Abstract
Copper chaperones bind intracellular copper and ensure proper trafficking to downstream targets via protein–protein interactions. In contrast to the mechanisms of copper binding and transfer to downstream targets, the mechanisms of initial copper loading of the chaperones are largely unknown. Here, we demonstrate that antioxidant protein 1 (Atox1 in human cells), the principal cellular copper chaperone responsible for delivery of copper to the secretory pathway, possesses the ability to interact with negatively charged lipid headgroups via distinct surface lysine residues. Moreover, loss of these residues lowers the efficiency of copper loading of Atox1 in vivo, suggesting that the membrane may play a scaffolding role in copper distribution to Atox1. These findings complement the recent discovery that the membrane also facilitates copper loading of the copper chaperone for superoxide dismutase 1 and provide further support for the emerging paradigm that the membrane bilayer plays a central role in cellular copper acquisition and distribution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Copper is an essential element for life, serving as a redox cofactor for many biological processes such as oxidative phosphorylation, antioxidant defense, neurotransmitter biosynthesis and iron transport (Linder and Goode 1991). However, the same redox properties that enable utility in biology also render the element toxic, either by catalyzing formation of damaging radicals via Fenton chemistry or by adventitious incorporation into nonspecific ligands replete in the cell (Bremner 1998; Halliwell and Gutteridge 1990). The requirement for the cell to accommodate both facets of copper chemistry is reflected by a spectrum of pathologies manifesting from copper deficiency (i.e. Menkes disease) to excess (Wilson disease) (Daniel et al. 2004; Gupta and Lutsenko 2009). Thus, to properly utilize copper while preventing toxicity, cells have evolved homeostatic distribution mechanisms that regulate ion flux (via copper transporters) and confine metal activity through coordination with cytosolic copper chaperones that, in turn, escort copper ions to target destinations (O’Halloran and Culotta 2000; Rosenzweig 2002).
Antioxidant protein 1 (Atx1 in yeast), the first copper chaperone to be discovered in Saccharomyces cerevisiae, was originally identified as a suppressor of strains lacking the protein superoxide dismutase (SOD) (Lin and Culotta 1995). While the mechanisms of its protective role against oxidative stress are unclear, the intersection of Atx1 with copper trafficking pathways arose from the discovery that the protein contributes to copper incorporation into the multicopper oxidase Fet3p via the secretory pathway, independent of its antioxidant function (Lin et al. 1997). Subsequent genetic and biochemical experiments have revealed that Atox1 (human ortholog of Atx1) delivers copper to P1B-ATPases (Hamza et al. 1999; Larin et al. 1999; Walker et al. 2002), which in turn translocate copper across membranes from the cytosol and into the lumen of the Golgi, intracellular compartments and vesicles (Pufahl et al. 1997; Lutsenko et al. 2008). Because of its central role in copper shuttling, it is not surprising that Atx1 is highly conserved, with orthologs identified in human, flies, worms and plants (Klomp et al. 1997; Southon et al. 2004; Wakabayashi et al. 1998; Himelblau et al. 1998).
Atox1 binds copper via two cysteine residues in a conserved CxxC motif (Arnesano et al. 2002). Subsequent biochemical and structural studies have shed light on possible copper transfer mechanisms to downstream metal binding domains within the target Cu-ATPases. NMR structures of Atx1 or Atox1 with a downstream metal binding domain reveal a copper-dependent heterocomplex whose structure suggests that copper transfer occurs via ligand exchange reactions that involve a tricoordinate Cu(I) species with cysteine ligands from both donor and acceptor proteins (Arnesano et al. 2001a; Banci et al. 2006, 2009).
While the mechanisms of copper transfer to downstream partners have been extensively studied, far less is known about how Atox1 proteins, as well as the other copper chaperones, acquire their cargo. Copper ion import into the eukaryotic cell is primarily achieved through membrane-bound transporters of the CTR family (Dancis et al. 1994; Kim et al. 2008). A central question that has remained unanswered is how chaperones find and receive copper after its initial translocation across the plasma membrane through the action of CTR proteins. Addressing this question, two diametrically opposed models have been proposed, that chaperones may obtain their cargo either through direct interactions with the transporters or from a readily exchangeable cytosolic copper pool that is fueled through nonspecific release of copper from the import transporters. Circumstantial evidence has been reported in support of both mechanisms (Xiao and Wedd 2002; Maryon et al. 2013), but definitive answers have remained elusive. A common problem with both proposed mechanisms is that they rely on 3D-random walk searches by the chaperones. The associated low success rate of productive molecular encounters raises questions of whether either mechanism on its own could sustain a tightly controlled homeostatic network of proteins that simultaneously functions to deliver copper to proper targets and to prevent nonspecific binding of copper to inappropriate intracellular components. Mitigating many of the problems inherently linked to 3D-random walks, recent evidence has implicated a role of the bilayer in facilitating the copper trafficking pathway of the copper chaperone for SOD (CCS) toward its target Cu/Zn SOD (SOD1) (Pope et al., submitted). Building on this discovery, we here explore whether the second principal cellular copper chaperone, Atox1, can engage the membrane bilayer as well and if any such ability has an impact on its physiological copper loading status.
Materials and Methods
Cloning and Mutagenesis of Atox1
The cDNA encoding Atox1 was excised from a pET21b Atox1 construct (kindly provided by Dr. Amy Rosenzweig, Northwestern University, Evanston, IL, USA) using NdeI and XhoI and subsequently cloned into the pET15b vector (Novagen, Madison, WI, USA), which introduces an N-terminal HIS6 tag followed by a thrombin cleavage site. All Atox1 mutations were generated by QuikchangeTM PCR mutagenesis protocols (Stratagene, La Jolla, CA, USA). For tetracycline-inducible expression in mammalian cells, Atox1 cDNA sequences were amplified by PCR and subcloned into the KpnI-ApaI cloning sites within the pcDNA5 FRT/TO CAT plasmid (Invitrogen, Carlsbad, CA; kindly provided by Dr. Svetlana Lutsenko, Johns Hopkins University, Baltimore, MD, USA). Subsequent mutation of the nonessential Cys41 residue to serine (C41S) was conducted by QuikchangeTM PCR mutagenesis. The presence of all desired mutations as well as construct fidelity were verified by automated DNA sequencing methods.
Protein Expression and Purification of Atox1 Mutants
pET15b Atox1 constructs were transformed into BL21(DE3) competent cells and plated on Luria broth (LB) plates containing 100 μg/ml ampicillin. Single colonies of each construct were used to inoculate 1 l of LB/Amp medium at 37 °C. When culture growth reached an OD600 of 0.4–0.6, cells were cold-shocked at 4 °C for 45 min, followed by induction with 0.3 mM IPTG for 18 h at 18 °C with shaking at 220 rpm. Cells were harvested by centrifugation at 3,500×g for 30 min, followed by washing with 10 mM MOPS (pH 7.4) and 300 mM NaCl (buffer A) and storage of induced cells at −80 °C.
For protein purification, induced cells were resuspended in 50 ml buffer A with one cOmpleteTM EDTA-free protease inhibitor cocktail tablet (Roche, Indianapolis, IN, USA). Cell lysis was achieved by two passes through a cell disruptor at 25,000 psi (TS Benchtop model; Constant Systems Inc, Kennesaw, GA, USA), and lysates were clarified by centrifugation at 40,000 rpm for 45 min at 10 °C in a Beckman type 45Ti rotor (Beckman Coulter, Brea, CA, USA). Imidazole (30 mM) and 5 mM 2-mercaptoethanol were added to the supernatant, which was subsequently passed though a gravity column at 4 °C packed with TALON® cobalt metal affinity resin (Clontech, Mountain View, CA, USA), preequilibrated with 10 mM MOPS (pH 7.4), 300 mM NaCl, 30 mM imidazole and 5 mM 2-mercaptoethanol (buffer B). Columns were washed with 20 column volumes of buffer B, and HIS6 Atox1 proteins were eluted in 25 ml of buffer A with 300 mM imidazole and 5 mM 2-mercaptoethanol. Eluates were then concentrated to ~1 ml via a 3,000 MWCO Ultracel-3 device (Millipore, Billerica, MA, USA), loaded in 500-μl batches onto a Superdex 75 10/300 GL column (GE Healthcare, Waukesha, WI, USA) and eluted with 10 mM MOPS (pH 7.4), 100 mM NaCl, 0.2 mM TCEP and 10 % glycerol. Fractions containing pure, monomeric Atox1 proteins were frozen in liquid nitrogen and stored at −80 °C. Protein concentrations were determined by the Pierce 660 nm reagent (Thermo Scientific, Waltham, MA, USA) using BSA as a standard.
For copper loading experiments, pooled eluates from the cobalt resin were digested with 200 U thrombin (MP Biochemicals, Santa Ana, CA, USA) for 60 h at 4 °C while simultaneously dialyzing against 50 mM MOPS (pH 7.4) and 150 mM NaCl (buffer C) to remove imidazole. Digested samples were then reapplied onto a cobalt column (equilibrated with buffer C), and the flowthrough and washes (containing digested Atox1) were pooled and concentrated as above. Proteins were then subjected to gel filtration as above, and fractions containing pure thrombin-digested Atox1 proteins eluted in buffer C were frozen in liquid nitrogen and stored at −80 °C.
Liposome Floats
POPA, POPC, POPG, POPS and brain polar and yeast polar lipid extracts were purchased as chloroform stocks (Avanti Polar Lipids, Alabaster, AL, USA); and lipid films were prepared by first evaporating chloroform, then washing the lipids with hexane and finally drying the lipids under an argon stream. Liposomes were generated by hydrating lipid films in 10 mM MOPS (pH 7.4) and 100 mM NaCl (buffer D) and subjecting them to five rounds of rapid freeze–thaw cycles with liquid nitrogen.
To assess membrane binding, 400 µg liposomes was mixed with 30 μg HIS6 Atox1 proteins in a 100 μl final volume for 1.5–2 h at 25 °C. Sucrose density gradients were created by transferring the mixtures into 11 × 34-mm polycarbonate ultracentrifuge tubes (Beckman Coulter), overlaid with 500 μl buffer D + 38 % sucrose, followed by 150 μl buffer D + 10 % sucrose and finally 150 μl buffer D + 0.004 % bromophenol blue (to help distinguish the 0–10 % sucrose interface). Gradients were centrifuged at 40,000 rpm for 25 min at 10 °C in a Beckman TLS-55 rotor. We then carefully harvested 75 μl from the 10 % sucrose layer, which contained proteoliposomes, without disturbing interfaces.
Harvested samples were precipitated in 25 % trichloroacetic acid, with 0.5 μg BSA added as an internal reference for precipitation efficiency, and spun for 15 min, 20,000×g, 10 °C. Pellets were washed twice with 200 μl hexane to extract any precipitated lipids and dissolved in 10 μl 1 M Tris (pH 8.8). Recovered Atox1 samples thus subjected to sucrose gradients in the presence or absence of liposomes (background control) were loaded onto a 15 %/5 %C Tricine-SDS-PAGE gel, which included a standard curve of 0.2, 0.5 and 1 μg BSA to serve as a normalization factor for Coomassie staining between gels. Coomassie R-250-stained gels were imaged on an Alpha Innotech (San Leandro, CA, USA) FluorChem HD2 Imager. Unless indicated otherwise, all images were analyzed using ImageJ software (NIH, Bethesda, MD, USA). Background subtraction utilized the 50-pixel rolling ball radius light background option, and integrated density measurements for each band were recorded and subsequently corrected for differences in staining (between gels) via the BSA standard curves.
Cell Culture and Mammalian Protein Expression
HEK293T cells (HEK293TREx strain; kindly provided by Dr. Svetlana Lutsenko, Johns Hopkins University, Baltimore, MD) were maintained in MEM + GlutaMAXTM medium, supplemented with 10 % FBS, 1 % nonessential amino acids and 1 % Pen-Strep (Invitrogen). For protein expression, cells cultured on 100-mm dishes at 70–75 % confluence were transiently transfected with 14 μg pcDNA5 FRT/TO Atox1 plasmids with 21 μl Turbofect (Thermo Scientific), according to the manufacturer’s protocol. Cells were simultaneously induced via addition of 500 ng/ml doxycycline hyclate (Sigma, St. Louis, MO, USA) to the growth medium for 36–38 h prior to cell harvesting and lysis.
Cys-Targeted Labeling and Neutravidin ELISA
Induced cells, transiently transfected with pcDNA5 FRT/TO Atox1 constructs or vehicle control, were washed with ice-cold PBS and lysed in 600 μl 50 mM MOPS (pH 7.4), 150 mM NaCl, 0.2 mM TCEP, 0.1 % IGEPAL CA-630 and 10 % glycerol. For copper-stimulated and copper-chelated growth conditions, 200 μM CuCl2 or 50 μM bathocuproine disulfonate (BCS, Sigma) was added to the growth medium 2 or 20 h, respectively, prior to cell lysis. Cell lysates were then clarified by centrifugation at 20,000×g, 10 min at 10 °C, and subsequently diluted to 750 μg/ml total protein concentration (as determined by Pierce 660 nm reagent, using BSA standards). A portion of the diluted lysates was reserved for analysis of protein expression by Western blot using rabbit polyclonal Atox1 (Novus Biologicals, Littleton, CO, USA) and rat monoclonal α-tubulin (Millipore) antibodies. The remaining lysate was reacted with 0.8 mM EZ-LinkTM maleimide-PEG11-biotin (Thermo Scientific) for 4 h at 10 °C.
Following ex vivo labeling, the reacted lysates were diluted to 0.5 μg/ml in ELISA buffer (PBS + 0.05 % Tween-20 + 0.5 % BSA) and 100-μl aliquots of each sample were bound in triplicate to a neutravidin-coated 96-well plate (G-Biosciences, St. Louis, MO, USA) overnight at 4 °C. Wells were then washed three times with ELISA buffer and incubated with 100 μl polyclonal Atox1 primary antibody solution (at 1:1,500 dilution) for 1 h at room temperature. After washes and incubation with goat-anti-rabbit HRP secondary antibody (1:2,000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA), washed wells were developed with a 100-μl solution of 0.4 mg/ml o-phenylenediamine (Sigma) in 50 mM citrate, 50 mM sodium phosphate (pH 5.0) with 0.1 % H2O2 for 30 min in the dark. The developing reactions were quenched with 100 μl 2.5 M H2SO4, and absorbance readings at 490 nm were carried out in a Power Wave XS2 plate reader (BioTek, Winooski, VT, USA).
Copper Loading and ICP-MS Analysis
Thrombin-digested Atox1 mutants (100 μM) were reduced in the presence of 0.5 mM DTT for 30 min at 10 °C. One equivalent of CuSO4 was then incubated with reduced protein for 1.5 h at 10 °C, analogous to established protocols (Pufahl et al. 1997; Anastassopoulou et al. 2004). Mixtures were then transferred into 3.5 K MWCO mini-dialysis units (Pierce, Rockford, IL, USA) and dialyzed against buffer C to remove unbound copper. Protein concentrations of the harvested samples were recalculated using a small fraction of material, while the remaining sample was digested in nitric acid (final concentration 3 %) and diluted to ensure copper ion concentration in the parts per billion range. All samples included 5 ppb yttrium internal standard (Inorganic Ventures, Lakewood, NJ, USA). Copper concentrations within the samples were determined by comparison to a standard curve of 0–200 ppb copper (Inorganic Ventures), with a correlation coefficient >0.9999. All measurements were conducted on a X-Series II ICP-MS (Thermo Fisher) at the Quantitative Bioelemental Imaging Center at Northwestern University (Evanston, IL, USA).
Statistics
Data are expressed as mean ± SEM. For lipid binding experiments of Atox1 mutants, data are represented as mean band intensity of a particular mutant in the presence of lipids minus its corresponding mean band intensity in the absence of lipids, while error bars represent SEM values with proper error propagation analysis. Unless indicated otherwise, statistical significance was determined by one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison posttest using the GraphPad Prism6 software (GraphPad, La Jolla, CA, USA). p < 0.05 was considered statistically significant.
Results
Atox1 Binds to Membranes
To assess whether the membrane bilayer could play a role in copper acquisition by the chaperone Atox1, membrane binding experiments were conducted with purified protein, utilizing sucrose density liposome floats. An initial screen using chemically defined synthetic phospholipid mixtures revealed that, relative to the no lipid background, Atox1 showed preferential association with negatively charged moieties (Fig. 1). One-way ANOVA of the liposome float data showed no statistically significant binding of Atox1 toward the neutral POPC headgroups relative to background. In contrast, all tested liposome mixtures comprising 20 % POPC-80 % negatively charged headgroups (POPA, POPS and POPG) showed statistically significant association with Atox1. Moreover, the observed interaction between Atox1 and lipids was not merely electrostatic in nature, as evidenced by differences in association between the various negative lipids, where both PA and PG headgroups exhibited an ~5-fold increase in Atox1 over background, while the larger PS headgroup recovered an ~2.8-fold increase (Fig. 1, lanes 3–5). Consequently, both steric and electrostatic properties seemed to govern the association between Atox1 and lipids. While informative, this initial lipid binding screen was extended to more biologically relevant lipid substrates. Data shown in lanes 6 and 7 of Fig. 1 revealed statistically significant associations of the chaperone toward both yeast polar lipid extracts (~3-fold over background) and mammalian brain polar lipid extracts (~2-fold over background), respectively.
Atox1 binds membranes as assessed by densitometric analysis of liposome flotation assays. Bars represent mean ± SEM. Statistical significance relative to protein-only control (Atox1 only, n = 35) was determined by one-way ANOVA with Dunnett’s posttest. For 100 % PC lipids (PC), n = 3; for all other samples n = 6−9. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Specific Surface Residues of Atox1 Impart Association with Membranes
Figure 2a depicts the solution structure of apo-Atox1 colored with the electrostatics of charged surface residues (PDB code 1TL5) (Anastassopoulou et al. 2004). Adjacent to the copper binding site, which consists of cysteine residues Cys12 and Cys15 (colored orange) (Hung et al. 1998), is a lysine residue, Lys60, which is conserved among Atox1 homologs (Hatori and Lutsenko 2013). In addition, there exists a cluster of positive residues on the surface of Atox1, comprising residues Lys25, Lys56 and Lys57 as well as possibly Arg21, that lies on the same face as the copper binding site. To further investigate the determinants of the observed preferential interaction between Atox1 and negatively charged lipid substrates, alanine point mutations of these noted positive residues were constructed and assessed for their potential contribution toward membrane association. Each protein was purified as a properly folded, primarily monomeric species similar to wild type, as assessed by size exclusion chromatography (data not shown).
Atox1 contains positive residues that mediate membrane association. a NMR structure of apo-Atox1 (PDB code 1TL5) was rendered in Chimera with electrostatic surfaces of charged residues (red = negative, blue = positive). The molecule is oriented with copper binding residues Cys12 and Cys15 (orange) on top, with the nonconserved cysteine residue (Cys41) in green. b Densitometric analysis of recovered Atox1 mutants from 20 % POPC-80 % POPG liposome floats using ImageJ software. Triple mutant refers to combined K25A K56A K57A mutation. c Densitometric analysis of recovered Atox1 mutants from brain polar lipid (left) or yeast polar lipid (right) extracts, using AlphaView software (Protein Simple). For b and c, bars represent mean ± SEM. Statistical significance relative to wild type (n = 9−11) was determined for each mutant (n = 6−10) via one-way ANOVA with Dunnett’s posttest. *p < 0.05, ***p < 0.001, ****p < 0.0001
To minimize ambiguities, liposome floats were first conducted with mutant Atox1 proteins using the 80 % PG lipid substrate since it exhibited the greatest association with Atox1 WT (Fig. 1). Of the single-point mutants, mutation of Lys57 to alanine exhibited the greatest reduction (~22 % of wild type) in Atox1 association with PG lipids (Fig. 2b). Similarly, the K56A single-point mutation also exhibited a statistically significant decrease in membrane binding (~36 % of wild type), and effects of a simultaneous loss of both of these positive charges appeared slightly additive (K56A K57A; Fig. 2b, lane 7). Contrasting with K56 and K57, mutation of K25A alone did not yield a statistically significant decrease in liposome association. However, when combined with the K56A K57A double mutation, the further loss of positive charges in the K25A K56A K57A mutation (triple; Fig. 2b, lane 8) resulted in nearly complete abrogation of liposome binding (~4 % relative to wild type). Thus, the molecular determinant of the observed Atox1 interaction with membranes appears to reside in the lysine-rich ridge centered around Lys57.
Extending the experiments that used highly negative synthetic lipid substrates, mutants were further assayed for binding to more biologically relevant membranes. Figure 2c reveals that for both brain and yeast polar lipid extracts, the combined triple lysine mutant exhibited a statistically significant decrease in membrane association relative to wild type (~45 %). In contrast, the K60A single mutation did not result in a statistically significant change in association either toward natural lipid extracts or toward 80 % PG membranes. This behavior served as an internal negative control, suggesting that not all positively charged residues of the Atox1 protein were involved in interactions with the headgroup region of the membrane bilayer.
Membrane Binding-Deficient Mutants Reduce Copper Loading of Atox1 In Vivo
Atox1 contains three cysteine residues: Cys12 and Cys15 (comprising the conserved CxxC copper binding motif) as well as a nonconserved Cys41 that does not bind copper (Hung et al. 1998; Narindrasorasak et al. 2004). Upon copper ion binding, two of the three cysteine residues (Cys12 and Cys15) become refractory to modification, thus generating an inverse relationship between copper binding and incorporation of chemical probes such as maleimide. Confirming this property of the chaperones by mass spectrometry after labeling purified apo-Atox1 and Cu-Atox1 with maleimide (data not shown), we proceeded to determine whether the observed Atox1 association with membranes translated to a functional role for cellular copper acquisition. This approach seemed suitable because changes in copper status of Atox1 in vivo should affect the fraction of molecules that are amenable to chemical modification after cell lysis. Accordingly, the various Atox1 mutants were transfected in HEK293T cells and indirectly assayed for copper loading by ex vivo cys-targeted labeling with the maleimide-PEG11-biotin reagent. To avoid ambiguities, these experiments were carried out in an Atox1 C41S background since, regardless of assay design, modifications of the nonessential cysteine would contribute to background activities that are unrelated to copper binding. Quantitation of labeled Atox1 molecules within cell lysates was achieved via a sandwich ELISA, where labeled lysates were first bound onto a neutravidin-coated surface, then probed with Atox1 antibodies and subsequent horseradish peroxidase secondary antibodies. (An inverted approach of binding reacted lysates to Atox1 antibody-coated surfaces then probing for captured biotin yielded highly nonspecific background levels and was thus deemed unfeasible.)
To test the ability of this cys-labeling approach to report differences in cellular copper loading, Atox1 WT was first transfected into the HEK293T cell line and induced under various copper media conditions (Fig. 3a). Compared to basal medium (containing ~3 μM copper as measured by ICP-MS), stimulating cells with 200 μM CuCl2 for 2 h resulted in a ~45 % decrease in maleimide incorporation, suggesting an increase in copper-bound Atox1 molecules. Similarly high concentrations of exogenous copper were used in a previous report to demonstrate a copper-dependent coimmunoprecipitation between Atox1 and either ATP7A or ATP7B from cell lysates (Hamza et al. 1999). Moreover, pulsing cells with a high amount of extracellular copper appeared appropriate to ensure sufficient copper incorporation into the overexpressed proteins tested in our study. Conversely, chelating copper in the growth medium with 50 μM BCS for 20 h resulted in an ~2-fold increase in cysteine labeling, indicating that under such low copper conditions Atox1 was mostly copper-free.
Atox1 membrane binding mutants show decreased cellular copper loading. a Extent of cys labeling of Atox1 C41S transfected into HEK293T cells grown under basal, high copper (200 μM CuCl2) or copper-depleted (50 μM BCS) conditions, assessed by neutravidin ELISA. Bars represent mean ± SEM absorbance values, adjusted for relative expression levels as determined by Western blot and normalized to basal conditions. Statistical significance (n = 3) was determined by two-tailed unpaired Student’s t test. b Representative expression profile of Atox1 mutant proteins (in C41S background) transfected into HEK293T cells. Six micrograms of mock-transfected (Con) or Atox1 mutant lysates was loaded onto 16 %/6 %C Tricine-SDS-PAGE gels and subjected to Western blotting using α-tubulin antibody (upper panel) or Atox1 antibody (lower panel). c, d Extent of cys labeling of Atox1 mutants (in C41S background) transfected into HEK293T cells grown under 200 μM CuCl2 (c) or 50 μM BCS (d), as assessed by neutravidin ELISA. Bars represent mean ± SEM absorbance values, adjusted for relative expression levels and normalized to wild type (WT). Statistical significance relative to wild type was determined for each mutant (n = 8−9 for c, n = 3 for d) via one-way ANOVA with Dunnett’s posttest. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. For BCS-treated cells (d), the familywise p value was 0.3159 and deemed not statistically significant
Having established the applicability of this approach in detecting relative differences in copper loading of Atox1 molecules, the various Atox1 membrane binding mutants were next transfected into HEK293T cells. Figure 3b represents a typical expression profile of the Atox1 mutants, as determined by Western blot. All mutants expressed at comparable levels under both Cu-stimulated or Cu-depleted growth conditions, with deviations less than 25 % of wild type. This finding is consistent with other reports that examined the cellular levels of Atox1 under various copper conditions (Hamza et al. 1999). Nevertheless, to account for any differences in expression, raw ELISA absorbance values were adjusted to relative Atox1 mutant expression levels, as determined by concomitant Western blots.
When stimulated with 200 μM CuCl2, statistically significant increases in maleimide incorporation were observed among some of the Atox1 mutants (Fig. 3c). In particular, the K57A mutant exhibited a 25 % increase in label incorporation. Further loss of positive charge within the lysine-rich ridge of Atox1 yielded an increasing amount of labeled protein (and thus greater apo-Atox1) as the K56A K57A double mutant and K25A K56A K57A triple mutant showed ~1.4- and ~2-fold increases, respectively, relative to WT. In addition, the K60A mutant demonstrated a twofold increase in maleimide incorporation, demonstrating a defect in copper loading that was independent of the ability of the chaperone to engage the bilayer surface (Fig. 2b, c).
To address whether these mutations affect the reactivity of maleimide chemistry, a similar analysis was conducted under copper-chelated conditions. As demonstrated in Fig. 3a, pretreating cells with the copper chelator BCS rendered most of the transfected wild-type Atox1 molecules susceptible to label incorporation. When the various Atox1 mutants were subjected to such conditions, no differences in cys-directed labeling were observed (Fig. 3d), thereby suggesting that increases in maleimide incorporation observed under copper-stimulated conditions (Fig. 3c) were due to a reduced ability to acquire copper.
Atox1 Mutants Do Not Exhibit Impaired Copper Binding In Vitro
Mutations involving the lysine-rich ridge as well as K60A exhibited decreased copper incorporation in stimulated cells. While these data were consistent with the idea that the membrane may play a role in copper loading of Atox1, an alternative explanation was that the mutations affected the ability of Atox1 to bind copper at a generic level. To address this issue, the various Atox1 mutants were purified and incubated with 1 equivalent of copper in vitro. After removal of unbound copper via dialysis, the copper content of each mutant was assessed by ICP-MS. As depicted in Fig. 4, there were no gross defects in copper loading in vitro among the various Atox1 mutants. In fact, the K56A K57A mutant exhibited a statistically significant 25 % increase in copper loading relative to wild type, but the reasons for that behavior were unknown.
Atox1 mutants bind copper in vitro. Purified Atox1 mutants were incubated with 1 equivalent of CuSO4, and bound copper was quantitated by ICP-MS analysis. Bars represent mean ± SEM of the ratio of mol bound copper to mol Atox1 protein after dialysis. Statistical significance relative to wild type (WT) was determined for each mutant (n = 3) via one-way ANOVA with Dunnett’s posttest. **p < 0.01
Discussion
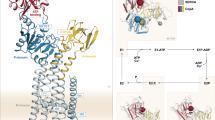
Atox1 is a small copper binding protein that at the structural level adopts a ferredoxin-like fold and features a CxxC copper binding motif at a solvent-accessible loop. The fold and copper binding site are widely conserved among copper homeostasis proteins, which, in addition to Atox1 and its orthologs, include the N-terminal metal binding domains of target P1B-type ATPases, domain I of CCS and the prokaryotic chaperone CopZ (Boal and Rosenzweig 2009). Proximal to the copper binding site, Atox1 orthologs that mediate copper transfer to the secretory pathway feature a unique, conserved, positively charged patch (Fig. 5) that has been proposed to complement the negative surface found in target metal binding domains of the eukaryotic Cu-ATPases and to facilitate chaperone docking and copper transfer (Huffman and O’Halloran 2000). In support of this idea, removal of positive charges in yeast Atx1 results in a reduction of copper incorporation into Fet3, as well as decreased copper-dependent interaction between Atx1 and Ccc2a via two-hybrid analysis (Portnoy et al. 1999). Despite its appeal, this electrostatic complementarity model does not seem to be universal as the respective surfaces of the CopZ–CopA interacting pair are negative and relatively neutral (Fig. 5). This raises the question of whether the conservation of these positive patches may have additional reasons. Putting forward a novel idea, we propose that the conserved, positively charged patch allows Atox1 to associate loosely with negatively charged biological membranes and that this intrinsic binding affinity for bilayers makes an important contribution to initial copper acquisition by the chaperone. While not explicitly investigated in this study, future experiments are needed to delineate the roles of this positive surface with regard to copper shuttling to downstream targets, and such endeavors would require expression of various Atox1 mutants in an Atox1−/− background cell line.
Atox1 orthologs contain a characteristic positive surface. Coulombic surface coloring of Atox1-like proteins was rendered in Chimera, using the following parameters: −2 to +2 kcal/(mol e), dielectric constant = 20, distance from surface = 1.5 Å. The molecules are oriented with the copper binding residues on top and the α-helical face of the ferredoxin-like fold in front. The following PBD codes were used: Atox1 (1TL5), Atx1 (1FES), Ccs1 domain I (1QUP), ATP7A MBD4 (1AW0), ATP7B MBD4 (2ROP), Ccc2 MBD1 (1FVQ), CopZ (1P8G) and CopA MBD2 (1P6T)
Through the experiments represented by this study, we have demonstrated that residues forming a positively charged patch on the surface of the copper chaperone Atox1 mediate membrane binding of this chaperone. The membrane binding properties were partly electrostatic in nature, with highly negative (PG, PA) lipid substrates exhibiting preferential binding. Partial loss of charge within a patch centered on Lys57 resulted in a decrease in Atox1 binding to PG liposomes, while further loss of positive charge nearly completely abrogated association to such membranes (Fig. 2b). Although negative phospholipid moieties exist in all biological membranes, PG is not the most abundant (van Meer et al. 2008). Thus, liposome floats with more biologically relevant substrates were employed and revealed that loss of the positive lysine patch diminished Atox1 association (Fig. 2c). Not surprisingly, the extent of this decrease was less, which likely was directly attributable to the lower mol-fraction of negatively charged lipids in the natural lipid mixtures. No doubt, the use of highly negatively charged bilayer substrates or complex lipid mixes is less than ideal. However, biological membranes exhibit distinct subcellular lipid profiles as well as spatially and temporally dynamic domains (van Meer et al. 2008). This may be relevant in the case of Atox1, which may obtain copper through copper uptake transporters at the cell surface for later delivery to the copper pumps ATP7A and ATP7B that are localized in the trans-Golgi network (Petris et al. 1996; Hung et al. 1997). Keeping in mind that the plasma membrane and trans-Golgi network have different lipid profiles and that direct localization studies of Atox1 would be extremely challenging, given its size and low cellular abundance, the use of generic lipid mixes in our experiments seemed a reasonable first step in probing for a potential role of the bilayer in copper delivery to Atox1.
Consistent with the hypothesis that the bilayer is involved in copper distribution to Atox1, a decreased ability to bind membrane surfaces in vitro correlated with an increase in the fraction of cellular apo-Atox1 when cells were challenged with copper. However, two of the mutant Atox1 proteins, K56A and particularly K60A, deviated from the observed trends. Since Atox1 K56A was significantly impaired in its ability to engage PG liposomes in vitro (~64 % decrease relative to wild type), one might reasonably expect that this mutant would display a significant copper-loading defect in vivo, similar to the K57A single mutation that demonstrated a ~78 % decrease in PG association and ~25 % increase in cys labeling. However, this was clearly not the case because the copper-loading defect of this mutant was very mild (<5 %). Reversing the situation, the K60A mutation was silent in membrane binding changes in vitro yet proved to be the most disruptive for copper loading (Fig. 3c). Acknowledging these data, one could choose to dismiss the idea that the membrane plays a supporting role in copper distribution to Atox1. However, any such dismissal should be accompanied by the proposal of a reasonable alternative that explains how a copper binding site of ~1,000 Å3 can efficiently find copper ions by randomly parsing a volume of 3 × 1013 Å3 of protein-accessible cytosol in a small organism like yeast (considering mammalian cells, the odds are stacked even less favorably). Although it has been proposed that chaperones obtain their copper from a freely exchangeable, glutathione-bound copper pool (Maryon et al. 2013), such a process cannot be efficient given that any copper transfer between Atox1 and glutathione–copper complexes requires these molecular species to collide in an orientation that is conducive to copper transfer.
Based on the plausibility of arguments given above, it seems likely that the results obtained for the K56A and K60A mutants of Atox1 are related to a more complex mechanistic argument. In case of the K56A mutation, the choice of lipid substrate for in vitro binding assays may be a source of ambiguity, but this aspect cannot be easily remedied. A more specific argument can be made for the K60A mutation. This residue, which is characteristic among Atox1 orthologs and situated in the vicinity of the CxxC-copper binding site, has been shown to impart conformational dynamics that enhance copper binding, presumably by stabilizing the overall negative charge of the cysteinate moieties upon copper binding (Anastassopoulou et al. 2004; Arnesano et al. 2001b). Other studies also suggested that lysine movement toward the copper center lowers the pKa and hence nucleophilicity of the cysteinates (Badarau and Dennison 2011) and even modulates the chaperone’s retention of copper in the presence of bulk copper acceptors (Hussain et al. 2008). Lastly, structures of a heterocomplex between Atox1 and the metal binding domains of target ATPases ascribe roles of this lysine residue in making intermolecular contacts (Arnesano et al. 2001a; Banci et al. 2009). Preying on the latter, it is not inconceivable that this residue is also required for putative interactions with a copper transporter of the CTR family prior to copper transfer to Atox1. In support of this hypothesis, the Atox1 K60A mutant did not seem to have a copper binding defect when loaded in vitro. Aggregating this observation with the fact that Lys60 also did not seem to be important for the ability of Atox1 to associate with membranes, it seems likely that mutation of this residue interrupted critical intermolecular interactions that lie upstream of copper acquisition.
The key ideas and findings of our study are summarized in Fig. 6, which presents a schematic mechanistic model for copper loading of Atox1 that mitigates the very unfavorable odds of finding copper cargo through uncontrollable 3D-random walks. In our model, Atox1 loosely associates with membranes, which allows it to scan the membrane surface for the presence of copper-loaded copper transporters such as CTR1. Some support for such a model comes from studies that demonstrated that the C-terminal domain of the yeast Ctr1 exchanges copper with Atx1 in vitro (Xiao and Wedd 2002). Notably, the emerging role of the bilayer in acquisition of copper cargo is not limited to Atox1. Similar observations have recently been made with another principal copper chaperone, CCS, which transfers copper to SOD1 (Pope et al., submitted). While these findings represent a new paradigm for the area of copper biology, they dovetail the growing recognition that bilayers play very active roles in many biological processes such as signaling and membrane trafficking (Adamian et al. 2011; Hartman and Groves 2011; van Meer and de Kroon 2011). Without a doubt, the first step undertaken here opens an intriguing new chapter in the analysis of intracellular copper flux and homeostasis.
Model for copper acquisition by Atox1. To acquire copper ions that have been translocated across the membrane by the CTR family of proteins, the cytosolic chaperone Atox1 is proposed to engage the inner surface of the membrane bilayer via a patch of positive residues (blue). This association is hypothesized to ensure proper orientation of the metal binding site (orange) for efficient copper transfer
References
Adamian L, Naveed H, Liang J (2011) Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim Biophys Acta 1808(4):1092–1102
Anastassopoulou I, Banci L, Bertini I, Cantini F, Katsari E, Rosato A (2004) Solution structure of the apo and copper(I)-loaded human metallochaperone HAH1. Biochemistry 43(41):13046–13053
Arnesano F, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Huffman DL, O’Halloran TV (2001a) Characterization of the binding interface between the copper chaperone Atx1 and the first cytosolic domain of Ccc2 ATPase. J Biol Chem 276(44):41365–41376
Arnesano F, Banci L, Bertini I, Huffman DL, O’Halloran TV (2001b) Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1. Biochemistry 40(6):1528–1539
Arnesano F, Banci L, Bertini I, Ciofi-Baffoni S, Molteni E, Huffman DL, O’Halloran TV (2002) Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res 12(2):255–271
Badarau A, Dennison C (2011) Copper trafficking mechanism of CXXC-containing domains: insight from the pH-dependence of their Cu(I) affinities. J Am Chem Soc 133(9):2983–2988
Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, Pierattelli R, Rosato A, Voulgaris P (2006) The Atx1–Ccc2 complex is a metal-mediated protein–protein interaction. Nat Chem Biol 2(7):367–368
Banci L, Bertini I, Calderone V, Della-Malva N, Felli IC, Neri S, Pavelkova A, Rosato A (2009) Copper(I)-mediated protein–protein interactions result from suboptimal interaction surfaces. Biochem J 422(1):37–42
Boal AK, Rosenzweig AC (2009) Structural biology of copper trafficking. Chem Rev 109(10):4760–4779
Bremner I (1998) Manifestations of copper excess. Am J Clin Nutr 67(Suppl 5):1069S–1073S
Dancis A, Haile D, Yuan DS, Klausner RD (1994) The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J Biol Chem 269(41):25660–25667
Daniel KG, Harbach RH, Guida WC, Dou QP (2004) Copper storage diseases: Menkes, Wilsons, and cancer. Front Biosci 9:2652–2662
Gupta A, Lutsenko S (2009) Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Med Chem 1(6):1125–1142
Halliwell B, Gutteridge JM (1990) Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol 186:1–85
Hamza I, Schaefer M, Klomp LW, Gitlin JD (1999) Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc Natl Acad Sci USA 96(23):13363–13368
Hartman NC, Groves JT (2011) Signaling clusters in the cell membrane. Curr Opin Cell Biol 23(4):370–376
Hatori Y, Lutsenko S (2013) An expanding range of functions for the copper chaperone/antioxidant protein Atox1. Antioxid Redox Signal. doi:10.1089/ars.2012.5086
Himelblau E, Mira H, Lin SJ, Culotta VC, Penarrubia L, Amasino RM (1998) Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol 117(4):1227–1234
Huffman DL, O’Halloran TV (2000) Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J Biol Chem 275(25):18611–18614
Hung IH, Suzuki M, Yamaguchi Y, Yuan DS, Klausner RD, Gitlin JD (1997) Biochemical characterization of the Wilson disease protein and functional expression in the yeast Saccharomyces cerevisiae. J Biol Chem 272(34):21461–21466
Hung IH, Casareno RL, Labesse G, Mathews FS, Gitlin JD (1998) HAH1 is a copper-binding protein with distinct amino acid residues mediating copper homeostasis and antioxidant defense. J Biol Chem 273(3):1749–1754
Hussain F, Olson JS, Wittung-Stafshede P (2008) Conserved residues modulate copper release in human copper chaperone Atox1. Proc Natl Acad Sci USA 105(32):11158–11163
Kim BE, Nevitt T, Thiele DJ (2008) Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol 4(3):176–185
Klomp LW, Lin SJ, Yuan DS, Klausner RD, Culotta VC, Gitlin JD (1997) Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J Biol Chem 272(14):9221–9226
Larin D, Mekios C, Das K, Ross B, Yang AS, Gilliam TC (1999) Characterization of the interaction between the Wilson and Menkes disease proteins and the cytoplasmic copper chaperone, HAH1p. J Biol Chem 274(40):28497–28504
Lin SJ, Culotta VC (1995) The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci USA 92(9):3784–3788
Lin SJ, Pufahl RA, Dancis A, O’Halloran TV, Culotta VC (1997) A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 272(14):9215–9220
Linder MC, Goode CA (1991) Biochemistry of copper. Biochemistry of the elements, vol 10. Plenum Press, New York
Lutsenko S, Gupta A, Burkhead JL, Zuzel V (2008) Cellular multitasking: the dual role of human Cu-ATPases in cofactor delivery and intracellular copper balance. Arch Biochem Biophys 476(1):22–32
Maryon EB, Molloy SA, Kaplan JH (2013) Cellular glutathione plays a key role in copper uptake mediated by human copper transporter 1. Am J Physiol Cell Physiol 304(8):C768–C779
Narindrasorasak S, Zhang X, Roberts EA, Sarkar B (2004) Comparative analysis of metal binding characteristics of copper chaperone proteins, Atx1 and ATOX1. Bioinorg Chem Appl 2(1–2):105–123
O’Halloran TV, Culotta VC (2000) Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem 275(33):25057–25060
Petris MJ, Mercer JF, Culvenor JG, Lockhart P, Gleeson PA, Camakaris J (1996) Ligand-regulated transport of the Menkes copper P-type ATPase efflux pump from the Golgi apparatus to the plasma membrane: a novel mechanism of regulated trafficking. EMBO J 15(22):6084–6095
Pope CR, DeFeo CJ, Unger VM (submitted) Cellular distribution of copper to super oxide dismutase involves scaffolding by membranes. PNAS
Portnoy ME, Rosenzweig AC, Rae T, Huffman DL, O’Halloran TV, Culotta VC (1999) Structure–function analyses of the ATX1 metallochaperone. J Biol Chem 274(21):15041–15045
Pufahl RA, Singer CP, Peariso KL, Lin SJ, Schmidt PJ, Fahrni CJ, Culotta VC, Penner-Hahn JE, O’Halloran TV (1997) Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278(5339):853–856
Rosenzweig AC (2002) Metallochaperones: bind and deliver. Chem Biol 9(6):673–677
Southon A, Burke R, Norgate M, Batterham P, Camakaris J (2004) Copper homoeostasis in Drosophila melanogaster S2 cells. Biochem J 383(Pt 2):303–309
van Meer G, de Kroon AI (2011) Lipid map of the mammalian cell. J Cell Sci 124(Pt 1):5–8
van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev 9(2):112–124
Wakabayashi T, Nakamura N, Sambongi Y, Wada Y, Oka T, Futai M (1998) Identification of the copper chaperone, CUC-1, in Caenorhabditis elegans: tissue specific co-expression with the copper transporting ATPase, CUA-1. FEBS Lett 440(1–2):141–146
Walker JM, Tsivkovskii R, Lutsenko S (2002) Metallochaperone Atox1 transfers copper to the NH2-terminal domain of the Wilson’s disease protein and regulates its catalytic activity. J Biol Chem 277(31):27953–27959
Xiao Z, Wedd AG (2002) A C-terminal domain of the membrane copper pump Ctr1 exchanges copper(I) with the copper chaperone Atx1. Chem Commun (Camb) 6:588–589
Acknowledgments
We would like to thank Dr Amy Rosenzweig for providing an Atox1 clone. We also are indebted to Dr Svetlana Lutsenko for the gift of the HEK293TREx cell line and a matched expression vector. This work was supported by NIH grant P01GM067166 (V.M.U.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Flores, A.G., Unger, V.M. Atox1 Contains Positive Residues that Mediate Membrane Association and Aid Subsequent Copper Loading. J Membrane Biol 246, 903–913 (2013). https://doi.org/10.1007/s00232-013-9592-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-013-9592-1