correlated in the Lake Tanganyika species studied in this context (this study; Duftner et al. 2006; Koblmüller et al. 2006), implicating a much greater diversity in genetic factors regulating behavior and morphology (Kocher 2004) in the phylogenetically diverse species assemblage of the older lake. Apart from the inferred differences in behavioral and physiological constraints on dispersal, the species investigated here differ dramatically in further traits associated with diversification and speciation, such as social structure, mating and brood care systems, habitat use, and diet. Moreover, we suggest that diversification patterns and responses to selection pressures are differentially affected by lineage-specific phylogenetic constraints originating from the longstanding separate evolution of Lake Tanganyika cichlid lineages.
References
Abila R, Barluenga M, Engelken J, Meyer A, Salzburger W (2004) Population-structure and genetic diversity in a haplochromine fish cichlid of a satellite lake of Lake Victoria. Mol Ecol 13:2589–2602
Arnegard ME, Markert JA, Danley PD, Stauffer JR Jr, Ambali AJ, Kocher TD (1999) Population structure and colour variation of the cichlid fish Labeotropheus fuelleborni Ahl along a recently formed archipelago of rocky habitat patches in southern Lake Malawi. Proc Roy Soc B 266:119–130
Baric S, Salzburger W, Sturmbauer C (2003) Phylogeography and evolution of the Tanganyikan cichlid genus Tropheus based upon mitochondrial DNA sequences. J Mol Evol 56:54–68
Barluenga M, Stoelting K, Salzburger W, Muschick M, Meyer A (2006) Sympatric speciation in Nicaraguan crater lake cichlids. Nature 439:719–723
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300
Brandstätter A, Salzburger W, Sturmbauer C (2005) Mitochondrial phylogeny of the Cyprichromini, a lineage of open-water cichlid fishes endemic to Lake Tanganyika, Africa. Mol Phylogenet Evol 34:382–391
Bruford MW, Hanotte O, Brookfield JFY, Burke T (1993) Multilocus and single-locus DNA fingerprinting. In: Hoelzel AR (ed) Molecular genetic analysis of populations. Oxford University Press, New York, pp 287–336
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cohen AS, Lezzar K-E, Tiercelin J-J, Soreghan M (1997) New palaeogeographic and lake-level reconstructions of Lake Tanganyika: implications for tectonic, climatic and biological evolution in a rift lake. Basin Res 9:107–132
Danley PD, Markert JA, Arnegard ME, Kocher TD (2000) Divergence with gene flow in the rock-dwelling cichlids of Lake Malawi. Evolution 54:1725–1737
Dominey WJ (1984) Effects of sexual selection and life history on speciation: species flocks in African cichlids and Hawaiian Drosophila. In: Echelle AA, Kornfield I (eds) Evolution of fish species flocks. University of Maine Orono Press, Orono, pp 231–249
Duftner N, Koblmüller S, Sturmbauer C (2005) Evolutionary relationships of the Limnochromini, a tribe of benthic deepwater cichlid fish endemic to Lake Tanganyika, East Africa. J Mol Evol 60:277–289
Duftner N, Sefc KM, Koblmüller S, Nevado B, Verheyen E, Phiri H, Sturmbauer C (2006) Distinct population structure in a phenotypically homogeneous rock-dwelling cichlid fish from Lake Tanganyika. Mol Ecol 15:2381–2395
Egger B, Obermüller B, Phiri H, Sturmbauer C, Sefc KM (2006) Monogamy in the maternally mouthbrooding Lake Tanganyika cichlid fish Tropheus moorii. Proc R Soc Lond B 273:1797–1802
Fryer G (1959) The trophic interrelationships and ecology of some of the littoral communities of Lake Nyasa with special reference to the fishes, and a discussion on the evolution of a group of rock-frequenting Cichlidae. P Zool Soc London 132:153–281
Fryer G, Iles TD (1972) The cichlid fishes of the great lakes of Africa: their biology and evolution. Oliver & Bond, Edinburgh
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Fu Y-X, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Gasse F, Lédée V, Massault M, Fontes J-C (1989) Water-level fluctuations of Lake Tanganyika in phase with oceanic changes during the last glaciation and deglaciation. Nature 342:57–59
Genner MJ, Turner GF (2005) The mbuna cichlids of Lake Malawi: a model for rapid speciation and adaptive radiation. Fish Fisheries 6:1–34
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available at: http://www.unil.ch/izea/softwares/fstat.html
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Joyce DA, Lunt DH, Bills R, Turner GF, Katongo C, Duftner N, Sturmbauer C, Seehausen O (2005) An extant cichlid fish radiation emerged in an extinct Pleistocene lake. Nature 435:90–95
Kawanabe H (1981) Territorial behaviour of Tropheus moorei (Osteichthyes: Cichlidae). Afr Study Mon 1:101–108
Kellogg KA, Markert JA, Stauffer JRJ, Kocher TD (1995) Microsatellite variation demonstrates multiple paternity in lekking cichlid fishes from Lake Malawi, Africa. Proc Roy Soc B 260:79–84
Koblmüller S, Salzburger W, Sturmbauer C (2004) Evolutionary relationships in the sand-dwelling cichlid lineage of Lake Tanganyika suggest multiple colonization of rocky habitats and convergent origin of biparental mouthbrooding. J Mol Evol 58:79–96
Koblmüller S, Duftner N, Katongo C, Phiri H, Sturmbauer C (2005) Ancient divergence in bathypelagic Lake Tanganyika deepwater cichlids: mitochondrial phylogeny of the tribe Bathybatini. J Mol Evol 60:297–314
Koblmüller S, Sefc KM, Duftner N, Warum M, Sturmbauer C (2006) Genetic population structure as indirect measure of dispersal ability in a Lake Tanganyika cichlid. Genetica (in press)
Kocher TD (2004) Adaptive evolution and explosive speciation: the cichlid fish model. Nat Genet Rev 5:288–298
Kohda M, Yanagisawa Y (1992) Vertical distributions of two herbivorous cichlid fishes of the genus Tropheus in Lake Tanganyika, Africa. Ecol Freshw Fish 1:99–103
Kohda M, Yanagisawa Y, Sato T, Nakaya K, Niimura Y, Matsumoto K, Ochi H (1996) Geographical colour variation in cichlid fishes at the southern end of Lake Tanganyika. Environ Biol Fish 45:237–248
Konings A (1998) Tanganjika-cichlids in their natural habitat. Cichlid Press, El Paso, TX
Kornfield I, Smith PF (2000) African cichlid fishes: model systems for evolutionary biology. Annu Rev Ecol Syst 31:163–196
Kuhner MK, Yamato J, Felsenstein J (1998) Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149:429–434
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Lee W-J, Conroy J, Huntting Howell W, Kocher TD (1995) Structure and evolution of teleost mitochondrial control regions. J Mol Evol 41:54–66
Markert JA, Arnegard ME, Danley PD, Kocher TD (1999) Biogeography and population genetics of the Lake Malawi cichlid Melanochromis auratus: habitat transience, philopatry and speciation. Mol Ecol 8:1013–1026
Meyer A, Montero C, Spreinat A (1994) Evolutionary history of the cichlid fish species flocks of the East African great lakes inferred from molecular phylogenetic data. Arch Hydrobiol Beiheft Ergebnisse Limnol 44:407–423
Morley JI, Balshine S (2002) Faithful fish: territory and mate defense favour monogamy in an African cichlid fish. Behav Ecol Sociobiol 52:326–331
Morley JI, Balshine S (2003) Reproductive biology of Eretmodus cyanostictus, a cichlid fish from Lake Tanganyika. Environ Biol Fish 66:169–179
Muñoz-Fuentes V, Green AJ, Negro JJ, Sorenson MD (2005) Population structure and loss of genetic diversity in the endangered white-headed duck, Oxyura leucocephala. Conserv Genet 6:999–1015
Parker A, Kornfield I (1996) Polygynandry in Pseudotropheus zebra, a cichlid fish from Lake Malawi. Environ Biol Fish 47:345–352
Pereyra R, Taylor MI, Turner GF, Rico C (2004) Variation in habitat preference and population structure among three species of the Lake Malawi cichlid genus Protomelas. Mol Ecol 13:2691–2697
Poll M, 1986. Classification des cichlidae du lac Tanganika. Tribus, genres et aspeces. Acad Roy Belg. Memories de la classe des sciences, -8O -2O serie, T. XLV-Fascicule 2:1–163
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248
Ribbink AJ, Marsh AC, Marsh BA, Sharp BJ (1983) A preliminary survey of the cichlid fishes of the rocky habitats in Lake Malawi. S Afr J Zool 18:149–310
Rico C, Turner GF (2002) Extreme microallopatric divergence in a cichlid species from Lake Malawi. Mol Ecol 11:1585–1590
Rico C, Bouteillon P, van Oppen MJH, Knight ME, Hewitt GM, Turner GF (2003) No evidence for parallel sympatric speciation in cichlid species of the genus Pseudotropheus from north-western Lake Malawi. J Evol Biol 16:37–46
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Rossiter A (1995) The cichlid fish assemblages of Lake Tanganyika: ecology, behavior and evolution of its species flocks. Adv Ecol Res 26:187–252
Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Rüber L, Verheyen E, Meyer A (1999) Replicated evolution of trophic specializations in an endemic cichlid fish linage from Lake Tanganyika. Proc Natl Acad Sci USA 96:10230–10235
Rüber L, Meyer A, Sturmbauer C, Verheyen E (2001) Population structure in two sympatric species of the Lake Tanganyika cichlid tribe Eretmodini: evidence for introgression. Mol Ecol 10:1207–1225
Salzburger W, Meyer A (2004) The species flock of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften 91:277–290
Salzburger W, Meyer A, Baric S, Verheyen E, Sturmbauer C (2002) Phylogeny of the Lake Tanganyika species flock and its relationship to the Central and East African Haplochromine cichlid fish fauna. Syst Biol 51:113–135
Salzburger W, Mack T, Verheyen E, Meyer A (2005) Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol Biol 5:17
Schliewen UK, Klee B (2004) Reticulate sympatric speciation in Cameroonian crater lake cichlids. BMC Front Zool 1:5
Schliewen UK, Tautz D, Pääbo S (1994) Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 368:629–632
Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152:1079–1089
Schneider S, Roessli D, Excoffier L (2000) Arlequin 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland
Schupke P (2003) Cichlids of Lake Tanganyika, Part 1: The species of the genus Tropheus. Aqualog, Rodgau, Germany
Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol 19:198–207
Seehausen O, van Alphen JJM (1999) Can sympatric speciation by disruptive sexual selection explain rapid evolution of cichlid diversity in Lake Victoria? Ecol Lett 2:262–271
Shaw PW, Turner GF, Idid MR, Robinson RL, Carvalho GR (2000) Genetic population structure indicates sympatric speciation of Lake Malawi pelagic cichlids. Proc Roy Soc B 267:2273–2280
Smith PF, Kornfield I (2002) Phylogeography of Lake Malawi cichlids of the genus Pseudotropheus: significance of allopatric colour variation. Proc Roy Soc B 269:2495–2502
Snoeks J (2001) Cichlid diversity, speciation and systematics: examples from the Great African Lakes. In: Coleman R (ed) Cichlid research: state of the art. J Aquaricult Aquat Sci 9:150–166
Sturmbauer C (1998) Explosive speciation in cichlid fishes of the African Great Lakes: a dynamic model of adaptive radiation. J Fish Biol 53:18–36
Sturmbauer C, Dallinger R (1995) Diurnal variation of spacing and foraging behavior in Tropheus moorii (Cichlidae) in Lake Tanganyika. Netherlands J Zool 45:386–401
Sturmbauer C, Meyer A (1992) Genetic divergence, speciation and morphological stasis in a lineage of African cichlid fishes. Nature 358:578–581
Sturmbauer C, Verheyen E, Rüber L, Meyer A (1997) Phylogeographic patterns in populations of cichlid fishes from rocky habitats in Lake Tanganyika. In: Kocher TD, Stephen CA (eds) Molecular Systematics of Fishes. Academic Press, pp 97–111
Sturmbauer C, Baric S, Salzburger W, Rüber L, Verheyen E (2001) Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes. Mol Biol Evol 18:144–154
Sturmbauer C, Hainz U, Baric S, Verheyen E, Salzburger W (2003) Evolution of the tribe Tropheini from Lake Tanganyika: synchronized explosive speciation producing multiple evolutionary parallelism. Hydrobiologia 500:51–64
Sturmbauer C, Koblmüller S, Sefc KM, Duftner N (2005) Phylogeographic history of the genus Tropheus, a lineage of rock-dwelling cichlid fishes endemic to Lake Tanganyika. Hydrobiologia 542:335–366
Swofford DL (2001) PAUP* 4.0b10: phylogenetic analysis using parsimony. Sinauer, Sunderland, MA
Taylor MI, Verheyen E (2001) Microsatellite data reveals weak population substructuring in Copadichromis sp. ‘virginalis kajose’, a demersal cichlid from Lake Malawi, Africa. J Fish Biol 59:593–604
Taylor MI, Rüber L, Verheyen E (2001) Microsatellites reveal high levels of population substructuring in the species-poor Eretmodine cichlid lineage from Lake Tanganyika. Proc Roy Soc B 268:803–808
Turner GF, Seehausen O, Knight ME, Allender CJ, Robinson RL (2001) How many species of cichlid fish are there in African lakes? Mol Ecol 10:793–806
van Oppen MJH, Turner GF, Rico C, Deutsch JC, Ibrahim KM, Robinson RL, Hewitt GM (1997a) Unusually fine-scale genetic structuring found in rapidly speciating Malawi cichlid fishes. Proc Roy Soc B 264:1803–1812
van Oppen MJH, Rico C, Deutsch JC, Turner GF, Hewitt GM (1997b) Isolation and characterization of microsatellite loci in the cichlid fish Pseudotropheus zebra. Mol Ecol 6:387–388
Verheyen E, Rüber L, Snoeks J, Meyer A (1996) Mitochondrial phylogeography of rock-dwelling cichlid fishes reveals evolutionary influence of historical lake level fluctuations in Lake Tanganyika, Africa. Philos Trans Roy Soc B 351:797–805
Verheyen E, Salzburger W, Snoeks J, Meyer A (2003) Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 300:325–329
Yanagisawa Y, Nishida M (1991) The social and mating system of the maternal mouthbrooder Tropheus moorii (Cichlidae) in Lake Tanganyika. Jap J Ichthyol 38:271–282
Zardoya R, Vollmer DM, Craddock C, Streelman JT, Karl S, Meyer A (1996) Evolutionary conservation of microsatellite flanking regions and their use in resolving the phylogeny of cichlid fishes (Pisces: Perciformes). Proc Roy Soc B 263:1589–1598
Acknowledgments
We thank L. Mwape, C. Mwelwa, and their team at the Mpulungu Station of the Ministry of Agriculture, Food and Fisheries, Republic of Zambia, and M. Nabuyanda for their help during fieldwork. Stephan Koblmüller and two anonymous reviewers provided insightful comments on the manuscript. Some of the data were collected in the course of master’s theses by Cornelia Rhomberg, Andreas Gallmetzer, and Beate Kirchberger. K.M.S., C.S., and S.B. were supported by the Austrian Science Foundation (P14724, P15239, and P17380) and the Jubiläumsfond der Österreichischen Nationalbank; W.S. was supported by a DOC fellowship from the Austrian Academy of Sciences and a Marie Curie fellowship from the EU.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Rafael Zardoya]
Rights and permissions
About this article
Cite this article
Sefc, K.M., Baric, S., Salzburger, W. et al. Species-Specific Population Structure in Rock-Specialized Sympatric Cichlid Species in Lake Tanganyika, East Africa. J Mol Evol 64, 33–49 (2007). https://doi.org/10.1007/s00239-006-0011-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-006-0011-4