Abstract
The transient receptor potential vanilloid subtype 1 (TRPV1) is a member of the TRP family gated by vanilloids, heat, and protons. Structurally, TRPV1 subunits have a modular architecture underlying different functionalities, namely stimuli recognition, channel gating, ion selectivity, subunit oligomerization, and regulation by intracellular signaling molecules. Considering modular organization and recent structural information in the ion channel field, we have modeled a full-length TRPV1 by assembly of its major modules: the cytosolic N-terminal, C-terminal, and membrane-spanning region. For N-terminal, we used the ankyrin repeat structure fused with the N-end segment. The membrane domain was modeled with the structure of the eukaryotic, voltage-gated Kv1.2 K+ channel. The C-terminus was cast using the coordinates of HCN channels. The extensive structure–function data available for TRPV1 was used to validate the models in terms of the location of molecular determinants of function in the structure. Additionally, the current information allowed the modeling of the vanilloid receptor in the closed and desensitized states. The closed state shows the N-terminal module highly exposed and accessible to adenosine triphosphate and the C-terminal accessible to phosphoinositides. In contrast, the desensitized state depicts the N-terminal and C-terminal modules close together, compatible with an interaction mediated by Ca2+–calmodulin complex. These models identify potential previously unrecognized intra- and interdomain interactions that may play an important functional role. Although the molecular models should be taken with caution, they provide a helpful tool that yields testable hypothesis that further our understanding on ion channels work in terms of underlying protein structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
TRPV1 is one member of the vanilloid subfamily of mammalian TRP channels (Montell et al. 2002; Pedersen et al. 2005; Venkatachalam and Montell 2007; Ramsey et al. 2006). This channel is involved in the detection and integration of painful chemical and thermal stimuli (Caterina et al. 2000). Receptor functional analysis demonstrated that TRPV1 is a nonselective cation channel activated by heat (≥42°C), low pH and capsaicin, playing a key role in nociception, as well as in calcium homeostasis, hyperalgesia and neurogenic inflammation (Caterina et al. 1997; Caterina and Julius 2001; Garcia-Martinez et al. 2002; Ji et al. 2002; Tominaga and Tominaga 2005). The general topology of the vanilloid channel includes an intracellular N- and C-terminal region of variable length and six transmembrane domains with an additional short amphipathic stretch between transmembrane regions 5 and 6. The N-terminal region contains six ankyrin repeats, and the C-terminal has a coiled–coil segment just adjacent to the sixth transmembrane segment that has been implicated in the subunit tetramerization and channel gating (Garcia-Sanz et al. 2004, 2007; Valente et al. 2008).
The activation of TRPV1 channel by heat, pH, voltage and ligands, as well as the cellular processes that modulate its activation, is consistent with a TRPV1 architecture comprising a number of functional protein modules. This organization has been suggested by extensive structure–function relationships. Hence, the temperature sensor has been located at the C-terminus of the protein, along with the interaction with phosphoinositides and calmodulin (Brauchi et al. 2007). The vanilloid binding site seems to be structured by the transmembrane segments S2–S4 (Jordt and Julius 2002; Johnson et al. 2006; Gavva et al. 2004), and the proposed voltage sensor seems to be defined by a single arginine residue in the S4 segment (Matta and Ahern 2007; Voets et al. 2007). Conversely, the pH sensor has been mapped in the extracellular, amphipathic segment connecting S5 and S6 (Jordt et al. 2000). The N-terminus domain harbors ankyrin repeats that are able to bind multiple ligands and modulate the channel activity (Lishko et al. 2007). The ankyrin domain competitively binds adenosine triphosphate (ATP) and calmodulin, which play a role modulating capsaicin-induced receptor tachyphylaxis (Lishko et al. 2007). In addition, this domain is involved in complexing with cytosolic proteins (Morenilla-Palao et al. 2004).
Despite the wealth of structure–function information on the TRPV1, the structural data available are restricted to the ankyrin domain (Lishko et al. 2007), which represents a handicap for a molecular interpretation of the functional data. The lack of structural information reflects the difficulty of overproducing and crystallizing protein membranes. However, an alternative approach that could be used is to build up structural models taking advantage of the modular organization of the proteins and the evolutionary conservation of such modules from prokaryotes to eukaryotes (Minor 2007). Thus, molecular or structural homologous proteins or protein domains could be utilized as valuable templates to model the architecture of eukaryotic ion channels. A potential problem is that the transference of information from available structures to similar, but not exactly the same proteins is inherently biased to the starting template (Minor 2007), and even with good homology models having the overall backbone fold correct, the exact positioning of side chains and loops could be compromised. In addition, the structure of an incomplete protein does not provide structural information on the absent domains, except for the rough location of the N-terminal and C-terminal connections. Nevertheless, these homology-based models, although with limitations, provide valuable testable hypothesis susceptible of function-directed structural refinement.
Here, we have used this approach to model the full-length TRPV1 channel. We took advantage of the solved structure of the eukaryotic Kv1.2 channel, which exhibits some molecular analogy with TRPV1, particularly in the organization of the transmembrane domain. In addition, several structure–function studies on TRPV1 suggested a similar structure for the pore domain of TRPV1 and voltage-gated K+ channels (Ferrer-Montiel et al. 2004). The structural homology is further substantiated by the voltage-dependent gating of TRPV1. The model was completed with the structural arrangement of the two distinct and independent modules present in the TRPV1 structure, namely the cytosolic N- and C-termini. We report the putative molecular forms of the closed and desensitized states of the TRPV1 channel. These models are compatible with the available functional data, thus providing the first testable structural templates of a full-length TRP channel.
Materials and Methods
Sequence Alignment of the Transmembrane Domains of TRPV1 and Kv1.2
The automatic multiple sequence alignment of the TRPV1 and Kv1.2 transmembrane regions was performed with CLUSTALW at the European Bioinformatics Institute (Labarga et al. 2007) site (http://www.ebi.ac.uk) using Gonnet matrices (Gonnet et al. 1992). Manual alignment of the transmembrane region was accomplished by the alignment editor BioEdit v7.0.9 (Hall 1999) using PAM250 matrices. The resulting alignment had 9.5% identity and 31.5% similarity.
Modeling Full-length TRPV1 Channel
The protein modules were modeled by homology using the structural templates of remote or close homologues available with the highest resolution possible at the Brookhaven Protein Data Bank (http://www.rcsb.org/pdb). The transmembrane region was cast using the spatial coordinates of the atomic structure of the rat potassium channel Kv1.2 (Long et al. 2007) (PDB code 2R9R at 2.4 Å resolution). The N-end domain of the N-terminus was modeled using the human thymidylate kinase (PDB code 1E9C at 1.6 Å resolution) as a template, while the ankyrin domain was molded using its recently solved atomic structure (PDB code 2PNN at 2.7 Å resolution) by Lishko et al. (2007). The C-terminal domain was modeled as two distinct domains: i) the coiled–coil TRP domain near the channel gate was previously cast using the coordinates of a parallel tetrameric four helix bundle (Yadav et al. 2006; Garcia-Sanz et al. 2007) (PDB code 1WL5 at 2.17 Å resolution); and, ii) the regulatory domain was modeled using the C-terminal region of the hyperpolarized, cyclic nucleotide-gated channel HCN2 in the presence of cGMP (Garcia-Sanz et al. 2004) (PDB code 1Q3E at 1.9 Å resolution).
The homology modeling of the individual domains was performed in the Swiss-Model Protein Modeling Server (Schwede et al. 2003) at ExPASy Molecular Biology site (http://kr.expasy.org/). The partial models obtained after homology modeling were cut and the residues were renumbered to match exactly the human TRPV1 sequence.
Domain Assembly
Because there is no direct structural information to connect the different parts of the TRPV1 (N-terminal, C-terminal and transmembrane domains) to construct the whole model, the assembly of the entire TRPV1 was made following the physical, biochemical, and functional information available. First, the C-terminal was positioned following the four S6 transmembrane regions according to the predicted coiled–coil structure, already described as essential for subunit oligomerization (Garcia-Sanz et al. 2004), and channel gating activation (Garcia-Sanz et al. 2007; Valente et al. 2008). Second, the N-terminal domain coupling was based on the connection of the ankyrin repeats crystal structure with the S1 transmembrane region by using an unordered peptide. Third, the relative position between N- and C-terminal in the cytosol was explored by positioning the domains in two plausible conformations: a closed state with noninteracting N-and C-terminal domains; and a putative desensitized state, where these domains interact via calmodulin. In both cases a large cytosolic cavity is produced, and the dimension of the starting models agrees well with that reported for TRPV1 channel revealed by electron microscopy (Moiseenkova-Bell et al. 2008). Peptide bond connection as well as structure visualization and checking were carried out by WHAT IF (Vriend 1990) and Swiss PDB viewer version 3.7 (Guex and Peitsch 1997). The final molecular graphic representations were created by PyMOL 0.99rc6 (http://www.pymol.org).
Side-chain Optimization
The initial orientation and optimization of the side chains were achieved in two steps: first, residues making van der Waals clashes were selected and fitted with “Quick and Dirty” algorithms; and second, models were energy minimized in vacuum (100 steps of steepest descent and 100 conjugate gradient, cutoff of 10 Å for nonbonded interactions) with Insight II (Biosym/MSI), previously to the molecular dynamics (see below). The edition of the structure was accomplished with Swiss PDB viewer (Guex and Peitsch 1997) and Insight II (Accelrys Software Inc., http://www.accelrys.com/). The starting models were initially evaluated with PROCHECK (Laskowski et al. 1996) showing a Ramachandran plot with 84.1% of the residues in most favorable regions and 15% in additional allowed regions. In addition, the model was tested in terms of energy with FoldX (Guerois et al. 2002; Schymkowitz et al. 2005) at the CRG site: http://foldx.crg.es. The force field of FoldX evaluated the properties of the structure, such as its atomic contact map, the accessibility of the atoms and residues, the backbone dihedral angles, and the hydrogen bond and electrostatic networks within the protein.
Molecular Dynamics Simulation
To relax the TRPV1 models, the system was embedded into a palmitoyl oleoyl phosphatidyl choline (POPC) lipid bilayer, and solvated in a water box (TIP3 model) considering the presence of 0.2 M NaCl. All water molecules with oxygen atoms closer than 3.8 Å to a nonhydrogen atom of the protein, as well as lipid molecules with at least one atom closer that 1.3 Å to a nonhydrogen atom of the protein, were removed. The lipid–protein assembly was done with VMD (Humphrey et al. 1996). Topology predictions allowed the appropriate positioning of the transmembrane region to the bilayer. The entire system was subjected to an equilibration process previous to the molecular dynamics simulation. The equilibration consisted in an initial minimization with fixed backbone atoms, followed by a minimization with restrained carbon alpha atoms and a short molecular dynamics (10 ps), to reduce initial bad contacts and to fill empty cavities. Then, the full system, under periodic bordering conditions in the three coordinate directions, was simulated at 310 K for 1 ns (“Langevin dynamics” and “Constant pressure” with restraints, “Constant pressure” without restraints, and then “Constant pressure” with reduced damping coefficients). The final stage of the simulation was performed with parameters that approximate more closely to free dynamics (“LangevinDamping” parameter reduced and “LangevinPistonDecay” increased), while still maintaining an NPT ensemble, for 2-ns simulation. All dynamic simulations were performed by NAMD (Phillips et al. 2005) with the force field CHARMM27 (Mackerell et al. 2004). The cutoff used for long-range interactions was 10 Å.
Results
Modular Nature of the TRPV1 Channel
Eukaryotic channel proteins are complex assemblies of central subunits organized around a central aqueous pore. Each channel subunit is in turn composed of several structural domains that endow the protein of a specific functionality. Structure–function studies have shown that TRPV1 is modular, containing at least three major protein domains: i) a cytosolic N-terminus region holding an ankyrin domain and involved in the modulation of the channel activity and in the interaction with cytosolic proteins (Morenilla-Palao et al. 2004; Lishko et al. 2007; Jung et al. 2002), ii) a transmembrane region encompassing segments S1–S6 having the vanilloid binding site, and the voltage and pH sensors (Jordt and Julius 2002; Johnson et al. 2006; Voets et al. 2007; Long et al. 2005; Jordt et al. 2000); and, iii) a C-terminus that includes a tetramerization domain, and the temperature sensor (Garcia-Sanz et al. 2004; Brauchi et al. 2007). To obtain a full-length structural model of TRPV1 we first modeled each domain separately, and then linked the three molded structures to have an initial template that was further structurally optimized and validated taking into account the plethora of structure–function relationships available.
Modeling of the Transmembrane S1–S6 Region
The template selected to model the transmembrane domains was the recently published structure of the voltage-dependent Kv1.2 rat potassium channel (Long et al. 2007) because they share an identical six transmembrane segment topology, and arrange in a homomeric quaternary structure (Latorre et al. 2007). In addition, structure–function relationships suggest that TRPV1 and Kv channels share an overall molecularly-related pore domain and a voltage sensor (Ferrer-Montiel et al. 2004; Pedersen et al. 2005; Ramsey et al. 2006; Cromer and McIntyre 2008; Latorre et al. 2007). However, as a result of the low amino acid similarity between TRPV1 and Kv1.2, both sequences cannot be automatically aligned to build the homology-based model because significant errors are produced. These alignment inaccuracies are further enhanced because of deletion and/or insertions in the water-exposed loops that connect the transmembrane segments (Fig. 1). Thus, a cautious manual adjustment of the computer-generated multiple sequence alignment is required to ensure that conserved residues are positioned, and to move gaps to loop regions that connect the transmembrane domains (Chang et al. 2005) (Fig. 1). An invaluable guide for manually fitting the sequences alignment was a previous study that reported a prediction of lipid-facing and burial residues in transmembrane regions of TRPV1 channel (Adamian and Liang 2006). This investigation used the transmembrane detection algorithm TMHMM to assign the transmembrane regions and the position of residues governing helical-helical and helical-lipid interactions (Krogh et al. 2001; Elofsson and von Heijne 2007). The secondary structure prediction was also used to guide the alignment (Fig. 1). The global alignment allowed the construction of a model where hydrophobic and charged/polar residues fitted reasonably well with the Kv1.2 template (Fig. 1). It was only necessary to perform minor rotamer adjustments to avoid strong van der Waals clashes, and that hydrogen bond networks were correctly formed.
Alignment between human TRPV1 and rat Kv1.2 transmembrane regions. The automatic alignment was refined by hand using the prediction of buried and lipid exposed residues for transmembrane region of TRPV1 channel. Identity percentage was 9.5%, and similarity was 31.5%. Underlined sequence show predicted α-helices for TRPV1 and crystallographic α-helices for Kv1.2. Differences in the S2–S3 loop and in pore helix are boxed. Highlighted positions in the pore helix reveal the limitations for the alignment in this region (see text)
The Kv1.2 channel depicts a significantly shorter intracellular S2–S3 loop than TRPV1 (Fig. 2a), and thus we could not use crystallographic coordinates to model its structure, which is shown as unordered. However, we imposed several functional constrains, in particular in the region close to the bilayer interface. This region is involved in the binding of vanilloids. The proposed molecular determinants in S3 are R491, Y511, and S512 (Jordt and Julius 2002; Johnson et al. 2006). Other residues involved in ligand interaction are L547, W549 and T550 in the S4 segment (Gavva et al. 2004). The modeled S2–S3 loop positions R491 in S2 toward the membrane interface interacting with the polar group of Q561 in S4, and in turn, hydrogen bonds with Y436 in S1 (Fig. 2b). These interactions fulfill two important requirements for stability: first, compensate a positive charge in a quite hydrophobic environment; second, anchors the S2–S3 loop containing the binding site for capsaicin and derivatives near the membrane interface. In this configuration, the residues Y511 and S512 that seem to be involved in capsaicin binding (Jordt and Julius 2002) are located at the beginning of S3 exposed to the cytosolic environment, but close to the membrane interface (Fig. 2a). Additional determinants of vanilloid binding are located in the S4 segment. Residue L547 is facing the lipid environment suggesting a marginal role in agonist binding, while W549 is oriented to a cavity structured by S1–S3, consistent with their involvement in capsaicin interaction. Residue T550 seems to play a more structural role by hydrogen bonding with T597 of the adjacent subunit. Mutation of this residue could therefore alter the vanilloid binding site consistent with the functional effect reported (Gavva et al. 2004).
Modeled TRPV1 transmembrane domain. (a) Global view of the TRPV1 S1 to S6 transmembrane domain. The model exhibits a voltage sensor module (left) and a pore module (right). The S2–S3 loop involved in the vanilloids binding is presented in a unordered conformation (black arrow). Details of important polar and hydrophobic interactions in S1–S4 (b) and S5–S6 (c) domains (see text). Hydrogen bonds are depicted as magenta discontinuous lines. The potassium ions located in the selectivity filter are drawn as orange spheres
Residue S502 that has been reported to be located in a consensus sequence for PKA, PKC, and CAMKII is solvent exposed and therefore fully accessible to be phosphorylated. The phosphorylation state of this residue is known to affect the binding affinity of vanilloids (Jung et al. 2004). Inspection of the modeled structure is consistent with the hypothesis that the phosphorylation state of S502 may change the conformation of the capsaicin binding site. This impact is because of a putative interaction of this region with polar and charged residues (Q424, D425, K426, etc.) located in the spatially close N-terminal regulatory domain.
The transmembrane domain also holds a voltage sensor that, at variance, with that of voltage-activated channels, seems to lack most of the positively charged residues in the S4 segment (Fig. 1). This lower positive charge is consistent with the weak voltage dependency exhibited by TRPV1, with charge movements during gating of ≤1e (Premkumar and Ahern 2000; Voets et al. 2004). Inspection of the TRPV1 S4 identifies a single arginine (R557) in the N-end of the S4 segment. The model suggests the R557 interacting with E570 and Q560, residues located in the S4–S5 linker (Fig. 2b). This arrangement is compatible with a role of the S4–S5 segment in voltage-gated channel gating (Voets et al. 2007; Matta and Ahern 2007). Imposition of a strong electric field would break this interaction and pull outward the S4 segment, which in turn would drag the linker, giving rise to pore opening. The central role of the S4–S5 segment is further underscored by the presence of positive residues K571, R575 and R579, that are oriented toward the intracellular moiety, close to the bilayer interface where they could be interacting with negatively-charged lipids, thus contributing to the stabilization of the linker in the closed state of the channel. This organization is in agreement with functional data that endow a role of K571, R575 and R579 in voltage sensing (Voets et al. 2007; Long et al. 2005), and lipid interaction (Brauchi et al. 2007).
The pore domain of TRPV1 is the region that shows the highest resemblance to Kv channels (Fig. 1). Indeed, this region was previously modeled using as a template the coordinates of the prokaryotic KcsA channel (Ferrer-Montiel et al. 2004). The most conspicuous difference is found in the size of the loop connecting the S5 and S6 transmembrane segment. This domain contains important determinants of function, such as the pH sensor (E600), the selectivity filter that defines the permeation properties, as well as a glysosylation site (N604) (Wirkner et al. 2005). As depicted in Fig. 2, the high resolution structure of the Kv1.2 channels seems to be an accurate template to cast the pore module of TRPV1. The overall structure shows that the wall of the ionic pore is formed by the S5 and S6 transmembrane helices, where the S5 of one subunit is interacting with the S4 of the adjacent monomer. The S6 holds at its C-end a set of hydrophobic residues that may structure the gate of the channel. Connecting both helices, there is a long loop that was partially modeled as an α-helix taking into consideration secondary structure prediction. The external loop connects with pore helix, which is followed by the extended configuration that holds the signature sequence TXGMG that structures the channel selectivity filter. The pore helix of TRPV1 and the Kv1.2 exhibit significant differences, in particular by the presence of E637 and K640 in the vanilloid receptor that are hydrophobic residues in Kv1.2 (Fig. 1). The K640 is located in the C-terminal of the pore helix, facing S5 and S6, and buried in an unfavorable hydrophobic environment. Thus, we compensate the positive charge of K640 by salt bridging it with the E637 that lies in the same face of the α-helix. This arrangement is supported by the observation that charge swapping at these positions gives rise to functional channels (Garcia-Martinez et al. 2000).
The structural conformation of the pore domain depicts residue E600, a molecular determinant of pH sensing and divalent cation binding (Jordt et al. 2000; Ahern et al. 2005). The model shows the residue E600 hydrogen bonds with Y632 located in the helix pore. Furthermore, it is exposed to the extracellular milieu and surrounded by the acidic residues D601, D605, D625 and E652 (Fig. 2c), forming a negatively-charged cluster that could be readily altered by protonation of E600. This organization of E600 is consistent with a role as a pH sensor. Another residue involved in pH sensing is T634 in the pore helix, located in the model close to the aqueous pore and facing the S5 region of the adjacent monomer, suggesting a role of the helix pore as an active component in channel gating (Ryu et al. 2007).
Another cluster of negatively-charged residues is located at the vestibule of the aqueous pore, near the signature sequence that forms the selectivity filter. These acidic amino acids are water-exposed forming a negative potential sink that attracts cations at the pore entryway (Ferrer-Montiel et al. 2004). Mutation of E637, D647, and E649 altered the sensitivity of the channel to divalent cations and ruthenium red (Garcia-Martinez et al. 2000; Welch et al. 2000; Ahern et al. 2005). Taken together, the proposed model of the membrane-spanning domain reasonably provides a structural framework, consistent with the functional observations and suggests additional experimental work to determine the involvement of additional residues in channel gating.
Modeling the Cytosolic N-terminal Domain
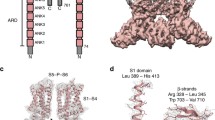
The whole N-terminus of TRPV1 seems composed of three distinct regions: i) an N-end stretch comprising the first 111 amino acids; ii) the ankyrin repeats involved in protein-protein interactions with cytosolic proteins and binding ATP (Morenilla-Palao et al. 2004; Lishko et al. 2007); and iii) a regulatory domain that connects the N-terminus to the membrane region. Of the three regions, the ankyrin domain is the only one that has been crystallized bound to ATP and its structure solved (Lishko et al. 2007), and therefore, we use it to build the conformation of the entire N-terminus (Fig. 3a).
Models of TRPV1 N-terminal and C-terminal domains. (a) The N-terminal of TRPV1 was modeled as three distinct regions, the N-end stretch (green, 1–111), the ankyrin repeats (cyan, 112–364), and the linker domain (red, 365–433), connecting to S1 transmembrane region. The black arrow points to the Pro-rich region in the N-end stretch. (b) The C-terminal was modeled as three different regions: the TRP domain (red, 684–721), the heat sensor (magenta, 727–752), and the regulatory domain (767–810) comprising the overlapping calmodulin (blue) and the PIP2 (cyan) binding sites
The first 111 residues have been described as an unstructured, proline-rich stretch of residues that are not crucial for capsaicin activated currents (Jung et al. 2002). A BLAST search with this segment against the PDB database gave partial similarities with the proximal proline stretch (amino acids 1–35) or with the distal segment encompassing residues 95–111 of the human thymidylate kinase. Because of this partial homology, we decided to model this stretch using as a template the structure of the kinase and connect it to the ankyrin domain. The structure fitted quite reasonably well with the C-end region of the N-terminus. The proximal N-end contains a proline rich motif comprising residues R32 to P38 that was predicted as an unstructured, extended conformation (Fig. 3a). Interestingly, this stretch contains a typical consensus sequence for SH3 domain recognition type I (RxxPxxP) that suggests that this region may participate in protein–protein interactions characteristics of membrane complexes. It is also tempting to hypothesize that this “putative” SH3 domain may be involved in the Src-kinase-mediated phosphorylation of Y200, a process that triggers the exocytotic insertion of TRPV1 channels into the cell surface in response to NGF-induced sensitization (Zhang et al. 2005).
The domain encompassing amino acids 365–433 connects N-terminus to the S1 transmembrane segment. The structure of this domain is presented in the model as virtually unstructured as a result of the lack of similarity with any protein available in the public databases (Fig. 3a). A small part was modeled taking a portion of the cytoplasmic beta-subunit associated to the Shaker potassium channel Kv1.2. The role of this region in the model is to be a connector of the ankyrin domain to the transmembrane region. In addition, the lack of structure–function data precludes a potential refinement of this structural conformation. Nonetheless, because the ankyrin domain seems to be involved in the conformational change that leads to a desensitized state, the flexibility of this domain may be important for the structural rearrangements triggered by ligand-induced desensitization.
Modeling the Cytosolic C-terminal Domain
We have previously modeled the C-terminus domain of TRPV1 (Fig. 3b) as two distinct regions, namely the TRP domain (E684-R721) and a regulatory region (767–810) connected by a flexible linker (Garcia-Sanz et al. 2004, 2007; Valente et al. 2008). The TRP domain is predicted to fold as a coiled–coil structure that seems essential for subunit oligomerization (Garcia-Sanz et al. 2004), and contributes to define the activation energy of channel gating (Garcia-Sanz et al. 2007). The coiled–coil TRP domains of the four subunits are arranged in a four-helix bundle structure, giving rise to a homotetramer (Fig. 4). Examination of the four-helix bundle shows that the core “a” and “d” positions of the coil mediate most of the intersubunit interactions, and mutations that disrupt or alter these interactions should have a profound impact on the activation energy of channel gating (Garcia-Sanz et al. 2007), and suggest that the preservation of compatible intersubunit and intrasubunit interactions in the TRP domain is critical for channel gating. Thus, it is suggested an important role of this segment in coupling the conformational change induced by the activating stimulus to gate opening. In addition, this segment holds residues R702 and K711 that are externally oriented consistent with a potential interaction with the polar heads of PIP2. It should be noted that a simultaneous interaction of R702/K711 and R575/R579 with PIP2, as described in Brauchi et al. (2007) for the homologous menthol-activated TRPM8 channel, is not supported by our model because the two set of amino acids are located quite distant. Nonetheless, it is plausible that they could interact with the lipid bilayer in conformationally different channel states, i.e., closed and open.
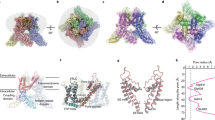
Tetrameric arrangement of the full human TRPV1 in the closed state. (a) Side view of the ribbon structural model of two opposite monomers of TRPV1 channel. The other two monomers are not shown for clarity. (b) Upper view of the channel. Potassium ions are shown as orange spheres. Contacts between monomers occur mainly in the membrane domain and at the C-terminus coiled–coil domain that lies around the central axis and surrounded by the N-terminus. (c) Global view of the TRPV1 model in the closed state, inserted into the lipid bilayer, after molecular dynamic simulation
The regulatory C-terminus was modeled taking into consideration the significant similarities in secondary structure prediction between TRPV1 and the HCN2 cyclic nucleotide binding domain (Zagotta et al. 2003). This domain was composed of two distinct sites corresponding to a region containing a putative PIP2 (amino acids 777–810) and calmodulin binding sites (residues 767–801) (Numazaki et al. 2003; Prescott and Julius 2003). The interaction of PIP2 with this region is controversial because mutations in C-terminal of TRPV1 indicate an inhibitory role for PIP2 (Prescott and Julius 2003). However, recent studies have found that PIP2 reduces tachyphylaxis (Liu et al. 2005; Stein et al. 2006; Lishko et al. 2007), consistent with an inhibition of the calmodulin binding to the C-terminal region that overlaps with the putative PIP2 binding site. In this context, our model also places the putative PIP2 binding site far from the membrane, at least in the modeled closed conformation. Thus, our proposed conformation is more consistent with those functional findings that locate the residues involved in PIP2 binding in the globular region of the TRP domain and the S4–S5 linker (Brauchi et al. 2007; Prescott and Julius 2003). Nevertheless, we can not rule out that the conformation change that leads to the open conformation approximates this domain to the membrane interface, although it seems unlikely as a result of the severe constrains that the N-terminus may imposed in the full-length, native receptor.
The temperature sensor of TRPV1 has been assigned to the C-terminus of the protein, specifically in the linker segment (amino acids 727–752) between the TRP domain and the calmodulin binding domain (Kwak et al. 2000; Brauchi et al. 2007). This region is proposed to be an unstructured conformation in the model, oriented to the cytosol and completely solvent exposed. This conformation does not provide any structural hint on the functional role assigned to this domain. It is plausible that temperature sensing may involve the interaction of the linker with other domains of the protein and/or molecules.
Modeling the Full-length TRPV1 Channel in the Closed State
Having modeled the three independent domains of TRPV1, we next connected them to produce a mold of the full-length protein (Fig. 4). Four of these subunits were arranged around central axis to produce the tetrameric channel in the closed state. The model shows that most intimate contacts between monomers occur in the membrane domain. In addition, significant interactions are also noticeable at the level of the coiled–coil domain at the C-terminus. The structure of the channel depicts that the C-terminal domains lie around the central axis, surrounded by the N-terminus. This organization suggests that both cytosolic domains may be interacting, which could contribute to stabilize the homotetramer. This tenet is consistent with the significant, but not complete, abrogation of channel assembly when the TRP domain is deleted (Garcia-Sanz et al. 2004). The organization of the N-terminus in the homotetramer is also compatible with its functional role. The fingers of the ankyrin domain are oriented externally where they could readily interact with cytosolic proteins. A similar orientation is outlined for the phosphorylation regulatory sites S117 and Y200 (Bhave et al. 2002; Jin et al. 2004). In addition, the extended configuration of the N-terminus linker and the S2–S3 loop may allow the access of S502 to protein kinases.
Modeling the Full-length TRPV1 Channel in the Putative Desensitized State
Notably, a CaM-mediated interaction of the N- and C-termini has been proposed to occur in the desensitized state of the channel (Lishko et al. 2007). This hypothesis suggests that, upon PIP2 depletion by PLC, Ca2+–calmodulin binds to its N and C-termini sites provoking conformational change of the N-terminus producing closing of the pore gate (Lishko et al. 2007). Thus, we questioned whether our model could also be consistent with this proposition and modeled a putative desensitized state of the channel by changing the relative position of the N-terminus with respect to that of the S1–S6 region and the C-terminal (Fig. 5). We checked the size compatibility for ankyrins-CaM-C-terminus interactions by inserting a Ca2+-CaM complex (PDB code 2F3Z). Although the exact points of interaction between Ca2+-CaM and the ankyrin repeats or Ca2+-CaM and the C-terminal are unknown, the model shows that Ca2+-CaM could be positioned between the C-termini of two adjacent monomers. This location reproduces the interaction of Ca2+-CaM with the C-end domain encompassing residues 767–801, that holds a Ca2+-CaM binding site (Rosenbaum et al. 2004) (see Fig. 5). Furthermore, the model depicts that the ankyrin domain could interact with Ca2+-CaM in the proposed region (amino acids 189–222) that lies close to their ATP binding site (Lishko et al. 2007). In addition, the phosphorylation sites S117 (Bhave et al. 2002) and Y200 (Jin et al. 2004) point toward negatively-charged residues in the CaM, and the phosphorylated form may interfere with the Ca2+-dependent binding of CaM to this site, as suggested by Lishko et al. (2007). In support of this notion, TRPV1 phosphorylation potentiates its channel activity partially by preventing the channel desensitization (Bhave et al. 2002, 2003). Taken together, a putative desensitized model is consistent with a structural arrangement of N-terminal module lying close to the C-terminal module, and interacting with it through CaM. Note that this conformational change is compatible with the flexible structure proposed for the N-terminus linker that allows a more extended configuration (Fig. 5a).
Putative interaction of N- and C-terminal regions of TRPV1 via calmodulin. (a, b) Side and upper views of the modeled TRPV1 with a different spatial location of the N-terminal, which allows the interaction with C-terminal mediated by calmodulin (in magenta). The presumed flexibility of the N-terminal linker allows high mobility for the entire N-terminal to interact with the C-terminal or with other cytoplasmatic proteins. (a) Only two opposite monomers for clarity. (C) Global view of the TRPV1 model in the putative desensitized state inserted into the lipid bilayer after molecular dynamic simulation
The molecular dynamics of the full systems, including the entire TRPV1 channel, POPC membrane, explicit water and counter ions were stable through 2 ns of simulation. During the trajectories, several residues reorganized, forming salt-bridges and hydrogen bonds, improving intersubunit interactions and providing stability to N- and C-terminal regions. At the residue level, the most important deviations from the starting model are found in the most mobile parts of the protein: loops and terminal regions. In contrast, the membrane segments remain stable in all simulations. At domain level, the largest fluctuations were found in the N-terminal cytoplasmic regions of the protein in the closed state model, while minor motions were found in the desensitized model (Figs. 4 and 5).
Discussion
The elucidation of the protein structure of ion channels is a difficult task that has been achieved for few members of this large family. In the absence of atomic resolution, the use of computer modeling represents a useful tool for the generation of protein models that may help in understanding the wealth of structure–function relationships. These structural representations are testable experimental hypothesis amenable of refinement. Here, we used this approach to build a structural blueprint of the full-length TRPV1 in the closed and a putative, Ca2+-CaM–driven desensitized states. The TRPV1 model was constructed by assembling the three structural and functional modules that have been identified in the protein monomers. The cytosolic N-terminus was modeled by using the atomic coordinates of the ankyrin repeats that was fused to a proline region at its N-end and to the linker segment that connects it to the transmembrane domain. The core membrane spanning region was molded using as a template the atomic structure of the Kv1.2 channel (Long et al. 2005). The use of this architecture is justified by the overall topological similarities between both channels—namely, they are composed of six transmembrane segments and an amphipathic loop connecting the S5 and S6 helices. In addition, they exhibit a comparable modular arrangement consisting of a voltage sensor and pore modules. Furthermore, they assemble with fourfold symmetry around a central aqueous pore. The membrane part was bond to the TRP domain of the C-terminus, a 25 amino acid stretch that forms a coiled–coil and acts as an association domain essential for subunit tetramerization. Modeling the C-terminal was accomplished by the analogy of its secondary structure prediction to that of the HNC channel family.
This assembly is in agreement with the reported 19 Å structure of TRPV1 determined by single particle electron cryomicroscopy (Moiseenkova-Bell et al. 2008), a fourfold symmetry and two distinct regions: a compact domain corresponding to the transmembrane portion, and a large open (basket-like) domain corresponding to the cytoplasmic N- and C-terminal domains. The models also showed a large aqueous cavity in the cytosolic part, and a plausible interaction between N- and C-terminal domains. Interestingly, these interactions are compatible with additional interactions with cytoplasmic proteins, and could account for the TRPV channel in the desensitized state (see below).
The assembled full length was optimized to fulfill the currently available functional data. Phosphorylation sites have been checked to be water exposed and accessible to modification. Similarly, the finger domains of the ankyrin repeats were externally oriented, consistent with their interaction with cellular proteins (Morenilla-Palao et al. 2004; Stein et al. 2006). Furthermore, residues involved in vanilloid binding were positioned near the membrane interface where they could readily interact and stabilize the ligand. The model also suggests how the elusive voltage sensor could be structured, as it implies that a positively charged residue in the C-end of S4 seems to play an essential role stabilizing the S4–S5 linker. Voltage-induced displacement of this residue would be translated into a destabilization of S4–S5 linker position, which presumably will be pulled toward the bilayer. This movement would be transferred to the channel gate. The presence of positively-charged residues in the S4–S5 linker support this hypothesis because they help to stabilize this segment close to the inner leaflet of the bilayer, and an strong depolarization would push it toward the membrane. Although this is a reasonable hypothesis of the mechanism underlying voltage-induced gating, experimental data are needed to substantiate it. Nevertheless, the proposed model has contributed to identifying the putative molecular determinants of voltage sensing.
A recent study proposed a mechanism for the Ca-CaM–induced desensitization of TRPV1 (Lishko et al. 2007). This hypothesis proposed that in the closed stated, ATP and PIP2 are interacting with the channel. After channel opening, the level of intracellular Ca2+ increases, activating phospholipase C that depletes the PIP2 pool allowing Ca2+-CaM binding to the ankyrin domain and C-terminus, which brings together both domains leading to the stabilization of a desensitized state of the channel. Our structural model is consistent with this mechanism as it structures a Ca2+-CaM binding site with the contribution of both cytosolic channel domains.
Molecular dynamics simulations showed minor deviations at domain level in the model representing desensitized state, probably as a result of the formation of stable protein-protein interactions between close N-terminal–CaM–C-terminal domains after simulation, thus representing an inactive state. The more significant motions observed in the closed state model could be related to the ability of the TRPV1 to undergo different conformational changes associated with protein activation.
Homology modeling of membrane proteins takes advantage of the concept that active sites, binding pockets, motifs and general protein architecture have conserved structural features that have been preserved throughout evolution. The identification of one of these sites in archaea or bacteria implies the existence of their counterpart in mammalian proteins. This evolutionary conservation can be reasonably used to model the eukaryotic proteins. However, a note of caution should be sounded because throughout evolution, proteins can absorb changes in the sequence by accommodating their structure. Variation in side chain properties from one homologue to another may perturb the overall structure in an unpredicted manner. Even with good templates, it is quite challenging to build a model with high accuracy (Minor 2007). Nonetheless, the construction of structural forms of membrane proteins is an important strategy to gain insights on their architecture. These models depict experimentally testable hypothesis that can be improved through a structure–function–based iterative approach. Accordingly, future experiments suggested by the outlined receptor structure will be used to refine this initial blueprint.
References
Adamian L, Liang J (2006) Prediction of transmembrane helix orientation in polytopic membrane proteins. BMC Struct Biol 6:13
Ahern GP, Brooks IM, Miyares RL, Wang XB (2005) Extracellular cations sensitize and gate capsaicin receptor TRPV1 modulating pain signaling. J Neurosci 25:5109–5116
Bhave G, Zhu W, Wang H, Brasier DJ, Oxford GS, Gereau RW (2002) cAMP-dependent protein kinase regulates desensitization of the capsaicin receptor (VR1) by direct phosphorylation. Neuron 35:721–731
Bhave G, Hu HJ, Glauner KS, Zhu W, Wang H, Brasier DJ, Oxford GS, Gereau RW (2003) Protein kinase C phosphorylation sensitizes but does not activate the capsaicin receptor transient receptor potential vanilloid 1 (TRPV1). Proc Natl Acad Sci USA 100:12480–12485
Brauchi S, Orta G, Mascayano C, Salazar M, Raddatz N, Urbina H, Rosenmann E, Gonzalez-Nilo F, Latorre R (2007) Dissection of the components for PIP2 activation and thermosensation in TRP channels. Proc Natl Acad Sci USA 104:10246–10251
Caterina MJ, Julius D (2001) The vanilloid receptor: a molecular gateway to the pain pathway. Annu Rev Neurosci 24:487–517
Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389:816–824
Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D (2000) Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288:306–313
Chang C, Ray A, Swaan P (2005) In silico strategies for modeling membrane transporter function. Drug Discov Today 10:663–671
Cromer BA, McIntyre P (2008) Painful toxins acting at TRPV1. Toxicon 51:163–173
Elofsson A, von Heijne G (2007) Membrane protein structure: prediction vs reality. Annu Rev Biochem 76:125–140
Ferrer-Montiel A, Garcia-Martinez C, Morenilla-Palao C, Garcia-Sanz N, Fernandez-Carvajal A, Fernandez-Ballester G, Planells-Cases R (2004) Molecular architecture of the vanilloid receptor. Insights for drug design. Eur J Biochem 271:1820–1826
Garcia-Martinez C, Morenilla-Palao C, Planells-Cases R, Merino JM, Ferrer-Montiel A (2000) Identification of an aspartic residue in the P-loop of the vanilloid receptor that modulates pore properties. J Biol Chem 275:32552–32558
Garcia-Martinez C, Humet M, Planells-Cases R, Gomis A, Caprini M, Viana F, De La PE, Sanchez-Baeza F, Carbonell T, De FC, Perez-Paya E, Belmonte C, Messeguer A, Ferrer-Montiel A (2002) Attenuation of thermal nociception and hyperalgesia by VR1 blockers. Proc Natl Acad Sci USA 99:2374–2379
Garcia-Sanz N, Fernandez-Carvajal A, Morenilla-Palao C, Planells-Cases R, Fajardo-Sanchez E, Fernandez-Ballester G, Ferrer-Montiel A (2004) Identification of a tetramerization domain in the C terminus of the vanilloid receptor. J Neurosci 24:5307–5314
Garcia-Sanz N, Valente P, Gomis A, Fernandez-Carvajal A, Fernandez-Ballester G, Viana F, Belmonte C, Ferrer-Montiel A (2007) A role of the transient receptor potential domain of vanilloid receptor I in channel gating. J Neurosci 27:11641–11650
Gavva NR, Klionsky L, Qu Y, Shi L, Tamir R, Edenson S, Zhang TJ, Viswanadhan VN, Toth A, Pearce LV, Vanderah TW, Porreca F, Blumberg PM, Lile J, Sun Y, Wild K, Louis JC, Treanor JJ (2004) Molecular determinants of vanilloid sensitivity in TRPV1. J Biol Chem 279:20283–20295
Gonnet GH, Cohen MA, Benner SA (1992) Exhaustive matching of the entire protein sequence database. Science 256:1443–1445
Guerois R, Nielsen JE, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320:369–387
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Ji RR, Samad TA, Jin SX, Schmoll R, Woolf CJ (2002) p38 MAPK activation by NGF in primary sensory neurons after inflammation increases TRPV1 levels and maintains heat hyperalgesia. Neuron 36:57–68
Jin X, Morsy N, Winston J, Pasricha PJ, Garrett K, Akbarali HI (2004) Modulation of TRPV1 by nonreceptor tyrosine kinase, c-Src kinase. Am J Physiol Cell Physiol 287:C558–C563
Johnson DM, Garrett EM, Rutter R, Bonnert TP, Gao YD, Middleton RE, Sutton KG (2006) Functional mapping of the transient receptor potential vanilloid 1 intracellular binding site. Mol Pharmacol 70:1005–1012
Jordt SE, Julius D (2002) Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell 108:421–430
Jordt SE, Tominaga M, Julius D (2000) Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc Natl Acad Sci USA 97:8134–8139
Jung J, Lee SY, Hwang SW, Cho H, Shin J, Kang YS, Kim S, Oh U (2002) Agonist recognition sites in the cytosolic tails of vanilloid receptor 1. J Biol Chem 277:44448–44454
Jung J, Shin JS, Lee SY, Hwang SW, Koo J, Cho H, Oh U (2004) Phosphorylation of vanilloid receptor 1 by Ca2+/calmodulin-dependent kinase II regulates its vanilloid binding. J Biol Chem 279:7048–7054
Krogh A, Larsson B, von HG, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kwak J, Wang MH, Hwang SW, Kim TY, Lee SY, Oh U (2000) Intracellular ATP increases capsaicin-activated channel activity by interacting with nucleotide-binding domains. J Neurosci 20:8298–8304
Labarga A, Valentin F, Anderson M, Lopez R (2007) Web services at the European bioinformatics institute. Nucleic Acids Res 35:W6–W11
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Latorre R, Brauchi S, Orta G, Zaelzer C, Vargas G (2007) ThermoTRP channels as modular proteins with allosteric gating. Cell Calcium 42:427–438
Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R (2007) The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron 54:905–918
Liu B, Zhang C, Qin F (2005) Functional recovery from desensitization of vanilloid receptor TRPV1 requires resynthesis of phosphatidylinositol 4,5-bisphosphate. J Neurosci 25:4835–4843
Long SB, Campbell EB, MacKinnon R (2005) Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309:903–908
Long SB, Tao X, Campbell EB, MacKinnon R (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450:376–382
Mackerell AD Jr, Feig M, Brooks CL III (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25:1400–1415
Matta JA, Ahern GP (2007) Voltage is a partial activator of rat thermosensitive TRP channels. J Physiol 585:469–482
Minor DL Jr (2007) The neurobiologist’s guide to structural biology: a primer on why macromolecular structure matters and how to evaluate structural data. Neuron 54:511–533
Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG (2008) Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc Natl Acad Sci USA 105:7451–7455
Montell C, Birnbaumer L, Flockerzi V (2002) The TRP channels, a remarkably functional family. Cell 108:595–598
Morenilla-Palao C, Planells-Cases R, Garcia-Sanz N, Ferrer-Montiel A (2004) Regulated exocytosis contributes to protein kinase C potentiation of vanilloid receptor activity. J Biol Chem 279:25665–25672
Numazaki M, Tominaga T, Takeuchi K, Murayama N, Toyooka H, Tominaga M (2003) Structural determinant of TRPV1 desensitization interacts with calmodulin. Proc Natl Acad Sci USA 100:8002–8006
Pedersen SF, Owsianik G, Nilius B (2005) TRP channels: an overview. Cell Calcium 38:233–252
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Premkumar LS, Ahern GP (2000) Induction of vanilloid receptor channel activity by protein kinase C. Nature 408:985–990
Prescott ED, Julius D (2003) A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science 300:1284–1288
Ramsey IS, Delling M, Clapham DE (2006) An introduction to TRP channels. Annu Rev Physiol 68:619–647
Rosenbaum T, Gordon-Shaag A, Munari M, Gordon SE (2004) Ca2+/calmodulin modulates TRPV1 activation by capsaicin. J Gen Physiol 123:53–62
Ryu S, Liu B, Yao J, Fu Q, Qin F (2007) Uncoupling proton activation of vanilloid receptor TRPV1. J Neurosci 27:12797–12807
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L (2005) The FoldX Web server: an online force field. Nucleic Acids Res 33:W382–W388
Stein AT, Ufret-Vincenty CA, Hua L, Santana LF, Gordon SE (2006) Phosphoinositide 3-kinase binds to TRPV1 and mediates NGF-stimulated TRPV1 trafficking to the plasma membrane. J Gen Physiol 128:509–522
Tominaga M, Tominaga T (2005) Structure and function of TRPV1. Pflugers Arch 451:143–150
Valente P, Garcia-Sanz N, Gomis A, Fernandez-Carvajal A, Fernandez-Ballester G, Viana F, Belmonte C, Ferrer-Montiel A (2008) Identification of molecular determinants of channel gating in the transient receptor potential box of vanilloid receptor I. FASEB J 22:3298–3309
Venkatachalam K, Montell C (2007) TRP channels. Annu Rev Biochem 76:387–417
Voets T, Droogmans G, Wissenbach U, Janssens A, Flockerzi V, Nilius B (2004) The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature 430:748–754
Voets T, Owsianik G, Janssens A, Talavera K, Nilius B (2007) TRPM8 voltage sensor mutants reveal a mechanism for integrating thermal and chemical stimuli. Nat Chem Biol 3:174–182
Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8:52–56
Welch JM, Simon SA, Reinhart PH (2000) The activation mechanism of rat vanilloid receptor 1 by capsaicin involves the pore domain and differs from the activation by either acid or heat. Proc Natl Acad Sci USA 97:13889–13894
Wirkner K, Hognestad H, Jahnel R, Hucho F, Illes P (2005) Characterization of rat transient receptor potential vanilloid 1 receptors lacking the N-glycosylation site N604. Neuroreport 16:997–1001
Yadav MK, Leman LJ, Price DJ, Brooks CL III, Stout CD, Ghadiri MR (2006) Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution. Biochemistry 45:4463–4473
Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425:200–205
Zhang X, Huang J, McNaughton PA (2005) NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. EMBO J 24:4211–4223
Acknowledgments
This work was supported by grants from the Spanish Ministry of Education and Science (MEC) (SAF2006-2580 to A. F. M.), the Fundación Ramón Areces (to A. F. M.), and the Generalitat Valenciana (GV/2007/025 to G. F. B.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fernández-Ballester, G., Ferrer-Montiel, A. Molecular Modeling of the Full-length Human TRPV1 Channel in Closed and Desensitized States. J Membrane Biol 223, 161–172 (2008). https://doi.org/10.1007/s00232-008-9123-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-008-9123-7