Abstract
Objective
To examine the effect of cytochrome P450 (CYP) 2C19 activity on the single-dose pharmacokinetics and pharmacodynamics of etizolam.
Methods
The subjects were 21 healthy Japanese volunteers. The two mutated alleles (CYP2C19*2 and CYP2C19*3) causing absent CYP2C19 activity were identified by a polymerase chain reaction method. Twelve subjects were extensive metabolizers (EMs) with no or one mutated allele, and nine subjects were poor metabolizers (PMs) with two mutated alleles. The subjects received a single oral 1–mg dose of etizolam, and blood samplings and evaluation of psychomotor function were conducted up to 24 h after dosing.
Results
The PMs had significantly larger total area under the plasma concentration–time curve (287±74 vs 178±122 ng·h/ml, p<0.05) and longer elimination half–life (14.8±4.2 vs 10.5±3.9 h, p<0.05) of etizolam than the EMs. The area under the score–time curve from 0 to 8 h of the Stanford Sleepiness Scale was significantly larger in the PMs than in EMs (28.9±5.2 vs 22.9±6.9 score·h, p<0.05).
Conclusion
The present study suggests that the single-dose pharmacokinetics and pharmacodynamics of etizolam are influenced by polymorphic CYP2C19 activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The thienodiazepine etizolam, 6–(o–chlorophenyl)–8–ethyl–1–methyl–1–methyl–4H–s–triazolo[3,4-c]–thieno[2,3-e]-[1,4]diazepine, shows pharmacological effects similar to those of diazepam [22]. This drug is widely used in the treatment of psychiatric disorders such as general anxiety disorder [5] and panic disorder [21]. Unpublished data of the manufacturer [16] suggest that etizolam is metabolized mainly by hydroxylation of the methyl group and that of the ethyl group. There have been no data on the bioavailability of this drug.
The cytochrome P450 (CYP) enzymes catalyzing etizolam metabolism have not been fully clarified. Our previous study [2] showed that itraconazole, a potent inhibitor of CYP3A4, inhibits the metabolism of etizolam in healthy volunteers, providing in vivo evidence that etizolam is at least partly metabolized by CYP3A4. Our recent study [14] demonstrated that carbamazepine, an inducer of CYP3A4, induces the metabolism of etizolam, supporting the view that CYP3A4 is involved in etizolam metabolism. However, the degrees of inhibition and induction of etizolam metabolism mentioned above were lower than those of alprazolam [8, 26], midazolam [3, 18], and triazolam [23], which are metabolized predominantly by CYP3A4. These findings point to possible involvement of other CYP enzyme(s) in etizolam metabolism.
There is evidence for the involvement of CYP2C19, which shows a genetic polymorphism [11], in the metabolism of diazepam, flunitrazepam, and quazepam. A significant difference exists between extensive metabolizers (EMs) and poor metabolizers (PMs) of CYP2C19 in the single-dose pharmacokinetics of diazepam [4] and quazepam [7]. There is a significant correlation between CYP2C19 activity and the two metabolic indices of flunitrazepam [9]. Therefore, it would not be surprising if CYP2C19 is involved in the metabolism of etizolam as well.
The purpose of the present study was to examine the effect of CYP2C19 activity on the single-dose pharmacokinetics and pharmacodynamics of etizolam.
Subjects and methods
The subjects were 21 healthy Japanese volunteers. Nineteen subjects were males, and two subjects were females. The mean±SD age was 30.6±7.2 years, and the mean±SD body weight was 69.4±7.2 kg. Ten subjects were smokers (>10 cigarettes/day), and 11 subjects were nonsmokers. None had taken any drug for at least 1 week before the study. The study protocol was approved by the Ethics Committees of Yamagata University School of Medicine and Hirosaki University School of Medicine, and each subject gave his or her written informed consent to participate.
After an overnight fast at 9 A.M., a single oral 1–mg dose of etizolam (Depas, Mitsubishi Pharma, Osaka, Japan) as the tablet formulation was given with a cup of tap water. No food was allowed until 3 h after dosing. Blood samples (10 ml each) were taken before and at 0.25, 0.5, 1, 2, 3, 4, 6, 8, 10, 12, and 24 h after dosing. At the times of blood samplings, psychomotor function was assessed using the Stanford Sleepiness Scale (SSS) [13] and the Digit Symbol Substitution Test (DSST) adapted from the Wechsler Adult Intelligence Scale [27] in 90 s. The SSS is a semiquantitative 7–point self–rating scale with increasing score according to increasing feeling of sleepiness [13]. The DSST is a paper–and–pencil test involving coding skills [27]. The sujects were required to substitute digits for symbols, and the total number of correct substitutions was recorded. No food or drink containing caffeine was allowed throughout the study period.
DNA was isolated from peripheral leukocytes using a QIAamp DNA Blood Kit (Qiagen, Tokyo, Japan). The CYP2C19*1(*1) allele (wild–type allele), and the CYP2C19*2(*2) and CYP2C19*3(*3) alleles causing absent enzyme activity [6, 10], were identified by the polymerase chain reaction (PCR)–based method described by Goldstein and Blaisdell [10].
Plasma concentrations of etizolam was measured by the high–performance liquid chromatography method of Hikida et al. [12]. The lowest limit of detection [signal/noise ratio (S/N)=3] was 0.3 ng/ml, and that of quantification (S/N=6) was 0.6 ng/ml. The coefficients of variation (CV) at the concentrations of 1, 5, and 20 ng/ml were all less than 2.8%.
The elimination rate constant (k) was estimated by the linear regression analysis of at least four points in the terminal log–linear plasma concentration–time curve. The elimination t 1/2 was calculated by 0.693/k. This value should be regarded as apparent, since the blood sampling period of 24 h might be too short to estimate the exact value. The area under the plasma concentration–time curve (AUC) from 0 to 24 h (AUC0–24) was calculated by the trapezoidal rule. The AUC from 0 h to infinity (AUC0–∞), or total AUC, was calculated by AUC0–24 + C24/k, in which C24 is the plasma concentration at 24 h. The peak plasma concentration (Cmax) and time to Cmax (Tmax) were determined graphically.
For the SSS and DSST, the area under the score–time curve from 0 to 8 h (AUSC0–8) was calculated.
Statistical analyses were performed by Student's t–test and one–way analysis of variance (ANOVA) followed by the Tukey test, using SPSS 11.1 J for Windows. A p value of 0.05 or less was considered significant.
Results
Pharmacokinetic assessment
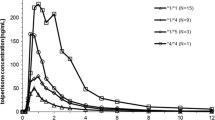
Five subjects were homozygous for the *1 allele, two were heterozygous for the *1 and *2 alleles, five were heterozygous for the *1 and *3 alleles, five were heterozygous for the *2 and *3 alleles, three were homozygous for the *2 allele, and one was homozygous for the *3 allele. Twelve subjects with no or one mutated allele were regarded as EMs, and nine subjects with two mutated alleles were regarded as PMs of CYP2C19.
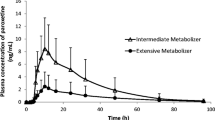
Plasma concentrations of etizolam were significantly higher in the PMs than in EMs from 8 to 24 h (p<0.05) (Fig. 1). The PMs had significantly (p<0.05) larger AUC0–24 and total AUC, and longer elimination t 1/2 of etizolam (Table 1).
The group with two mutated alleles showed significantly larger AUC0–24 (p<0.01) and total AUC (p<0.05), and longer elimination t 1/2 (p<0.05) than that with no mutated allele (Table 2). The group with one mutated allele had significantly larger AUC0–24 (p<0.05) than that with no mutated allele.
Plasma concentrations and pharmacokinetic parameters of etizolam were not different between the nonsmokers and smokers (data not shown).
Pharmacodynamic assessment
The PMs had significantly higher SSS scores than the EMs at 0.5 (p<0.01), 6 (p<0.05), and 10 h (p<0.05) (Fig. 2). The DSST scores were not significantly different between the two groups at any time point (data not shown).
The AUSC0–8 of SSS was significantly larger in the PMs than in EMs (Table 1). The AUSC0–8 of DSST was not significantly different between the two groups (Table 1).
Discussion
The activity of CYP2C19 is bimodally distributed in the population, and individuals with absent enzyme activity are classified as PMs, while the rest are termed as EMs [11]. The PM phenotype is inherited as an autosomal recessive trait and occurs with a frequency of 18–23% in Japanese and 3–5% in Caucasians [11]. The *2 and *3 alleles are the major mutated alleles causing absent enzyme activity [11]. These mutated alleles explain the PMs of Japanese completely [11, 15]. The activity of CYP2C19 decreases significantly according to the increase of the number of mutated alleles, suggesting a gene dose effect [6].
There is evidence that CYP2C19 is involved in the single-dose pharmacokinetics of three benzodiazepines. Bertilsson et al. [4] reported that PMs showed significantly lower clearance and longer elimination t 1/2 of diazepam than EMs. Gafni et al. [9] showed that CYP2C19 activity correlated with the demethylation and 3–hydroxylation capacity of flunitrazepam, though the AUCs of flunitrazepam and its metabolites were not different between EMs and PMs. The authors recently reported that PMs had significantly higher Cmax and larger AUC of quazepam than EMs [7]. On the other hand, it has been shown that CYP2C19 is not involved in the metabolism of estazolam [1] or triazolam [25]. It has been suggested that CYP2C19 does not play a major role in etizolam metabolism as a whole [19], though this enzyme could be involved in the formation of the minor α–hydroxy metabolite [24].
The authors did not determine the sample size beforehand by a power calculation. However, a post hoc power calculation revealed that the actual number of subjects was sufficient to detect significant differences between EMs and PMs. For example, the power to detect a difference of 5.3 h in the elimination t 1/2 (50% of the mean value of the EM group) at the significance level of 0.05 was calculated as 0.81 [28].
In the present study, the PMs had significantly higher plasma concentrations of etizolam than the EMs at several time points. The PMs showed significantly larger AUC and longer elimination t 1/2 of etizolam than the EMs. These results suggest that CYP2C19 is involved in the metabolism of etizolam at least to some extent. Therefore, etizolam is the fourth benzodiazepine whose metabolism is catalyzed by CYP2C19.
Qin et al. [20] reported that the AUC and elimination t 1/2 of diazepam increased significantly according to the increase of mutated CYP2C19 alleles, suggesting a significant gene dose effect of CYP2C19 on diazepam metabolism. In the present study, there were significant differences in the pharmacokinetic parameters of etizolam between the group with no mutated allele and that with one or two mutated alleles. However, no significant difference was found between the group with one mutated allele and that with two mutated alleles. Therefore, it remains unclear whether there is a significant gene dose effect of CYP2C19 on etizolam metabolism.
In line with the pharmacokinetic results, the CYP2C19 phenotype had significant effects on psychomotor function. The SSS scores were significantly higher in the PMs than in EMs at some time points. Also, the AUSC of SSS was significantly larger in the former group than in the latter group. These results suggest increased hypnotic effects of etizolam in PMs even after a single dose. In clinical practice where multiple dosing is the rule rather than the exception, the difference between PMs and EMs increases because of accumulation. Therefore, it may be advisable to adjust the etizolam dose according to CYP2C19 activity. This may be particularly the case in Asians in whom the frequency of PMs is considerably high [11]. Meanwhile, in the studies on diazepam [4, 20] and flunitrazepam [9] psychomotor function was not assessed, and in the study on quazepam [7] no significant difference was found between PMs and EMs. Therefore, the clinical significance of CYP2C19 polymorphism in the treatments with these benzodiazepines remains unclear.
Finally, there were three drawbacks in this study. First, the subjects were not phenotyped or genotyped for CYP3A4, which is also involved in etizolam metabolism [2, 14]. Therefore, it is possible that the majority of the subjects had low CYP3A4 activity by chance, and that the CYP2C19 polymorphism has a major impact on etizolam metabolism in such a specific group only. Second, the metabolites of etizolam were not measured and, therefore, the specific pathway(s) catalyzed by CYP2C19 remained unclarified. Third, the genetic polymorphism affecting the CYP1A2 induction by cigarette smoking [17] was not analyzed, and because of this the effect of cigarette smoking and/or CYP1A2 activity on etizolam metabolism could not be examined in detail.
In conclusion, the present study suggests that the single-dose pharmacokinetics and pharmacodynamics of etizolam are influenced by polymorphic CYP2C19 activity.
References
Aoshima T, Fukasawa T, Otsuji Y, Okuyama N, Gerstenberg G, Miura M, Ohkubo T, Sugawara K, Otani K (2003) Effects of the CYP2C19 genotype and cigarette smoking on the single oral dose pharmacokinetics and pharmacodynamics of estazolam. Prog Neuropsychopharmacol Biol Psychiatry 27:535–538
Araki K, Yasui–Furukori N, Fukasawa T, Aoshima T, Suzuki A, Inoue Y, Tateishi T, Otani K (2004) Inhibition of the metabolism of etizolam by itraconazole in humans: evidence for the involvement of CYP3A4 in etizolam metabolism. Eur J Clin Pharmacol 60:427–430
Backman JT, Olkkola KT, Ojala M, Laaksovirta H, Neuvonen PJ (1996) Concentrations and effects of oral midazolam are greatly reduced in patients treated with carbamazepine or phenytoin. Epilepsia 37:253–257
Bertilsson L, Henthorn TK, Sanz E, Tybring G, Sawe J, Villen T (1989) Importance of genetic factors in the regulation of diazepam metabolism: relationship to S–mephenytoin, but not debrisoquin, hydroxylation phenotype. Clin Pharmacol Ther 45:348–355
Casacchia M, Bolino F, Ecari U (1990) Etizolam in the treatment of generalized anxiety disorder: a double–bind study versus placebo. Curr Med Res Opin 12:215–223
de Morais SM, Goldstein JA, Xie HG, Huang SL, Lu YQ, Xia H, Xiao ZS, Ile N, Zhou HH (1995) Genetic analysis of the S–mephenytoin polymorphism in a Chinese population. Clin Pharmacol Ther 58:404–411
Fukasawa T, Yasui–Furukori N, Aoshima T, Suzuki A, Tateishi T, Otani K (2004) Single oral dose pharmacokinetics of quazepam is influenced by CYP2C19 activity. Ther Drug Monit 26:529–533
Furukori H, Otani K, Yasui N, Kondo T, Kaneko S, Shimoyama R, Ohkubo T, Nagasaki T, Sugawara K (1998) Effect of carbamazepine on the single oral dose pharmacokinetics of alprazolam. Neuropsychopharmacology 18:364–369
Gafni I, Busto UE, Tyndale EF, Kaplan HL, Sellers EM (2003) The role of cytochrome P450 2C19 activity in flunitrazepam metabolism in vivo. J Clin Psychopharmacol 23:169–175
Goldstein JA, Blaisdell J (1996) Genetic tests which identify the principal defects in CYP2C19 responsible for the polymorphism in mephenytoin metabolism. Methods Enzymol 272:210–218
Goldstein JA, de Morais SMF (1994) Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 4:285–299
Hikida K, Inoue Y, Nouchi E, Ohkura Y (1990) Determination of etizolam in human serum or plasma using automated column–switching high–performance liquid chromatography. Jpn J Clin Chem 19:354–359
Hoddes E, Zarcone V, Smythe H, Phillips R, Dement WC (1973) Quantification of sleepiness: a new approach. Psychophysiology 10:413–416
Kondo S, Fukasawa T, Yasui–Furukori N, Aoshima T, Suzuki A, Inoue Y, Tateishi T, Otani K (2005) Induction of the metabolism of etizolam by carbamazepine in humans. Eur J Clin Pharmacol 63:185–188
Kubota T, Chiba K, Ishizaki T (1996) Genotyping of S–mephenytoin 4'–hydroxylation in an extended Japanese population. Clin Pharmacol Ther 60:661–666
Mitsubishi Pharma (2003) Unpublished data in package insert
Nakajima M, Yokoi T, Mizutani M, Kinoshita M, Funayama M, Kamataki T (1999) Genetic polymorphism in the 5'–flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humans. J Biochem 125:803–808
Olkkola KT, Backman JT, Neuvonen PJ (1994) Midazolam should be avoided in patients receiving the systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther 55:481–485
Otani K, Yasui N, Kaneko S, Ohkubo T, Osanai T, Sugawara K, Hayashi K, Chiba K, Ishizaki T (1997) Effects of genetically determined S–mephenytoin 4–hydroxylation status and cigarette smoking on the single–dose pharmacokinetics of oral alprazolam. Neuropsychopharmacology 16:8–14
Qin XP, Xie HG, Wang W, He N, Huang SL, Xu ZH, Ou–Yang DS, Wang YJ, Zhou HH (1999) Effect of the gene dosage of CYP2C19 on diazepam metabolism in Chinese subjects. Clin Pharmacol Ther 66:642–646
Savoldi F, Somenzini G, Ecari U (1990) Etizolam versus placebo in the treatment of panic disorder with agoraphobia: a double–blind study. Curr Med Res Opin 12:185–190
Tsumagari T, Nakajima A, Fukuda T, Shuto S, Kenjo T, Morimoto Y, Takigawa Y (1978) Pharmacological properties of 6–(o–chrorophenyl)–8–ethyl–1–methyl–4H–s–triazolo[3,4–c]thieno[2,3–e][1,4]diazepine(Y–7131), a new anti–anxiety drug. Arzneimittelforschung 28:1158–1178
Varhe A, Olkkola KT, Neuvonen PJ (1994) Oral triazolam is potentially hazardous to patients receiving systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther 56:601–607
Venkatakrishnan K, Greenblatt DJ, von Moltke LL, Shader RI (1998) Alprazolam is another substrate for human cytochrome P450–3A isoforms. J Clin Psychopharmacol 18:256
Yasui N, Otani K, Ohkubo T, Osanai T, Sugawara K, Chiba K, Ishizaki T, Kaneko S (1997) Single–dose pharmacokinetics and pharmacodynamics of oral triazolam in relation to cytochrome P4502C19 (CYP2C19) activity. Ther Drug Monit 19:371–374
Yasui N, Kondo T, Otani K, Furukori H, Kaneko S, Ohkubo T, Osanai T, Nagasaki T, Sugawara K (1998) Effect of itraconazole on the single oral dose pharmacokinetics and pharmacodynamics of alprazolam. Psychopharmacology 139:269–273
Wechsler D (1981) Wechsler Adult Intelligence Scale–Revised. Harcourt Brace Jovanovich, New York, NY
Zar JH (1999) Biostatistical analysis, 4th edn. Prentice–Hall, Upper Saddle River, NJ
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fukasawa, T., Yasui–Furukori, N., Suzuki, A. et al. Pharmacokinetics and pharmacodynamics of etizolam are influenced by polymorphic CYP2C19 activity. Eur J Clin Pharmacol 61, 791–795 (2005). https://doi.org/10.1007/s00228-005-0032-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-005-0032-8