Abstract
LINE-1 (L1) elements constitute the major family of retrotransposons in mammalian genomes. Here we report the first investigation of L1 evolution in New World monkeys (NWM). Two regions of the second open-reading frame were analyzed by two methods in three NWM species, the squirrel monkey (Saimiri sciureus), the tamarin (Saguinus oedipus), and the spider monkey (Ateles paniscus). Since these three species diverged, L1 has amplified in the Saimiri and Saguinus lineages but L1 activity seems to have been strongly reduced in the Ateles lineage. In addition, the active L1 lineage has evolved rapidly in Saimiri and Saguinus, generating species-specific subfamilies. In contrast, we found no evidence for a species-specific subfamily in Ateles, a result consistent with the low L1 activity in this species for the last ~25 My.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
L1 (LINE-1) elements (Fig. 1A) are mammalian long interspersed repeats that replicate autonomously by retrotransposition (reviewed by Furano 2000). They have been evolving and replicating since before the mammalian radiation (~100 My ago) and their accumulation in mammalian genomes accounts for up to 25% of the DNA mass of these genomes. L1 retrotransposition generates mostly defective copies, which remain in the genome and diverge from each other at the pseudogene rate (Hardies et al. 1986; Pascale et al. 1993; Voliva et al. 1983). With time, novel replication-competent L1 variants arise and some can generate families of hundreds to thousands of copies. Although more than one family of closely related variants (referred to as subfamilies) can be concurrently active (Boissinot et al. 2000; Furano et al. 1994; Pascale et al. 1993; Smit et al. 1995), ultimately only one prevails (Cabot et al. 1997). This process is exemplified by the successive emergence of a single lineage of replicatively dominant L1 families during the last 60 My of human evolution (Boissinot et al. 2001; Smit et al. 1995).
A Structure of a typical full-length human L1 element. The 5′ untranslated region (5′UTR) has a regulatory function; open reading frame (ORF) I encodes an RNA-binding protein; ORFII encodes the L1 cDNA replicase containing highly conserved endonuclease (EN) and reverse transcriptase (RT) domains; the 3′UTR contains a conserved G-rich polypurine motif. Genomic copies of L1 end in an A-rich stretch (open rectangle) (Furano 2000). The A and B regions correspond to the two regions studied in this paper. B Evolutionary relationships between the primate species used in these studies (Goodman et al. 1998).
The persistence of L1 activity in all mammalian orders studied so far (Burton et al. 1986) is puzzling because L1 insertions can cause genetic defects (Kazazian 1998) and L1 activity in general can pose a serious genetic load to its host (Boissinot et al. 2001). How serious depends, in part, on the rate at which L1 amplifies. In murine rodents the L1 amplification rate changes over evolutionary time; periods of high and low amplification rates alternate (Pascale et al. 1993; Verneau et al. 1998). Apparent L1 replication rates also differ between species; the rate of genetic defects due to L1 insertions that cause disease in human is 30 times lower than in the house mouse (Ostertag and Kazazian 2001), and it has been suggested that L1 amplification has recently decreased in humans and their immediate primate antecedents (Boissinot et al. 2000; Consortium 2001).
As L1 elements have been characterized in only a few nonhuman primate species (DeBerardinis and Kazazian 1998; Mathews et al. 2003; Stanhope et al. 1993), most of our knowledge of primate L1 is based on human studies. Here we report the first analysis of L1 evolution in an additional group of nonhuman primates, New world monkeys (NWMs; infraorder Platyrrhini, following the classification of Goodman et al. [1998]). NWMs are restricted to Central and South America, where they have evolved and diversified since their split from Catarrhini primates (Old World monkeys, apes, and human), ~35 My ago (Fig. 1B). We collected sequences from two regions of L1 in three NWM species that diverged from each other between 20 and 25 My ago. L1 amplified and evolved in the tamarin (g. Saguinus) and squirrel monkey (g. Saimiri) lineages. In contrast, both processes were drastically reduced in the spider monkey lineage (g. Ateles). We also found that two L1 lineages coexisted in the common ancestor of these three species but only one of them has persisted until the present time.
Materials and Methods
Sources of Samples
DNA samples of squirrel monkey (Saimiri sciureus), spider monkey (Ateles paniscus), rhesus monkey (Macaca mulatta), and lemur (Lemur catta) were obtained from individuals kept in captivity, respectively, at the University of München, the Wuppertal Zoological Garden, the Leipzig Zoological Garden, and the Münster Zoological Garden. The tamarin (Saguinus oedipus) and human (Homo sapiens) DNAs were purchased from the Coriell Institute for Medical Research. The phylogenetic relationship between these species is shown in Fig. 1B.
PCR Amplification and Cloning
PCR primers were cognate to a highly conserved region of the second open reading frame (ORFII; Region A in Fig. 1A). Primers GAGAAATGCAAATCAAAACCA (forward) and GCTGCATAGTATTCCATTGT (reverse) amplify a 396-bp fragment located near the 3′ extremity of ORFII (from position 5426 to position 5822 on alignment ALIGN_000165 from the EMBL-ALIGN database [Boissinot and Furano 2001]). This region encompasses a cysteine-rich motif (Moran et al. 1996) that is conserved in all L1 and L1-like elements (Clements and Singer 1998). The conservation of this part of ORFII was confirmed by obtaining amplification with the same primers in a wide range of mammals including mouse, rabbit, dog, cow, pig, and elephant shrew (data not shown). PCRs were performed in capillary tubes in an Idaho Technology Air-Thermo Cycler using the reagents provided by the company. Primers were used at a final concentration of 0.5 µM. Fifty nanograms of genomic DNA was amplified in a total volume of 25 µl using the following conditions: 94°C for 0 s, 50°C for 0 s, and 72°C for 10 s, for 30 cycles. PCR products were purified with the QIAquick PCR purification kit (QIAGEN) and cloned into the pGEM vector (Promega). Positive clones were sequenced in both directions using the Sequenase version 2.0 sequencing kit (Amersham).
When this first method failed to yield young L1 elements from Ateles, we used a cloning protocol designed to select for young L1 elements (Cantrell et al. 2000; Casavant et al. 2000). This method depends on the higher likelihood of intact ORFs in younger than in older L1 elements. Degenerate primers were used to amplify a portion of the conserved reverse transcriptase domain of ORFII (Region B in Fig. 1A). The PCR product was then cloned in a modified bacterial cloning vector (pBS II KS+ vector; Stratagene) that does not contain a functional LacZ reading frame but can produce a functional LacZ fusion peptide if an L1 ORF amplified with the degenerate primers is inserted appropriately. When electroporated into E. coli strain TG1, plasmids containing an L1 fragment with an intact ORF yielded blue colonies, while L1 elements with an interrupted ORF yielded white colonies. Clones from blue colonies were then sequenced. The modified vector was kindly provided by Dr. Holly Wichman (University of Idaho, Moscow).
DNA Blotting and Hybridization Conditions
DNAs were digested with NlaIII. About 50 ng of each DNA digest was electrophoresed on a 6% acrylamide gel and electroblotted to Zeta-probe membranes. Alternatively, DNA dot-blots were applied manually on Zeta-probe membranes. Membranes were hybridized to family-specific [32p]GTP-labeled oligonucleotide in the presence of competitor oligonucleotides (described by Verneau et al. 1997).
Sequence Analysis
Sequences were aligned manually. Phylogenetic analyses were performed using the PHYLIP 3.5c computer package (Felsenstein 1993). The percentage nucleotide substitution between sequences was calculated using Kimura’s (1980) two-parameter method. Trees were built with the neighbor-joining method (Saitou and Nei 1987) and their robustness was assessed using a bootstrap procedure. The aligned sequences (in fasta format) are available by anonymous FTP from helix.nih.gov/usr/ftp/pub/avf/Align_NWM_ Region_A_aIl_seq.
Results
Analysis of Region A
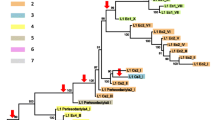
Twenty-six PCR-derived clones from each of Saimiri sciureus, Saguinus oedipus, and Ateles paniscus and six clones from Macaca mulatta were sequenced. The sequences were aligned together with the consensus sequence of the most recently evolved L1 subfamily in human, the Ta-1 subfamily. Insertions were removed from the alignment to keep the sequences in frame. Phylogenetic trees (Fig. 2) were built with the neighbor-joining (NJ) method based on the Kimura two-parameter distance, using the observed transition-to-transversion ratio of 2.1:1. The Saimiri and the Saguinus trees (trees A and B in Fig. 2, respectively) have the typical cascade structure expected if L1 evolution is driven by a single major lineage (Clough et al. 1996). In both cases the cascade structure ultimately leads to a group of closely related sequences suggestive of recent L1 amplification in both species. In Saimiri the divergence of the five most similar sequences is 1.36% (SE, 0.42%), compared to 2.46% (SE, 0.51%) for the five most similar Saguinus sequences. Thus, L1 may have been active more recently in Saimiri than in Saguinus. Using a NWM pseudogene rate of 0.21%/My (derived from Table 8.2 of Li 1997), we calculated that the most recent L1 inserts in our Saimiri and Saguinus samples are ~1.3 and ~3.4 My old, respectively. These are very rough estimates because of the short length of the sequences analyzed here.
Phylogenetic trees of L1 elements based on region A of ORFII. Trees were built using the neighbor-joining method based on Kimura’s two-parameter distances. Sequences AL022150-2 and AL022150-3 were used as outgroups because they have 75 of the 84 characters in region A that distinguish the modern human L1 family from the L1P5 family (Smit et al. 1995), an ancestral L1 family that amplified before the split between New World monkeys and Old World monkeys and apes. Boxes indicate the sequences that have the characters diagnostic of the four hybridization probes. Trees built using (A) Saimiri sciureus sequences, (B) Saguinus oedipus sequences, and (C) Ateles paniscus sequences.
The topology of the Ateles tree (tree C; Fig. 2) differs from that of the Saimiri and Saguinus trees: the cascade structure is less obvious and the tree does not show any group of closely related sequences as observed on trees A and B. Some NWM sequences share a number of characters that were not present in humans. Probe 1 (Fig. 3) is specific to several of these characters and hybridizes to the three NWMs DNAs but not to human, macaque, or lemur DNA (Fig. 4). Therefore L1 sequences that have these changes amplified after the split between NWMs and OWMs and apes. The solid gray boxes in Fig. 2 indicate the sequences that have this NWM specific character. Interestingly, 73% (19/26) of the Saimiri sequences and 65% (17/26) of the Saguinus sequences have this character, while only 8% (2/26) of the Ateles sequences have it.
Alignment of primate L1 subfamilies consensus sequences. The sequence at the top of the alignment corresponds to an “ancestral” sequence we reconstructed based on the alignment in Fig. 1 of Smit et al. (1995). These authors defined nine major L1 families in human based on the last 683 bp of ORFII and the 3′UTR arranged from the oldest one (L1M4) to the youngest one (L1P1). We used their alignment to infer at each position the ancestral state of the last common ancestor which would be the nucleotide found in the sequences of the families that predate family L1P4, the last family to amplify before the split between NWMs and OWMs and apes. The age of L1P4 is known because an L1 element belonging to family L1P4 is found in the γ1-globin gene in human and spider monkey but not in galago, tarsier, and rabbit. The human sequence (Homo Ta-1) is the consensus sequence of the most recently derived L1 subfamily in human. The Macaca, Saguinus, and Saimiri consensus sequences were derived from the five most closely related sequences in each species. The L1nwm-old consensus was derived from the four sequences in the dashed-line box in Fig. 2C. Numbered boxes correspond to the oligonucleotide probes used for hybridization.
Hybridization of various L1 probes to blot of primates DNA. Each panel shows the result of overnight hybridization with a different probe as described under Materials and Methods. The sequences of the probes are shown in Fig. 3. The top three panels are Southern blots of NlaII-digested DNAs and the bottom panel is a dot blot. Lc—Lemur catta; So—Saguinus oedipus; Ss—Saimiri sciureus; Ap—Ateles paniscus; Mm—Macaca mulatta; Hs—Homo sapiens. Probe 1, hybridization with competitor at 45°C; probe 2, no competitor, at 40°C; probe 3, with competitor, at 50°C; probe 4, with competitor, at 45°C.
The foregoing observations suggest that far more of the NWM specific L1 elements have been inserted in the genome of Saguinus and Saimiri than in Ateles since these species diverged ~25 Mya. To confirm the lack of young L1 elements in Ateles, more clones were collected, but this time we screened specifically for younger elements. E. coli colonies were transferred to Hybond-N nylon membranes (Amersham) using the procedure described in (Buluwela et al. 1989). The membranes were hybridized to the NWMs specific probe (probe 1 in Fig. 3). Twelve positive clones were picked and sequenced. Despite this additional cloning, we failed to find a group of young L1 sequences in Ateles as we found in Saguinus and Saimiri. The divergence of these additional sequences ranged from 3.4 to 14.1% and the five most similar sequences are 5.14% (SE = 0.79%) divergent. This apparent lack of L1 sequences younger than 8 My in Ateles suggests that the amplification of L1 elements was strongly reduced in the Ateles lineage.
Figure 5A shows a phylogenetic tree of the NWM L1 lineages. For simplicity, we selected the most similar sequences from each species; trees built with other groups of sequences (not shown) were similar to the one shown here. We added to this data set two human and two macaque sequences and three sequences representative of a group of divergent sequences identified in Fig. 2C by a dashed gray box (called L1nwm-old in Fig. 5A). This L1 tree mimics the species tree shown in Fig. 1B, suggesting that L1 has amplified in each species leading to species-specific clusters of sequences. The Ateles, Saguinus, and Saimiri specific L1 clusters are supported by relatively high bootstrap values but the relationship among these three clusters is not fully resolved.
Phylogenetic relationship between NWM L1 lineages. A Neighbor-joining tree based on Kimura’s two-parameter distances. The Ateles sequences that begin with N are derived from clones selected with probe 1 as described under Results. The number at a particular node indicates its percentage of appearance in 500 bootstrap replicates. Only values >75% are indicated. B Parsimony tree of consensus sequences. The tree is rooted using the ancestral sequence shown in Fig. 2. The branch lengths are proportional to the number of characters supporting each branch. CpG dinucleotides were excluded from this analysis because in most cases it is not possible to determine the ancestral state at these hypermutable sites.
The divergent sequences cloned from Ateles cluster together and this group (L1nwm-old) is supported by a high bootstrap value. The L1nwm-old clade branches outside the (Ateles (Saguinus, Saimiri)) clade and therefore probably split from the main L1 lineage before the three species diverged from each other but after the split between NWMs and OWMs and apes. This was confirmed by hybridization with probe 2 (Fig. 4), which hybridized to the three NWM DNAs but not to other primate DNAs. The difference among the intensities of hybridization to the three species indicates that L1nwm-old amplified differentially in the three species. Thus, L1nwm-old apparently remained active (at least in Saguinus and Ateles) after the three species diverged from each other. This is supported by the fact that the least divergent L1nwm-old sequences differ by only 8%, which corresponds to 19 My. Although this is a very rough estimate, it is consistent with the hypothesis that L1nwm-old remained active in Ateles after Ateles diverged from the Saguinus/Saimiri clade, about 25 My ago. The fact that L1nwm-old sequences were recovered only from the DNA of Ateles is not really surprising because the abundance of young elements in Saimiri and Saguinus would dominate the population of PCR products, making the recovery of old L1 lineages problematic.
We then examined the evolution of the active L1 lineage in NWMs. We first determined the consensus sequences (Fig. 3) of the most recently active subfamily in each species and then the evolutionary relationships between these consensus sequences (Fig. 5B). The parsimony tree in Fig. 5B reveals that the active lineage of L1 has evolved at very different rates in NWMs. In Saimiri and Saguinus, L1 has evolved into species-specific subfamilies that are characterized, respectively, by eight and six diagnostic characters. The species specificity of these families was verified by hybridization with probes 3 (Saimiri specific) and 4 (Saguinus specific; Figs. 2 and 4). In contrast, the consensus built with Ateles sequences shows only one (non-CpG) character that would be specific to Ateles. However, this character is also found sporadically in older subfamilies and therefore might be a hypervariable site. Therefore, the most recently active L1 lineage has accumulated more changes in Saimiri and Saguinus than in Ateles.
Analysis of Region B
To confirm the difference between L1 amplification and evolution in the NWMs studied here, we used another method to distinguish old and new L1 families. As described by Cantrell et al. (2000), PCR products of region B were cloned in frame into lacZ. Clones with intact reading frames should yield blue colonies, while clones with interrupted reading frames should yield white ones. The fraction of blue colonies was significantly higher (p = 0.000, Fisher’s exact test) in Saimiri (63.4%; 298 colonies examined) and Saguinus (62.2%; 399 colonies) than in Ateles (35.4%; 223 colonies). This result confirms the above finding that Saimiri and Saguinus contain a larger proportion of young (i.e., with an intact reading frame) L1 elements than Ateles. This observation is consistent with the lack of recent L1 amplification in the Ateles lineage deduced from the analysis of region A. Twenty-three blue colonies were picked, sequenced (9 from Ateles and 7 each from Saimiri and Saguinus), and used in a phylogenetic analysis (Fig. 6). This NJ tree shows that most Saguinus and Saimiri sequences are closely related and that they cluster into species-specific clades. On the other hand, Ateles sequences are very divergent from each others and do not constitute a monophyletic lineage. This tree topology substantiates our conclusions from examining region A of ORFII (Fig. 5B), namely, that species-specific lineages have evolved in Saimiri and Saguinus but not in Ateles.
Phylogenetic tree of L1 elements based on region B of ORFII. The tree was built using the neighbor-joining method based on Kimura’s two-parameter distances. The number at a particularnode indicates its percentage of appearance in 500 bootstrap replicates. Only values >75% are indicated. The tree is rooted with the consensus of ancestral subfamily L1PA8.
Discussion
Here we investigated the evolutionary history of L1 retrotransposons in NWM in which L1 evolution had not yet been examined. We found that episodes of L1 amplification occurred at different times in three NWM species. Whereas L1 amplified recently in Saimiri and in Saguinus, L1 activity seems to have been strongly reduced in the Ateles lineage for the last ~10 My. In both Saimiri and Saguinus the active L1 lineage has evolved and has generated new species-specific subfamilies. In contrast, we could not find convincing evidence for new L1 subfamilies in the Ateles lineage. In fact, our results suggest that the last amplification of a discernable L1 family in this species occurred more than 19 Mya. These results were obtained by analyzing two different regions of ORFII with different methods. As both methods were PCR-based, any bias would favor yielding younger elements (more likely to retain nonmutated PCR primer sites). Therefore, we are confident that our observations are not experimental artifacts and correspond to real differences in the rate of evolution and amplification of L1 in Ateles, Saguinus, and Saimiri.
Either L1 activity is so low in Ateles that any active family will have too few copies to be discernible above the background of ancestral L1 elements or L1 is now extinct in this species. Demonstrating the extinction of L1 is not easy. Casavant et al. (2000) suggested that such a conclusion should satisfy three criteria: (1) no taxon-specific band observed by Southern blot; (2) no recently inserted elements isolated by methods designed to enrich for young elements; and (3) no genomewide pattern of L1 distribution observed by in situ hybridization. Using these criteria, these authors showed that L1 is probably extinct in a group of South American rodents (genus Oryzomys) (Casavant et al. 2000).
In our case, the second criterion is clearly met but not the first and the third. The reason could be that these criteria are more likely to be met by rapidly evolving rodents than by the far more slowly evolving primates (Li 1997). For instance, Fig. 4 reveals little, if any, hybridization by probe 1 to taxon-specific bands in Saguinus genomic DNA, although L1 has evolved and amplified in this species. Similarly, bands corresponding to old subfamilies are not going to vanish as rapidly as in rodents because their restriction sites will not degenerate as fast as in rodents (as shown for the L1nwm-old clade, probe 2 in Fig. 4) As L1 evolution requires L1 replication, it is not surprising that species-specific L1 clades arose in Saguinus and Saimiri but not in Ateles. In the former two species, L1 amplification (and thus replication) was robust compared to Ateles, where L1 amplification was very low. Indeed, the most recently active NWM L1 family is found in Saimiri, where the active L1 lineage has experienced the largest number of changes.
Rather than going extinct in Ateles, L1 may just be in a low-activity phase. Alternating episodes of high and low activity typify L1 evolution in murine rodents (Pascale et al. 1993; Verneau et al. 1998). Among other possibilities, cyclic repression of L1 activity and subsequent escape by a novel L1 family could explain the episodic nature of L1 amplification. The host could adapt to deleterious L1 activity by repressing it directly or indirectly (e.g., altering a host factor required for L1 replication). As L1 replication is error prone (Furano 2000), a low level of L1 replication could generate and maintain a repository of variants but not produce a discernible family, especially if numerous variants are contemporaneously active. Any variant that can bypass host repression (or regain effective interaction with putative essential host factors) could produce a full-blown L1 amplification.
Primate L1 evolution may be consistent with this model. First, at times, L1 activity has been seriously deleterious as judged by strong purifying selection against only full-length L1 elements (those capable of replication [Boissinot et al. 2001]). Presumably, the deleterious effect of L1 was due to high L1 activity. Second, L1 concurrently underwent adaptive evolution. This suggests that the host responded to the deleterious L1 activity (Boissinot and Furano 2001). Both the purifying selection against L1 elements and their adaptive evolution greatly decreased in the African apes (Boissinot and Furano 2001). Interestingly, the accumulation of both L1 and SINE (thought to be retroposed by L1) elements also dramatically decreased about the time that African apes emerged 14–16 Mya (Consortium MGS 2002). As for the currently active human L1Pa1 family, it may be too early to judge whether this family represents a departure from the current phase of low L1 activity.
In NWMs a single major L1 lineage has evolved and is responsible for most of the recent L1 amplification. However, the common ancestor of the three NWM species contained another L1 lineage (L1nwm-old) that was also active and probably generated inserts after the three species diverged from each other, as indicated by the hybridization experiment (Fig. 4). L1nwm-old seems to have been active until at least 19 Mya. As the three NWM species diverged ~25 Mya, L1nwm-old and the major active lineage could have coexisted for at least 6 My and probably longer because the major lineage was clearly differentiated before the split among the three species. Thus, two active L1 lineages coexisted for several million years during the evolution of NWMs.
The persistence of more than one active L1 lineage for any length of time is unusual in mammals, although this conclusion is based on an admittedly small number of species. In humans only a single active lineage comprising five major families has persisted for at least the last 25 My (Boissinot et al. 2000; Smit et al. 1995). Although several differentiated subsets (subfamilies) within these families have been concurrently active, one ultimately attained replicative dominance (A.V. Furano, unpublished observations; Boissinot et al. 2001). The same situation occurred in old world rats and mice (murine rodents) (Furano 2000). However, as L1 families have not been nearly as well catalogued in murine rodents as in humans, this conclusion may need to be modified.
Nonetheless, until the present work, the only unequivocal exception to the single L1 lineage model was found in the new world rodent, Peromyscus (the deer mouse). Here two active lineages persisted and evolved for at least 10 My (Casavant et al. 1996). One possible explanation for the existence of a single L1 lineage is competition between active L1 elements for a limiting host factor(s) essential for L1 replication. As the competition would presumably be reduced during periods of low L1 activity, the latter condition could favor the coexistence of multiple active L1 lineages. In fact, multiple L1 lineages seem to typify nonmammalian species which do not support the high level of L1 activity possible at times in mammals (Eickbush and Furano 2002).
References
S Boissinot AV Furano (2001) ArticleTitleAdaptive evolution in LINE-1 retrotransposons. Mol Biol Evol 18 2186–2194 Occurrence Handle1:CAS:528:DC%2BD3MXptFCrs7k%3D Occurrence Handle11719568
S Boissinot P Chevret AV Furano (2000) ArticleTitleL1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17 915–928 Occurrence Handle1:CAS:528:DC%2BD3cXjvFygs7g%3D Occurrence Handle10833198
S Boissinot A Entezam AV Furano (2001) ArticleTitleSelection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 18 926–935 Occurrence Handle1:CAS:528:DC%2BD3MXktFemtLc%3D Occurrence Handle11371580
L Buluwela A Forster T Boehm TH Rabbitts (1989) ArticleTitleA rapid procedure for colony screening using nylon filters. Nucleic Acids Res 17 452 Occurrence Handle1:CAS:528:DyaL1MXptFClug%3D%3D Occurrence Handle2911481
FH Burton DD Loeb CF Voliva SL Martin MH Edgell III CA Hutchison (1986) ArticleTitleConservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol 187 291–304 Occurrence Handle1:CAS:528:DyaL28XhtVentbc%3D Occurrence Handle3009828
EL Cabot B Angeletti K Usdin AV Furano (1997) ArticleTitleRapid evolution of a young L1 (LINE-1) clade in recently speciated Rattus taxa. J Mol Evol 45 412–423 Occurrence Handle1:CAS:528:DyaK2sXms1Wls7Y%3D Occurrence Handle9321420
MA Cantrell RA Grahn L Scott HA Wichman (2000) ArticleTitleIsolation of markers from recently transposed LINE-1 retrotransposons. Biotechniques 29 1310–1316 Occurrence Handle1:CAS:528:DC%2BD3cXovFajtbs%3D Occurrence Handle11126134
NC Casavant AN Sherman HA Wichman (1996) ArticleTitleTwo persistent LINE-1 lineages in Peromyscus have unequal rates of evolution. Genetics 142 1289–1298 Occurrence Handle1:CAS:528:DyaK28XivVGjtbc%3D Occurrence Handle8846905
NC Casavant L Scott MA Cantrell LE Wiggins RJ Baker HA Wichman (2000) ArticleTitleThe end of the LINE?: Lack of recent L1 activity in a group of South American rodents. Genetics 154 1809–1817 Occurrence Handle1:STN:280:DC%2BD3czpsFOgug%3D%3D Occurrence Handle10747071
AP Clements MF Singer (1998) ArticleTitleThe human LINE-1 reverse transcriptase:effect of deletions outside the common reverse transcriptase domain. Nucleic Acids Res 26 3528–3535 Occurrence Handle1:CAS:528:DyaK1cXltlKqtrY%3D Occurrence Handle9671814
JE Clough JA Foster M Barnett HA Wichman (1996) ArticleTitleComputer simulation of transposable element evolution: random template and strict master models. J Mol Evol 42 52–58 Occurrence Handle1:CAS:528:DyaK28XntFOmsQ%3D%3D Occurrence Handle8576964
IHGS Consortium (2001) ArticleTitleInitial sequencing and analysis of the human genome. Nature 409 860–921 Occurrence Handle1:CAS:528:DC%2BD3MXhsFCjtLc%3D Occurrence Handle11237011
MGS Consortium (2002) ArticleTitleInitial sequencing and comparative analysis of the mouse genome. Nature 420 520–562 Occurrence Handle10.1038/nature01262 Occurrence Handle12466850
RJ DeBerardinis HH Jr Kazazian (1998) ArticleTitleFull-length L1 elements have arisen recently in the same 1-kb region of the gorilla and human genomes. J Mol Evol 47 292–301 Occurrence Handle1:CAS:528:DyaK1cXmt1antro%3D Occurrence Handle9732456
TH Eickbush AV Furano (2002) ArticleTitleFruit flies and humans respond differently to retrotransposons. Curr Opin Genet Dev 12 669–674 Occurrence Handle10.1016/S0959-437X(02)00359-3 Occurrence Handle1:CAS:528:DC%2BD38XosFOhsrc%3D Occurrence Handle12433580
J Felsenstein (1993) PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author, Department of Genetics, University of Washington Seattle
AV Furano (2000) ArticleTitleThe biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acids Res Mol Biol 64 255–294 Occurrence Handle1:CAS:528:DC%2BD3cXivVCnsL0%3D
AV Furano BE Hayward P Chevret F Catzeflis K Usdin (1994) ArticleTitleAmplification of the ancient murine Lx family of long interspersed repeated DNA occurred during the murine radiation. J Mol Evol 38 18–27 Occurrence Handle1:CAS:528:DyaK2cXpslKltg%3D%3D Occurrence Handle8151711
M Goodman CA Porter J Czelusniak SL Page H Schneider J Shoshani G Gunnell CP Groves (1998) ArticleTitleToward a phylogenetic classification primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol 9 585–598 Occurrence Handle1:STN:280:DyaK1czjsVGrsQ%3D%3D Occurrence Handle9668008
SC Hardies SL Martin CF Voliva III CA Hutchison MH Edgell (1986) ArticleTitleAn analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol Biol Evol 3 109–125 Occurrence Handle1:CAS:528:DyaL28Xhs1KjsL0%3D Occurrence Handle3444397
HH Jr Kazazian (1998) ArticleTitleMobile elements and disease. Curr Opin Genet Dev 8 343–350 Occurrence Handle1:CAS:528:DyaK1cXkvVWhu7k%3D Occurrence Handle9690999
M Kimura (1980) ArticleTitleA simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16 111–120 Occurrence Handle7463489
W-H Li (1997) Molecular evolution. Sinauer Associates Sunderland MA
LM Mathews SY Chi N Greenberg I Ovchinnikov GD Swergold (2003) ArticleTitleLarge differences between LINE-1 amplification rates in the human and chimpanzee lineages. Am J Hum Genet 72 739–748 Occurrence Handle10.1086/368275 Occurrence Handle1:CAS:528:DC%2BD3sXitFajs7s%3D Occurrence Handle12574987
JV Moran SE Holmes TP Naas RJ DeBerardinis JD Boeke HH Jr Kazazian (1996) ArticleTitleHigh frequency retrotransposition in cultured mammalian cells. Cell 87 917–927 Occurrence Handle1:CAS:528:DyaK28Xntlyqs70%3D Occurrence Handle8945518
EM Ostertag HH Jr Kazazian (2001) ArticleTitleBiology of mammalian L1 retrotransposons. Annu Rev Genet 35 501–538 Occurrence Handle10.1146/annurev.genet.35.102401.091032 Occurrence Handle1:CAS:528:DC%2BD38XlsVOg Occurrence Handle11700292
E Pascale C Liu E Valle K Usdin AV Furano (1993) ArticleTitleThe evolution of long interspersed repeated DNA (L1, LINE 1) as revealed by the analysis of an ancient rodent L1 DNA family. J Mol Evol 36 9–20 Occurrence Handle1:CAS:528:DyaK3sXls12ksA%3D%3D Occurrence Handle8433380
N Saitou M Nei (1987) ArticleTitleThe neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 Occurrence Handle1:STN:280:BieC1cbgtVY%3D Occurrence Handle3447015
AFA Smit G Tóth AD Riggs J Jurka (1995) ArticleTitleAncestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol 246 401–417
MJ Stanhope DA Tagle MS Shivji M Hattori Y Sakaki JL Slightom M Goodman (1993) ArticleTitleMultiple L1 progenitors in prosimian primates: Phylogenetic evidence from ORF1 sequences. J Mol Evol 37 179–189 Occurrence Handle1:CAS:528:DyaK3sXlvFOjtL0%3D Occurrence Handle8411207
O Verneau F Catzeflis AV Furano (1997) ArticleTitleDetermination of the evolutionary relationships in Rattus sensu lato (Rodentia: Muridae) using L1 (LINE-1) amplification events. J Mol Evol 45 424–436 Occurrence Handle1:CAS:528:DyaK2sXms1Wltrs%3D Occurrence Handle9321421
O Verneau F Catzeflis AV Furano (1998) ArticleTitleDetermining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proc Natl Acad Sci USA 95 11284–11289 Occurrence Handle10.1073/pnas.95.19.11284 Occurrence Handle1:CAS:528:DyaK1cXmt1agsL4%3D Occurrence Handle9736728
CF Voliva CL Jahn MB Comer III CA Hutchison MH Edgell (1983) ArticleTitleThe L1Md long interspersed repeat family in the mouse: Almost all examples are truncated at one end. Nucleic Acids Res 11 8847–8859 Occurrence Handle1:CAS:528:DyaL2cXmt1OksQ%3D%3D Occurrence Handle6324106
Acknowledgements
The authors thank the Wuppertal Zoological Garden, the Leipzig Zoological Garden, and the Münster Zoological Garden for providing DNA samples.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boissinot, S., Roos, C. & Furano, A.V. Different Rates of LINE-1 (L1) Retrotransposon Amplification and Evolution in New World Monkeys . J Mol Evol 58, 122–130 (2004). https://doi.org/10.1007/s00239-003-2539-x
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-003-2539-x