Abstract
Purpose
The aim of our study was to evaluate the impact of CYP3A4, CYP3A5, and ABCB1 polymorphisms on donepezil disposition and clinical outcome.
Methods
Fifty-four Italian patients diagnosed with probable mild to moderate Alzheimer’s disease, treated with donepezil (37 patients 5 mg/day, 17 patients 10 mg/day) were genotyped for CYP3A4 (*1B, *3, and *4), CYP3A5 (*2, *3, and *6) and ABCB1 (3435C>T, 2677G>T/A, and 1236C>T) polymorphisms. All patients were evaluated for the degree of cognitive impairment with Mini Mental State Examination (MMSE) screening test at baseline (before treatment) and after at least 3 months of donepezil treatment at stable dose, when the drug plasma levels were measured.
Results
Three patients carried one detrimental CYP3A4 allelic variant, and 12 carried one functional CYP3A5*1 allele. No statistically significant association was found between CYP3A4 or CYP3A5 genotypes and plasma donepezil concentrations, or between genotypes and clinical response (as measured by change in MMSE score). Nine ABCB1 haplotypes were observed, the most common being 1236C/2677G/3435C (46%) and 1236T/2677T/3435T (41%). Patients homozygous for the T/T/T haplotype had slightly though not significantly lower plasma donepezil concentration-to-dose ratios than those carrying other genotypes [median (95% CI) 0.18 (0.13–0.45) vs. 0.31 (0.30–0.44) mg/l/mg/kg, respectively]. These patients also showed a slightly better clinical response (as measured by change in MMSE score) than the other genotype groups [median (95% CI) 0 (−1.3 to 3.3) vs. −1.0 (−2.1 to 0.0), respectively].
Conclusions
Our data suggest that the CYP3A4 and CYP3A5 polymorphisms are unlikely to influence donepezil metabolism and/or clinical outcome. On the other hand, the ABCB1 polymorphisms may play a role in donepezil disposition and clinical outcome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acetylcholinesterase (AChE) inhibitors, such as donepezil, are the current pharmacological class indicated for the symptomatic treatment of moderate to mild Alzheimer’s disease (AD) [1]. There is a large variability in the clinical response to donepezil, partially related to interindividual differences in its pharmacokinetics, caused by factors such as age, concomitant diseases, and drug-drug interactions. A role may also be played by genetic polymorphisms affecting drug disposition.
Donepezil is metabolized mainly by the cytochrome P450 enzymes CYP2D6 and 3A [2]. The CYP2D6 gene is highly polymorphic (http://www.imm.ki.se/CYPalleles/cyp2d6.htm), and the population can be divided into phenotypes with deficient (poor metabolizers, PM), normal (extensive metabolizers, EM), or increased (ultrarapid metabolizers, UM) enzymatic activity.
Varsaldi et al. [3] observed a large interindividual variability in the plasma concentration-to-dose (C/D) ratios of donepezil among AD patients. This variability was associated with the CYP2D6 genotype, suggesting a role for this polymorphic enzyme in donepezil kinetics and clinical outcome. Co-administration of substrates or inhibitors of CYP3A (e.g., cimetidine, ketokonazole) has been shown to cause changes in the plasma concentrations of donepezil [4, 5], indicating a role for these enzymes in donepezil metabolism as well. A number of allelic variants have been described for the genes encoding CYP3A4 and CYP3A5 (http://www.imm.ki.se/CYPalleles/cyp3a.htm). Three CYP3A4 variants (CYP3A4*1B, CYP3A4*3, and CYP3A4*4) have been reported to alter enzyme activity and to affect the in vivo metabolism of commonly used drugs such as simvastatin, tacrolimus, and cyclosporine [6–8]. Only subjects carrying at least one CYP3A5*1 allele (about 10% of the population) express the CYP3A5 enzyme [9]. The main cause of the absence of CYP3A5 among a majority of Caucasians is the CYP3A5*3 allele [9, 10]. CYP3A5*6 and CYP3A5*2 are more rare variants coding for a nonfunctional protein [10].
P-glycoprotein (P-gp) has been recently recognized as an important determinant for the disposition of donepezil, influencing its efflux from the brain to the blood at the blood-brain barrier level [11]. The gene encoding this protein (ABCB1) is highly polymorphic [12, 13]. Three polymorphisms, 1236C>T, 2677G>T/A, and 3435C>T, which are in linkage disequilibrium [14, 15], have been studied extensively with respect to their effects on P-gp function and clinical relevance.
3435C>T is a silent SNP in exon 26, associated with a lower expression and function of the protein [16]. 2677G>T/A is a tri-allelic polymorphism in exon 21. Both variant alleles (A or T) result in an amino acid change, Ala893Thr and Ala893Ser, respectively, which alter expression and activity of P-gp [17, 18]. The studies that have evaluated the role of the 1236C>T SNP, a silent polymorphism in exon 12, in the pharmacokinetics of drugs such as fexofenadine have produced discordant results [15, 19–21]. The frequencies of these three ABCB1 SNPs have also been shown to differ among ethnic groups [22].
The aim of this study was to evaluate the impact of CYP3A4, CYP3A5, and ABCB1 polymorphisms on the steady-state plasma concentrations and therapeutic outcome of donepezil in a population of AD patients, taking CYP2D6 genotype status into account.
Methods
Study design and population
Fifty-four Italian AD outpatients of Caucasian ethnicity were enrolled in the study at the geriatric clinical unit Unità Operativa Autonoma (UOA) of the Ospedale Maggiore della Carità, Novara, Italy. Most patients (n = 42) had participated in a previous study designed to assess the impact of CYP2D6 polymorphisms on donepezil kinetics and therapeutic outcome [3]. The demographic and clinical characteristics of the patients are shown in Table 1. All participants had been diagnosed with probable mild to moderate AD according to the NINCDS-ADRDA Work Group Criteria for AD diagnosis [23]. They were evaluated for the degree of functional and cognitive impairment with the Mini-Mental State Examination (MMSE) screening test [24] and for daily living with the Clinical Dementia Rating (CDR), Instrumental Activity of Daily Living (ADL), and Clinician’s Interview Based Impression of Change-Plus (CIBIC-Plus) scales. The MMSE test is a psychometric test designed to quantitatively estimate the severity of cognitive impairment and to serially document cognitive changes in domains such as memory, orientation, language, and praxis [24]. Its score ranges from 0 to 30; subjects with a score of 24 or more are classified as cognitively normal. A negative change in the score reflects a worsening in the cognitive status. The MMSE score was corrected for age and educational level. Physical health, other concomitant drug therapies, and adverse reactions to donepezil were also recorded. Subjects with cardiac or hepatic impaired functions were excluded from the study. Tests to assess the liver function were performed at baseline (inclusion point) and repeated at the time of the second clinical evaluation. Fifteen patients received concomitant therapies with one or more substrates or weak inhibitors of CYP3A or P-gp (Table 1). The clinical evaluations were performed at baseline, once the patients were diagnosed with AD, before donepezil treatment (first assessment), and at the time of blood sampling for the donepezil concentration measurement (second assessment). All patients started donepezil therapy at 5 mg/day. After 4 weeks of treatment, the treating physicians decided whether to maintain the same dose (37 patients) or increase it to 10 mg/day (17 patients), based on clinical evaluations. At the second assessment, the patients had been treated with a stable dose of donepezil for at least 3 months. The mean treatment time before the second assessment was 9 months (range 3–40 months). The protocol was approved by the Research Ethics Committee at the Ospedale Maggiore della Carità of Novara (Italy) in accordance with the ethical standards laid down in the Declaration of Helsinki. Prior to inclusion in the study, all patients or their legal guardians gave their written informed consent.
Control group
In order to estimate whether the genotype and allele frequencies among the AD patients were consistent with those in the general population, 285 Italian healthy volunteers (Table 1) were also included in the study and genotyped for CYP3A4, CYP3A5, and ABCB1.
Determination of donepezil plasma concentrations
Twelve to 15 hours after the last drug administration (at 8–10 a.m.), a 5 ml blood sample was collected in heparinized tubes. Plasma was separated and stored at −20°C until analysis. Donepezil plasma concentrations were determined by high-performance liquid chromatography (HPLC) with UV absorbance detection, according to Yasui-Furukori et al. [25] with slight modifications, as described by Varsaldi et al. [3]. Mean recoveries were 85–96%, and the intra- and interday coefficients of variation were below 4.2 and 5.1%, respectively, at concentrations ranging between 1 and 100 mg/l. The lowest limit of quantification was 1.3 ng/l. No interfering peaks were observed despite the fact that various other drugs were coadministered with donepezil.
Genotyping methods
Genomic DNA was isolated from peripheral leukocytes using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) according to the guidelines of the manufacturer.
CYP3A4*3, CYP3A4*4, CYP3A5*2, CYP3A5*6, and ABCB1 polymorphisms were identified by real time PCR with TaqMan kits purchased from Applied Biosystems (Warrington, UK), according to the guidelines of the manufacturer (for CYP3A4*3, assay ID: C__27535825_20, for CYP3A4*4, assay ID: C__30634211_30, for CYP3A5*2, assay ID: C__30633862_10, for CYP3A5*6, assay ID: C__30203950_10; for ABCB1 1236C>T, assay ID: C__7586662_10; for ABCB1 3435C>T, assay ID: C__7586657_1_; and for ABCB1 2677G>A/T: forward primer GTA AGC AGT AGG GAG TAA CAA AAT AAC ACT, reverse primer GAC AAG CAC TGA AAG ATA AGA AAG AAC T, 2677G probe VIC-CCT TCC CAG CAC CT, 2677A probe FAM-CTT CCC AGT ACC TTC, 2677T probe FAM-CTT CCC AGA ACC TT). The CYP3A5*3 was identified by real time PCR according to Mirghani et al. [26]. The CYP3A4*1B (-290A>G) was analyzed by allele-specific PCR followed by digestion with restriction enzyme according to van Schaik et al. [27].
The CYP2D6 genotype was available for most patients from the previous study [3]. The newly included patients were genotyped with the same methods.
Statistical analysis
Data were analyzed by Kruskal-Wallis nonparametric rank analysis of variance to establish an overall difference between the genotypes, and by the Mann-Whitney t-test for comparisons of two groups. When allele and genotype frequencies between AD patients and controls were compared, the chi-squared test was used. Statistical analysis was performed using the GraphPad Prism 4 software (San Diego, CA, U.S.A.). A p value of 0.05 or lower was regarded as statistically significant.
Results
Genotypes
Three AD patients (5.6%) and 24 (8.4%) volunteers carried one detrimental CYP3A4 allele [AD patients: CYP3A4*1/*1B (n = 2) and CYP3A4*1/*3 (n = 1); volunteers: CYP3A4*1/*1B (n = 20) and CYP3A4*1/*3 (n = 4)].
Twelve AD patients (22.2%) were heterozygous for CYP3A5*1, while all the others (77.8%) were homozygous for the CYP3A5*3 allele. Among volunteers, 246 (86.3%) were homozygous for CYP3A5*3 and 4 (1.4%) carried the CYP3A5*3/*6 genotype. Five volunteers (1.7%) were homozygous and 30 (10.5%) were heterozygous for CYP3A5*1. No AD patient or volunteer carried CYP3A4*4 or CYP3A5*2 alleles. The frequencies of the allelic variants of CYP3A4 and CYP3A5 did not differ between patients and volunteers and were similar to those reported in other Caucasian populations (Table 2). The allele and genotype frequencies among both volunteers and patients were in equilibrium with the Hardy-Weinberg equation.
The allele frequencies of individual ABCB1 polymorphisms are given in Table 2. Nine haplotypes for ABCB1 were observed. The most common ABCB1 haplotypes among AD patients and volunteers were 1236C/2677G/3435C (with frequencies of 46 and 45%, respectively) and 1236T/2677T/3435T (41 and 42%, respectively, Table 3).
Fourteen (25.9%) AD patients could be classified as heterozygous CYP2D6 EM (CYP2D6*1/*3, n = 2; CYP2D6*1/*4, n = 10; CYP2D6*1/*5, n = 1; CYP2D6*1/*6, n = 1). Two (3.7%) patients carried two detrimental alleles (CYP2D6*3/*6, n = 1; CYP2D6*5/*5, n = 1) and were classified as PM, while two (3.7%) were found to carry extra copies of a functional CYP2D6 allele and were thus classified as UM.
Donepezil concentrations and the CYP3A4/3A5/ABCB1 genotypes
The plasma concentrations of donepezil ranged from 0.078 to 0.654 mg/l among patients receiving a donepezil dose of 5 mg/day and from 0.024 to 0.537 mg/l among those receiving 10 mg/day. Since the pharmacokinetics of donepezil is linear [28, 29], and there was a large variation in body weight (39–90 kg), the measured plasma concentrations were corrected for the dose and the body weight (C/D).
The C/D ratios were not associated with the CYP3A4 or CYP3A5 genotypes (p > 0.05), and a considerable overlap was observed in C/D ratios between subjects homozygous and heterozygous for both CYP3A4*1 and CYP3A5*3. The median C/D ratio among the 15 patients receiving concomitant CYP3A4 and/or P-gp substrates/weak inhibitors was slightly, not significantly (p = 0.21), higher than that among patients not receiving any CYP3A4- or P-gp-interacting drugs [median 0.41 (95% CI 0.29–0.51) (range 0.17–0.91) vs. 0.28 (95% CI 0.26–0.42) (range 0.02–0.95) mg/l/mg/kg, respectively].
Donepezil C/D was not significantly influenced by ABCB1 polymorphisms (P > 0.05) (Table 4). On the other hand, there was a trend for subjects homozygous for the T variants at all three sites to have lower donepezil C/Ds compared to other genotype groups (Table 4). Consistently, subjects homozygous for the haplotype 1236T/2677T/3435T had slightly, though not significantly (p = 0.176), lower plasma C/D of donepezil [median 0.18 (95% CI 0.13–0.45) (range 0.07–0.85) mg/l/mg/kg] compared to the other genotypes [median 0.31 (95% CI 0.30–0.44) (range 0.02–0.95) mg/l/mg/kg] (Fig. 1).
Clinical outcome and the CYP3A4/3A5 and ABCB1 genotypes
No statistically significant association was found between CYP3A4 or CYP3A5 genotypes and clinical response, as measured by the CIBIC-plus score or change in the MMSE score. However, patients carrying one CYP3A5*1 allele showed a slightly worse clinical response, measured as MMSE change from the first to the second assessment, compared to the CYP3A5*3/*3 group [median −1.8 (95% CI −3.9 to 1.8) (range −8.0 to 8.4) vs. −0.05 (−1.5 to 0.5) (−8.6 to 6.0) respectively, p = 0.519]. No impact of concomitant CYP3A4 and/or P-gp substrates/weak inhibitors was seen on the clinical outcome, as measured by the change in MMSE score (data not shown).
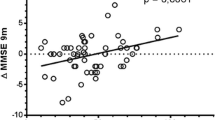
Subjects with ABCB1 T/T genotypes showed a trend towards a better clinical response, as measured by the change in the MMSE score, compared with other genotypes (Table 4). Consistently, the median change in MMSE score among patients homozygous for the ABCB1 1236 T/2677 T/3435 T haplotype was 0 (95% CI −1.3 to 3.3) (range −3.0 to 8.4) compared to −1.0 (95% CI −2.1 to 0.0) (range −8.6 to 6.0) among those with other genotypes (p = 0.138) (Fig. 2).
CYP2D6 genotype, donepezil plasma concentrations, and clinical outcome
The median C/D ratios of donepezil in UMs, homozygous EMs, heterozygous EMs and PMs were 0.13 (95% CI −0.57 to 0.82) (range 0.07–0.18), 0.31 (95% CI 0.29–0.43) (range 0.07–0.91), 0.30 (95% CI 0.21–0.49) (range 0.02–0.85), and 0.51 (95% CI –5.1 to 6.1) (range 0.07–0.95) mg/l/mg/kg, respectively. The heterozygous EM showed a significantly better clinical response to the therapy than homozygous EM as measured by change in the MMSE score (median 1.20 vs. −1.00, respectively; p = 0.03). The two UMs and the two PMs were among the subjects who experienced a most marked worsening in their cognitive functions (UM change in MMSE score −4.4 and −2.0, PM change in MMSE score −3.3 and −1.8, respectively).
Discussion
To our knowledge this is the first study to evaluate the impact of CYP3A4 and ABCB1 genotypes on the clinical outcome in donepezil therapy. The frequencies of CYP3A4, CYP3A5, and ABCB1 allelic variants detected in this study were similar among the two groups studied and consistent with earlier data on Caucasian populations (Table 2). No statistically significant differences in allele or genotype frequencies were detected between AD patients and volunteers, suggesting that CYP3A4/3A5 and ABCB1 polymorphisms are unlikely to represent a risk factor for susceptibility to Alzheimer’s disease. No AD patient or volunteer carried the CYP3A4*4 allele, indicating that this variant has a frequency <0.5%, and could therefore represent a rare single gene defect, rather than an actual polymorphism. Consistently, so far there are no data regarding this allele’s frequency in Caucasians. The CYP3A5*2 allele was detected in a previous study with a frequency of 1% [10], but in our study it was not found either among AD patients or volunteers, suggesting that this is also a very rare variant among Italians.
Kuehl et al. [30] have reported that the CYP3A4*1B and CYP3A5*1 alleles are in linkage disequilibrium in African Americans and suggested that this linkage disequilibrium might influence CYP3A activity towards exogenous and endogenous compounds. Since the frequency of CYP3A4*1B is lower in Caucasians than in African Americans, larger populations would be needed to demonstrate whether CYP3A4*1B and CYP3A5*1 are in linkage disequilibrium in Caucasians [31]. Furthermore, Ingelman-Sundberg et al. [32] suggested that the presence of a CYP3A5*1 allele, leading to expression of CYP3A5, could compensate for the reduction in CYP3A4 activity caused by the CYP3A4*1B allele (that causes decreased transcription [33]) and result in total CYP3A activity similar to that of CYP3A4*1A/CYP3A5*3 carriers. Previous studies in Germans [34] and Spaniards [35] reported frequencies of 4 and 5.4%, respectively, for the CYP3A4*1B/CYP3A5*1 allele combination. Consistently, in the present study, we found the allele combination CYP3A4*1B/CYP3A5*1 with a frequency of 3.7% among AD patients and 4.9% among volunteers (2 out of 2 AD patients and 14 out of 20 volunteers with CYP3A4*1B also carried CYP3A5*1).
Varsaldi et al. [3] showed that CYP2D6 polymorphism does influence donepezil kinetics and therapeutic outcome. In the present study, we confirmed these results in a somewhat larger population. Heterozygous EMs had a better clinical response than homozygous EMs as measured by change in the MMSE score (p = 0.03). UM subjects had the lowest median C/D ratio and PMs had the highest. However, only two patients with these genotypes were included in the study, and the data are thus very uncertain with respect to these groups.
In accordance with previous interaction studies [4, 5, 36], the concurrent administration of CYP3A4 and/or P-gp substrates and/or weak inhibitors was found to be associated with slightly, but not significantly, higher C/D ratios compared with patients with no concomitant interacting therapies.
The large interindividual variability in the C/D ratio of donepezil among AD patients was not related to the CYP3A genotype, and a considerable overlap was observed in C/D ratios between subjects homo- and heterozygous for both CYP3A4*1 and CYP3A5*3. Furthermore, the low frequency of CYP3A4 allelic variants in the Italian population suggests that polymorphisms in this gene are unlikely to play a fundamental role for donepezil.
In previous studies, reduced P-gp activity and higher exposure levels of drug substrates such as digoxin were associated with homozygosity for the three variant alleles of ABCB1, 3435T, 2677T, and 1236T [15, 37, 38]. Accordingly in our study, AD patients homozygous for the haplotype 1236T/2677T/3435T showed a tendency towards a better clinical response, measured by change in MMSE score, although the results did not reach statistical significance. Concurrently, these patients had slightly lower C/Ds of donepezil. The same trend was also seen when the three SNPs were analyzed separately (Table 4), suggesting that this haplotype might play a certain role in the clinical outcome and plasma levels of donepezil.
The low C/D in patients with tendency for better response in T/T/T homozygous subjects was somewhat surprising. However, Lamba et al. [39] showed that subjects homozygous for the ABCB1 2677T allele had higher CYP3A4 expression than subjects homozygous for the 2677G allele, suggesting a link between ABCB1 and CYP3A4 regulation. This would be in agreement with our findings of low C/Ds of donepezil among T/T/T homozygous subjects. Furthermore, these patients showed a tendency towards a better clinical response. A possible explanation could be that the SNPs in the ABCB1 gene, associated with decreased P-gp activity, may lead to increased donepezil CNS levels due to a reduced efflux of the drug from the CNS to the blood compartment. Such a phenomenon has been suggested for other P-gp substrates such as risperidone [40].
Limitations of this study are the small number of patients and the lack of a placebo control group. Thus, larger and preferably prospective studies are necessary to clarify the clinical relevance of the CYP3A4, CYP3A5, and ABCB1 genotypes on donepezil plasma concentrations and clinical outcome among AD patients.
Conclusions
Our results suggest that the CYP3A4 and CYP3A5 allele variants considered in the present study do not play a pivotal role in the variability in donepezil metabolism. Conversely, ABCB1 polymorphisms may contribute to the variability in donepezil disposition and clinical outcome.
References
Scarpini E, Scheltens P, Feldman H (2003) Treatment of Alzheimer’s disease: current status and new perspectives. Lancet Neurol 2(9):539–547
Barner EL, Gray SL (1998) Donepezil use in Alzheimer’s disease. Ann Pharmacother 32(1):70–77
Varsaldi F, Miglio G, Scordo MG, Dahl ML, Villa LM, Biolcati A, Lombardi G (2006) Impact of the CYP2D6 polymorphism on steady-state plasma concentrations and clinical outcome of donepezil in Alzheimer’s disease patients. Eur J Clin Pharmacol 62(9):721–726
Tiseo PJ, Perdomo CA, Friedhoff LT (1998) Concurrent administration of donepezil HCL and cimetidine: assessment of pharmacokinetic changes following single and multiple doses. Br J Clin Pharmacol 46(Suppl 1):25–29
Tiseo PJ, Perdomo CA, Friedhoff LT (1998) Concurrent administration of donepezil HCL and ketoconazole: assessment of pharmacokinetic changes following single and multiple doses. Br J Clin Pharmacol 46(Suppl 1):30–34
Wang A, Yu BN, Luo CH, Tan ZR, Zhou G, Wang LS, Zhang W, Li Z, Liu J, Zhou HH (2005) Ile118Val genetic polymorphism of CYP3A4 and its effects on lipid-lowering efficacy of simvastatin in Chinese hyperlipidemic patients. Eur J Clin Pharmacol 60(12):843–848
Hesselink DA, van Shaik RHN, van der Heiden IP et al (2003) Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther 74:245–54
Hesselink DA, van Gelder T, van Schaik RH, Balk AH, van der Heiden IP, van Dam T, van der Werf M, Weimar W, Mathot RA (2004) Population pharmacokinetics of cyclosporine in kidney and heart transplant recipients and the influence of ethnicity and genetic polymorphisms in the MDR-1, CYP3A4, and CYP3A5 genes. Clin Pharmacol Ther 76(6):545–556
Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, Nuessler AC, Neuhaus P, Klattig J, Eiselt R, Koch I, Zibat A, Brockmöller J, Halpert JR, Zanger UM, Wojnowski L (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11(9):733–9
van Schaik RH, van der Heiden IP, van den Anker JN, Lindemans J (2002) CYP3A5 variant allele frequencies in Dutch Caucasians. Clin Chem 48(10):1668–1671
Ishiwata K, Kawamura K, Yanai K, Hendrikse NH (2007) In vivo evaluation of P-glycoprotein modulation of 8 PET radioligands used clinically. J Nucl Med 48(1):81–87
Kroetz DL, Pauli-Magnus C, Hodges LM, Huang CC, Kawamoto M, Johns SJ, Stryke D, Ferrin TE, DeYoung J, Taylor T, Carlson EJ, Herskowitz I, Giacomini KM, Clark AG, Pharmacogenetics of Membrane Transporters Investigators (2003) Sequence diversity and haplotype structure in the human ABCB1 (MDR1, multidrug resistance transporter) gene. Pharmacogenetics 13(8):481–94
Ito S, Ieiri I, Tanabe M, Suzuki A, Higuchi S, Otsubo K (2001) Polymorphism of the ABC transporter genes, MDR1, MRP1 and MRP2/Cmoat, in healthy Japanese subjects. Pharmacogenetics 11(2):175–84
Horinouchi M, Sakaeda T, Nakamura T, Morita Y, Tamura T, Aoyama N, Kasuga M, Okumura K (2002) Significant genetic linkage of MDR1 polymorphisms at position 3435 and 2677: functional relevance to pharmacokinetics of digoxin. Pharm Res 19(10):1581–5
Johne A, Köpke K, Gerloff T, Mai I, Rietbrock S, Meisel C, Hoffmeyer S, Kerb R, Fromm MF, Brinkmann U, Eichelbaum M, Brockmöller J, Cascorbi I, Roots I (2002) Modulation of steady-state kinetics of digoxin by haplotypes of the P-glycoprotein MDR1 gene. Clin Pharmacol Ther 72(5):584–94
Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, Brinkmann U (2000) Functional polymorphisms of the human multidrug resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Soc USA 97(7):3473–3478
Verstuyft C, Schwab M, Schaeffeler E, Kerb R, Brinkmann U, Jaillon P, Funck-Brentano C, Becquemont L (2003) Digoxin pharmacokinetics and MDR1 genetic polymorphisms. Eur J Clin Pharmacol 58(12):809–12
Morita Y, Sakaeda T, Horinouchi M, Nakamura T, Kuroda K, Miki I, Yoshimura K, Sakai T, Shirasaka D, Tamura T, Aoyama N, Kasuga M, Okumura K (2003) MDR1 genotype related duodenal absorption rate of digoxin in healthy Japanese subjects. Pharm Res 20(4):552–6
Illmer T, Schuler US, Thiede C, Schwarz UI, Kim RB, Gotthard S, Freund D, Schäkel U, Ehninger G, Schaich M (2002) MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res 62(1):4955–4962
Kim RB, Leake BF, Choo EF, Dresser GK, Kubba SV, Schwarz UI, Taylor A, Xie HG, McKinsey J, Zhou S, Lan LB, Schuetz JD, Schuetz EG, Wilkinson GR (2001) Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther 70(2):189–199
Mathijssen RH, Marsh S, Karlsson MO, Xie R, Baker SD, Verweij J, Sparreboom A, McLeod HL (2003) Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res 9(9):3246–3253
Ozawa S, Soyama A, Saeki M, Fukushima-Uesaka H, Itoda M, Koyano S, Sai K, Ohno Y, Saito Y, Sawada J (2004) Ethnic differences in genetic polymorphisms of CYP2D6, CYP2C19, CYP3As and MDR1/ABCB1. Drug Metab Pharmacokinet 19(2):83–95
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of the Department of Health and Human Services Task Force on Alzheimer’s disease. Neurology 34(7):939–944
Folstein MF, Folstein SE, McHugh PR (1975) Mini-Mental State. A practical method for grading the cognitive state of patients for the clinician J Psychiatr Res 12(3):669–677
Yasui-Furukori N, Furuya R, Takahata T, Tateishi T (2002) Determination of donepezil, an acetylcholinesterase inhibitor, in human plasma by high-perfomance liquid chromatography with ultraviolet absorbance detection. J Chromatogr B Analyt Technol Biomed Life Sci 768(2):261–265
Mirghani RA, Sayi J, Aklillu E, Allqvist A, Jande M, Wennerholm A, Eriksen J, Herben VM, Jones BC, Gustafsson LL, Bertilsson L (2006) CYP3A5 genotype has significant effect on quintine 3-hydroxilation in Tanzanians, who have lower total CYP3A activity compared to a Swedish population. Pharmacogenet Genomics 16(9):637–45
van Schaik RH, de Wildt SN, van Iperen NM, Uitterlinden AG, van den Anker JN, Lindemans J (2000) CYP3A4-V polymorphism detection by PCR-restriction fragment length polymorphism analysis and its allelic frequency among 199 Dutch Caucasians. Clin Chem 46(11):1834–36
Rogers SL, Friedhoff LT (1998) Pharmacokinetic and pharmacodynamic profile of donepezil HCl following single oral doses. Br J Clin Pharmacol 46(suppl 1):1–6
Rogers SL, Cooper NM, Sukovaty R, Pederson JE, Lee JN, Friedhoff LT (1998) Pharmacokinetic and pharmacodynamic profile of donepezil HCl following multiple oral doses. Br J Clin Pharmacol 46(suppl 1):7–12
Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E (2001) Sequence diversity in CYP3A4 promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27(4):383–391
Lamba JK, Lin YS, Schuetz EG, Thummel KE (2002) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54(10):1271–1294
Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetics, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116(3):496–526
Rodriguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M (2005) Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun 338(1):299–305
Dally H, Bartsch H, Jäger B, Edler L, Schmezer P, Spiegelhalder B, Dienemann H, Drings P, Kayser K, Schulz V, Risch A (2004) Genotype relationship in the CYP3A locus in Caucasians. Cancer Lett 207(1):95–99
Sinues B, Vicente J, Fanlo A, Vasquez P, Medina JC, Mayayo E, Conde B, Arenaz I, Martinez-Jarreta B (2007) CYP3A5*3 AND CYP3A4*1B allele distribution and genotype combinations: differences between Spaniards and Central Americans. Ther Dug Monit 29(4):412–416
Nagy CF, Kumar D, Perdomo CA, Wason S, Cullen EI, Pratt RD (2004) Concurrent administration of donepezil HCl and sertraline HCl in healthy volunteers: assessment of pharmacokinetic changes and safety following single and multiple oral doses. Br J Clin Pharm 58(Suppl 1):25–33
Chowbay B, Cumaraswamy S, Cheung YB, Zhou Q, Lee EJ (2003) Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporine disposition in heart transplant recipients. Pharmacogenetics 13(2):89–95
Kurata Y, Ieiri I, Kimura M, Morita T, Irie S, Urae A, Ohdo S, Ohtani H, Sawada Y, Higuchi S, Otsubo K (2002) Role of human MDR1 gene polymorphism in bioavaliability and interaction of digoxin, a substrate of P-glycoprotein. Clin Pharmacol Ther 72(2):209–19
Lamba J, Strom S, Venkataramanan R, Thummel KE, Lin YS, Liu W, Cheng C, Lamba V, Watkins PB, Schuetz E (2006) MDR1 genotype is associated with hepatic cytochrome P450 3A4 basal and induction phenotype. Clin Pharmacol Ther 79(4):325–338
Gunes A, Spina E, Dahl M-L, Scordo MG (2008) ABCB1 polymorphisms influence steady-state plasma levels of 9-hydroxyrisperidone and risperidone active moiety. Ther Drug Monit 30(5):628–633
Sata F, Sapone A, Elizondo G, Miller SP, VP ZW, Raunio H, Crespi CL, Gonzalez FJ (2000) CYP3A4 allelic variants with amino acid substitutions in exons 7 and 12: evidence for an allelic variant with altered catalytic activity. Clin Pharmacol Ther 67(1):48–56
van Schaik RH, de Wildt SN, Brosens R, van Fessem M, van den Anker JN, Lindemans J (2001) The CYP3A4*3 allele: is it really rare? Clin Chem 47(6):1104–1106
Cascorbi I, Gerloff T, Johne A, Meisel C, Hoffmeyer S, Schwab M, Schaeffeler E, Eichelbaum M, Brinkmann U, Roots I (2001) Frequency of single nucleotide polymorpisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther 69(3):169–174
Acknowledgements
This work was supported by grants from the Swedish Society of Medicine, the Swedish Research Council, and the “Piedmont County Grants for Research” (2008, Turin, Italy). We thank Dr. Norio Yasui-Furukori (Hirosaki University, School of Medicine, Japan) for the generous gift of donepezil.
The study protocol was approved by the Research Ethics Committee at the “Ospedale Maggiore della Carità” of Novara (Italy) in accordance with the ethical standards laid down in the Declaration of Helsinki.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Magliulo, L., Dahl, ML., Lombardi, G. et al. Do CYP3A and ABCB1 genotypes influence the plasma concentration and clinical outcome of donepezil treatment?. Eur J Clin Pharmacol 67, 47–54 (2011). https://doi.org/10.1007/s00228-010-0883-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-010-0883-5