Abstract
Rafting on floating seaweeds facilitates dispersal of associated organisms, but there is little information on how rafting affects the genetic structure of epiphytic seaweeds. Previous studies indicate a high presence of seaweeds from the genus Gelidium attached to floating bull kelp Durvillaea antarctica (Chamisso) Hariot. Herein, we analyzed the phylogeographic patterns of Gelidium lingulatum (Kützing 1868) and G. rex (Santelices and Abbott 1985), species that are partially co-distributed along the Chilean coast (28°S–42°S). A total of 319 individuals from G. lingulatum and 179 from G. rex (20 and 11 benthic localities, respectively) were characterized using a mitochondrial marker (COI) and, for a subset, using a chloroplastic marker (rbcL). Gelidium lingulatum had higher genetic diversity, but its genetic structure did not follow a clear geographic pattern, while G. rex had less genetic diversity with a shallow genetic structure and a phylogeographic break coinciding with the phylogeographic discontinuity described for this region (29°S–33°S). In G. lingulatum, no isolation-by-distance was observed, in contrast to G. rex. The phylogeographic pattern of G. lingulatum could be explained mainly by rafting dispersal as an epiphyte of D. antarctica, although other mechanisms cannot be completely ruled out (e.g., human-mediated dispersal). The contrasting pattern observed in G. rex could be attributed to other factors such as intertidal distribution (i.e., G. rex occurs in the lower zone compared to G. lingulatum) or differential efficiency of recruitment after long-distance dispersal. This study indicates that rafting dispersal, in conjunction with the intertidal distribution, can modulate the phylogeographic patterns of seaweeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The dispersal ability of marine species is a major trait determining the genetic structure of their benthic populations (Weersing and Toonen 2009; Selkoe and Toonen 2011; Haye et al. 2014). In general, species with high dispersal ability (i.e., presence of planktonic larvae, swimming or crawling structures in adults) tend to have lower genetic structure due to higher gene flow between geographically distant populations compared to species with direct development (absence of larvae) or low mobility (e.g., Dawson et al. 2014; Haye et al. 2014). However, other factors such as oceanographic, geological, geographical and ecological features can also affect connectivity, and therefore, the distribution of genetic diversity (Palumbi 1994). In particular, on rocky shores, the tidal height where the organisms are distributed might influence the genetic structure of local populations, with species from medium and high tidal levels having greater genetic structure than species from the low intertidal or subtidal zone (Kelly and Palumbi 2010). This is frequently assumed to be due to the patchiness and greater variety of environmental stresses in the high- to mid-intertidal zones that may generate differential natural selection than in lower zones where the conditions tend to be more homogeneous. Several studies have reported this pattern, which tends to be more prevalent in seaweeds and sessile invertebrates (Engel et al. 2004; Billard et al. 2005; Valero et al. 2011; Krueger-Hadfield et al. 2013; Robuchon et al. 2014).
Seaweeds from intertidal or shallow subtidal habitats are considered good models for phylogeographic studies (Hu et al. 2016). This is due to the complex reproductive cycles (alternation of haploid and diploid phases) of numerous species from all seaweed divisions that may affect the genetic structure of their populations (Krueger-Hadfield and Hoban 2016), coupled with the low dispersal capacity of spores (Santelices 1980; Destombe et al. 1992). However, other mechanisms such as rafting permit dispersal over long distances (Thiel and Haye 2006; Muhlin et al. 2008; Fraser et al. 2009a; 2010; Coyer et al. 2011a, b). For example, some buoyant seaweeds, such as the bull kelp Durvillaea antarctica (Fraser et al. 2010) and the giant kelp Macrocystis pyrifera (Macaya and Zuccarello 2010a, b) can float over extensive distances (>1000 km) after detachment from the primary substratum, and occasionally even cross entire ocean basins (e.g., over 5000 km, between the coasts of New Zealand and Chile), disrupting the potential for genetic differentiation among distant populations (Thiel and Gutow 2005a; Fraser et al. 2010; Coyer et al. 2011b). Rafting may not only facilitate gene flow among benthic populations of floating species, but also of their epibiont communities (Thiel and Haye 2006).
Only a few studies have evaluated the effects of rafting on the genetic diversity and structure of epibionts (also called secondary rafters), focusing mostly on animals associated with floating kelps (see Nikula et al. 2010, 2011a, b, 2013; Cumming et al. 2014). In their recent review of phylogeographic studies on non-buoyant seaweeds associated with floating substrata, Macaya et al. (2016) have shown that most of these epibionts present low genetic structure and high genetic connectivity among populations. Nevertheless, in most cases the authors of the genetic studies only suggested this connectivity via rafting of floating seaweeds and could not completely exclude other vectors of dispersal (e.g., floating marine litter, see Kiessling et al. 2015).
A good choice to study the phylogeography of epibionts is conducting research in areas where there is extensive prior information about abundances and environmental factors that could affect the persistence of floating substrata, especially detached seaweeds. In particular, one of the oceans where there have been several studies on rafting and phylogeography of seaweeds is the South East Pacific coast (SEP, ~14°S to 56°S) (Thiel and Gutow 2005b; Fraser et al. 2010; Macaya and Zuccarello 2010a, b; see also for review Guillemin et al. 2016a). In this zone, phylogeographic studies of benthic species (invertebrates and seaweeds) have focused on testing the concordance between the proposed biogeographic boundaries (at 30°S and 42°S) and phylogeographic breaks (for recent reviews see Haye et al. 2014; Guillemin et al. 2016a). In particular, seaweed species with low dispersal ability presented notorious phylogeographic breaks, suggesting that evolutionary lineages constitute distinct phylogenetic species, as in the intertidal macroalgae Lessonia nigrescens (now separated into L. berteroana and L. spicata; Tellier et al. 2009; González et al. 2012) and Mazzaella laminarioides (Montecinos et al. 2012). On the other hand, seaweeds with high dispersal ability have shallow phylogeographic breaks and a low genetic structure, such as the floating kelp Macrocystis pyrifera (Macaya and Zuccarello 2010a, b). A distinct phylogeographic pattern (i.e., strong genetic structure and high values of genetic diversity) has been reported for the bull kelp Durvillaea antarctica, a species with a high dispersal potential by rafting, along the continental coast of Chile (Fraser et al. 2010). This has been attributed to ineffective long-distance dispersal, either due to low effectiveness in recruitment of new individuals in resident populations or because bull kelp supplies are highly variable in certain areas (Fraser et al. 2010). More than 40 species of seaweeds, mainly Rhodophyta, have been found attached to holdfasts of stranded specimens of D. antarctica along the continental coast of Chile (Macaya et al. 2016). Therefore, dispersal as secondary rafter could also modulate the genetic structure of these epiphytic algae as suggested for other non-buoyant seaweeds with low genetic differentiation among distant populations (Boo et al. 2014a; Guillemin et al. 2014).
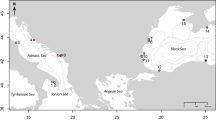
Gelidium lingulatum (Kützing 1868) and G. rex (Santelices and Abbott 1985) are two endemic red seaweeds from the SEP (Santelices 1990). They form monospecific beds at wave-exposed sites (Ortega et al. 2001), mainly in the intertidal zone, where they grow attached to rocks and calcareous shells, but also to holdfasts of large kelps (Santelices 1990; Macaya et al. 2016). Particularly, G. lingulatum is present at 1–2 m above MLLW (mean lower low water) and G. rex is most often found at lower intertidal heights, about 0–0.5 m above MLLW (Santelices 1986). The reported geographical distribution of G. lingulatum extends from Antofagasta (23°S) to Tierra del Fuego (56°S) (Ramírez and Santelices 1991; Hoffmann and Santelices 1997; Fig. 1). However, the current distribution of the species is not clearly established, because identification was based solely on morphological characters (Santelices 1990). Since G. lingulatum features high phenotypic plasticity and juveniles are morphologically very similar to other species of the genus, there is a certain risk of erroneous identifications. On the other hand, G. rex has morphological features facilitating identification in the field. This species is distributed more narrowly, between Coquimbo (30°S) and Concepción (36°S) (Hoffmann and Santelices 1997), although it has been suggested that its northern and southern distribution could extend to 16°S and 39°S, respectively (Santelices and Abbott 1985; Fig. 1).
Geographic location of the coast of Chile, showing the main biogeographic zones (provinces and districts) and breaks (30°S–33°S and 41°S–42°S) (modified from Camus 2001). The sampled distributions of Gelidium lingulatum and G. rex along the Chilean coast are also indicated (shaded bars), as well as the geographic distribution according to the literature (lines). Also, local strandings (stranded biomass and length) of floating bull kelp Durvillaea antarctica in different biogeographic districts are shown (correspondence between circle sizes and stranded biomasses, and kelp sizes and lengths are indicated) (extracted from López et al. 2017)
Both species of Gelidium have a reproductive cycle with an alternation of haploid and diploid phases (Hernández 1997), thalli that can re-attach to the substratum (Rojas et al. 1996), and their spores only survive for short time periods in the water so that the dispersal potential via spores is limited (Bobadilla and Santelices 2005). Also, these turf-forming seaweeds are ecologically important as settlement and nursery area for small invertebrates (González et al. 1991). Moreover, they are economically important for agar extraction (Matsuhiro and Urzúa 1990, 1991; Melo 1998). Within their geographic ranges individuals of both species are frequently found in holdfasts of floating bull kelp Durvillaea antarctica (i.e., >10% in the case of G. lingulatum and 1–10% in G. rex, Macaya et al. 2016), and while intrinsic dispersal ability is strongly limited in these species, dispersal on floating bull kelps could potentially enhance connectivity between their populations. However, this could also be modulated by the distribution across the tidal gradient, where more structure would be expected in G. lingulatum from the mid-intertidal zone compared to G. rex, which grows in the low intertidal zone.
Genetic studies in Gelidiales have revealed high species diversity within the group and also important limitations of the morphological identification of species (e.g., Nelson et al. 2006; Boo et al. 2013, 2014b, 2016). Currently, few phylogeographic studies are available for species from the genus Gelidium. For example, both G. canariense in the Canary Islands (Bouza et al. 2006) and G. elegans on the coast of Korea, China and Japan (Kim et al. 2012) have high genetic variability between populations, numerous private haplotypes, and low genetic connectivity. This high level of genetic structure among populations has also been observed for species with a wide geographical range, such as G. vagum (Yoon et al. 2014), G. crinale and G. pusillum (Kim and Boo 2012).
Using two molecular markers (COI and rbcL), the present study aimed to determine the geographical distribution of genetic diversity for two species from the genus Gelidium, which are partially co-distributed along the Chilean coast and occur at distinct tidal levels. Based on these results, this study also aimed to evaluate whether the observed phylogeographic patterns might be influenced by rafting dispersal via floating seaweeds.
Materials and methods
Biogeographical features of the study area
The SEP coast (~14°S to 56°S) is characterized by a linear topography and no major topographical discontinuities between 14°S and 42°S, south of where it becomes a coast characterized by the presence of channels and fjords (Camus 2001; Thiel et al. 2007; Försterra 2009). Ocean circulation in this area is mainly determined by the Humboldt Current with south–north orientation, and by the southward Cape Horn Current in the southernmost area (Thiel et al. 2007). Also, it is characterized by a latitudinal temperature gradient in surface waters (Tapia et al. 2014) where the occurrence of seasonally persistent upwelling events affect the biogeographic structure of the coastal zone (Lachkar and Gruber 2012; Aravena et al. 2014). Two major biogeographic provinces have been described for the continental coast of Chile: the Peruvian Province (18°S–30°S) and the Magellanic Province (42°S–56°S), which are separated by a broad transition zone, the Intermediate Area, between 30°S and 42°S (Camus 2001) (Fig. 1). A recent study had reported a strong pattern of stranded biomass and length of beach-cast bull kelps (Durvillaea antarctica) in different biogeographical districts (i.e., subdivisions of the biogeographic provinces), particularly within the Intermediate Area (López et al. 2017, Fig. 1), suggesting areas where the connectivity of their populations and that of their secondary rafters could be greater than in others.
Sampling of Gelidium lingulatum and G. rex
Species identification
Both species were identified using morphological traits as described by Santelices and Montalva (1983), Santelices and Stewart (1985), Vargas and Collado-Vides (1996), and Hoffmann and Santelices (1997). For G. lingulatum some difficulties in visual species identification were encountered due to its close morphological similarity with other co-occurring Gelidium species (e.g., G. chilense), particularly in the northern part of the described distribution range of the species. Fully developed individuals consist of a crawling and an erect portion. The creeping axes adhere to the substratum by short discoidal rhizoids, while the erect axes are cylindrical with tongue-like blades and sparsely branched at the base (Hoffmann and Santelices 1997) (Fig. 2). While G. lingulatum is supposed to occur as far north as 23°S (Ramírez and Santelices 1981), no individuals with the morphological characteristics of G. lingulatum were found in eight locations from the northern part of our study area (~20°S to 28°S, Fig. 2, Online Resource 1). In locations at 28°S (i.e., BURR and APOL), we found individuals with typical traits of the genus Gelidium, but not of G. lingulatum, and molecular characterization confirmed the distinctiveness from G. lingulatum (unpubl. data). This suggests that G. lingulatum does not occur north of 29°S, where other, morphologically similar species have been found; consequently, the distribution range of G. lingulatum seems to be more restricted than reported by Ramírez and Santelices (1991) and Hoffmann and Santelices (1997). On the other hand, a clear morphological distinction of G. rex is possible because of the cylindrical axes at the base and flattened middle and upper parts with toothed margins. Also, this species lacks branchlets along the main axis and has a rigid, crispate and cartilaginous thallus (Fig. 3). In addition, G. rex is the largest species from the genus Gelidium present in Chile (Santelices and Abbott 1985). No individuals of G. rex were found in surveys north of 28°S (Online Resource 1) and south of 34°S (Fig. 3).
Sampling locations
Individuals of G. lingulatum and G. rex were collected in winters and summers of 2012–2015 from natural populations in the mid–lower intertidal zone (0.5–1 m) of wave-exposed rocky shores. Sampling was performed in a total of 11 locations (28°S–34°S; 790 km of coastline) for G. rex, covering 68% of the described geographic range, and in a total of 20 locations (29°S–42°S; 1770 km of coastline) for G. lingulatum (45% of the initially described geographic range) (Table 1; Figs. 2, 3). For both G. lingulatum and G. rex, we collected at least 15 individuals per locality (for this study, an individual was composed of one or several erect axes that arise from stoloniferous thalli, Santelices 1986), except in those locations of low species abundance, such as in the north of the study range (Table 1). A total of 319 and 179 specimens were analyzed for G. lingulatum and G. rex, respectively.
Sample manipulation
For each sample, several branches of small and well-identified patches of G. lingulatum or G. rex were collected. Samples were only taken from patches that had a minimum distance of 1 m apart, since vegetative propagation occurs by prostrate stoloniferous thalli (Santelices 1986). Samples were carefully cleaned from epibionts, then stored in individual plastic bags filled with silica gel beads for rapid dehydration, and transported to the laboratory for further genetic analysis.
DNA extraction, PCR amplification, sequencing and sequences alignment
For each sample, a small piece of dry tissue (50 mg) was finely ground using the Tissue Lyser® (Rotsch, Hilden, Germany) at 240 rpm for 5 min. The subsequent DNA extraction was performed using the EZNA® Tissue DNA Kit (Bio-Tek OMEGA, Atlanta, USA), according to the manufacturer’s specifications.
Polymerase chain reaction (PCR) amplification of the partial Cytochrome Oxidase c subunit I gene (COI) was performed using primers designed by Saunders (2005) for red seaweeds (GazF1: 5′ TCAACAAATCATAAAGATATTGG 3′ and GazR2: 5′ ACTTCTGGATGTCCAAAAAAYCA 3′) and using the same conditions for PCR concentrations and program as Fraser et al. (2009b). Reactions were done using dNTPs and DNA polymerase GoTaq, Fermelo Biotec (Promega, Madison, USA), and PCR reactions were performed in a thermocycler Veriti (Applied Biosystems, Foster City, USA).
In order to compare the results of the COI marker with a marker with a slower evolving rate, a subset of 22 individuals (10 G. lingulatum and 12 G. rex) was selected for the rbcL sequencing, considering primarily the specimens having different COI haplotypes and trying to cover the maximum of the species distribution range. It is important to note that a single marker may not be representative of the species history, and therefore, the combination of multiple markers facilitates the detection of different processes occurring at different time scales (Ballard and Whitlock 2004). The chloroplast-encoded rbcL corresponds to the large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo). PCR amplifications of rbcL were performed using two primer combinations, F7–R753 (F7: 5′AACTCTGTAGAACGNACAAG 3′; R753: 5′ GCTCTTTCATACATATCTTCC 3′; Freshwater and Rueness 1994; Gavio and Fredericq 2002) and F645–RrbcSstart (F645: 5′ ATGCGTTGG AAAGAAAGATTCT 3′ and RrbcSstart: 5′ TGTGTTGCGGCCGCCCTTGTGTTAGTCTCAC 3′; Freshwater and Rueness 1994; Lin et al. 2001). The conditions for PCR concentrations and program were identical to Boo et al. (2013). The PCR reagents used were similar to those described for the COI marker.
PCR products were purified and then sequenced using the reverse amplification primers (GazR2 for COI, R753 and RrbcSstart for rbcL) by Macrogen Inc. (Seoul, South Korea: http://www.macrogen.com). Sequences were visualized and edited in Chromas v2.5.1 (Technelysium Pty Ltd 2016) and multiple sequence alignment was performed using CLUSTALW function of BioEdit 7.2.5 (Hall 1999) for each species and marker dataset. Final alignments were checked visually. The resulting datasets for G. lingulatum consisted of a 622-base pair (bp) alignment for the mitochondrial DNA region and of a 1484-bp alignment for the chloroplastic DNA region, while G. rex datasets consisted of a 628-bp alignment and a 1543-bp alignment, respectively. All haplotype sequences were deposited in GenBank (Accession Numbers KX961986–KX962024) and analyzed by BLAST analysis to identify matches with other sequences (Altschul et al. 1990).
Genetic diversity and genetic differentiation
Estimations of standard genetic diversity indices per location and per species
The following molecular diversity indices were computed at the species-level only for the rbcL dataset and at the species- and location-levels for COI, using Arlequin v 3.5.2.2 (Excoffier and Lischer 2010): the number of haplotypes (h), the number of private haplotypes (i.e., haplotypes found at a single sampled location, h priv), the number of polymorphic sites (S), haplotype diversity (H, based on haplotype frequency, the probability that two randomly chosen haplotypes are different; Excoffier and Lischer 2010) and nucleotide diversity (π, the probability that two randomly chosen homologous nucleotide sites are different, expressed as %π; Excoffier and Lischer 2010). For each COI dataset, considering the different sample sizes, a rarefaction method was used with the Contrib program (Petit et al. 1998) to calculate the standardized haplotype diversity at location (excluding the locations with less than 14 individuals) and overall (species-level). Since sample size of G. lingulatum (n = 319 samples) is about twice as large as that of G. rex (n = 179), we considered a sample size of rarefaction of 179 individuals for the case of G. lingulatum in order to compare between the two species.
Estimations of pairwise and overall φ ST
Population differentiation between populations of G. lingulatum and G. rex, and within species was inferred by calculating pairwise and overall (species-level) φ ST-statistics (F ST-like taking into account haplotype frequencies and amount of differences among haplotype pairs). Only locations with a minimum of 14 individuals were included in this analysis. Computing values and tests for significance were done using non-parametric permutation tests (1000 permutations, with Arlequin). Sequential Bonferroni correction was used for multiple comparisons.
Geographic structure
We evaluated whether locations of G. lingulatum and G. rex were geographically structured through spatial analysis of molecular variance (SAMOVA) test using SAMOVA v2.0 software (Dupanloup et al. 2002). Genetic differentiation was investigated using a hierarchical analysis of the genetic variance by partitioning F ST into F SC and F CT indicating the genetic differentiation of populations within groups and between groups, respectively. For each COI dataset, locations with less than 14 samples were excluded and each combination of groups was tested using 500 permutations.
Isolation by distance
The isolation-by-distance model (Slatkin 1993) was tested using a Mantel test in Arlequin with 1000 permutations, testing for a positive correlation between pairwise geographic distance (in km) and raw (D) average pairwise differences for COI datasets, excluding locations with less than 14 samples. Linearized population pairwise φ ST values could not be used in the Mantel test because several locations were genetically monomorphic for different haplotypes, and pairwise comparisons between such fixed populations gave an φ ST of 1.0. The geographical distance between location pairs was measured as distance along the coast for continental locations and taken as the straight-line distance for the island locations (Chiloé Island: MBRA, CUCA and SBA), using the ‘path ruler’ tool in Google Earth (http://earth.google.com/).
Haplotype network reconstruction and historical demography
To represent the genealogical relationship between haplotypes, a network of COI haplotypes was constructed for each species, using the median-joining algorithm implemented in NETWORK v5.0 (Bandelt et al. 1999). This method is based on a maximum parsimony algorithm to simplify the complex branching pattern and to represent the most parsimonious intraspecific phylogenies (Polzin and Daneshmand 2003).
To infer the historical demography of G. lingulatum and G. rex, we first calculated neutrality tests, Tajima’s D (Tajima 1989), and Fu’s F s (Fu 1997) statistics for each COI dataset, in order to detect significant past changes in population size. Significant departure from selection-drift equilibrium was tested by 1000 bootstrap replicates in Arlequin. Under the assumption of neutrality, negative values characterize populations in expansion while positive values, associated to the loss of rare haplotypes, are considered as a signature of recent bottlenecks.
As a complementary approach to infer the historical demography of each species, we compared the observed mismatch distributions of the number of differences between pairs of sequences to estimated values under a model of sudden pure demographic expansion (Rogers and Harpending 1992) and a model of spatial expansion (Excoffier 2004) using Arlequin. For each expansion model and each species, the fit between observed and estimated mismatch distributions was calculated through a generalized least squares approach and tested by 1000 permutations. A multimodal distribution generally indicates a population in demographic equilibrium, while a unimodal distribution is associated with a recent pure demographic expansion or a range expansion.
In the particular case of G. rex, in both approaches, the COI dataset was separated into three groups, according to the results of the geographic distribution of haplotypes (see “Results”), those locations from 28°S to 31°S (including FUAD named “G. rex north”), the same locations without FUAD (i.e., 28°S to 30°S named “G. rex north-f”), and those locations of the southern range (between ~31°S and 34°S, called “G. rex south”).
Results
Sequence characteristics
A 622-bp portion of COI was analyzed from 319 individual G. lingulatum, detecting 24 haplotypes with 21 polymorphic sites (Table 1). On the other hand, the 628-bp portion of COI sequenced for 179 individual G. rex revealed 11 haplotypes, with 10 polymorphic sites (Table 1). From the 10 individuals of G. lingulatum sequenced also for the rbcL marker (1484-bp alignment), three haplotypes were detected, differing by 2 polymorphic sites (Online Resource 2), while sequencing of 12 individuals G. rex (1543-bp portion of rbcL) revealed only one single haplotype.
A query of sequences for the COI haplotypes of G. rex, using a BLAST search, revealed a 100% identity between two of our haplotypes (GR4 and GR9, query cover: 516 bp) and a reference sequence identified as G. rex, from Tongoy Bay, Chile (30°15′S; 71°29′W; GenBank Accession Number: HM629875; Kim et al. 2011). Similarly, the unique rbcL haplotype recovered for G. rex (GR701) presented a 94–100% match with the two reference sequences for the species, both identified as G. rex from Tongoy Bay (GenBank Accession Number: AF305801, query cover: 1430 bp, Thomas and Freshwater 2001; GenBank Accession Number: HM629835, query cover: 1353 bp, Kim et al. 2011). The taxonomic unit G. lingulatum was absent from GenBank, but some of our haplotypes presented a 100% identity with sequences registered as Gelidium sp., from Chile (Chungungo: 29°26′S; 71°18′W and Caleta Horcón: 32°42′S; 71°29′W). The sequences from Chungungo matched with GL5 and GL702 haplotypes, for COI and rbcL, respectively, while sequences from Caleta Horcón matched with GL3 and GL703 haplotypes, respectively (GenBank accession numbers: COI, JX891593–JX891594; rbcL, JX89619–JX891622; query cover: COI, 511 bp; rbcL: 1354 bp; Boo et al. 2013).
Overall, for COI the nucleotide diversity (%π) was 0.352 ± 0.216 and 0.131 ± 0.112 in G. lingulatum and G. rex, respectively, while standardized haplotype diversity was 0.781 ± 0.013 and 0.628 ± 0.029 after rarefaction, being in both cases greater in G. lingulatum than in G. rex (see Table 1). On the other hand, for the rbcL marker, nucleotide diversity and standardized haplotype diversity in G. lingulatum were %π = 0.059 ± 0.051 and H = 0.711 ± 0.086, respectively. In the case of G. rex, no genetic diversity was observed for this marker.
Phylogeographical patterns
For G. lingulatum, 15 of 24 COI haplotypes were private (62.5%) (i.e., haplotypes found at a single location), most of them (12 haplotypes) being unique (i.e., haplotypes found only in one single individual), while for G. rex, 9 of 11 haplotypes were private (81.5%), but only two of them were unique.
A contrasting pattern among species was observed regarding the distribution of the frequent haplotypes. The three most frequent COI haplotypes of G. lingulatum were widespread and shared among geographically distant locations (GL3: 11 locations distributed along the complete study range ~29°S–42°S, 2000 km distance; GL5: 10 locations between ~29°S and 41°S, 1800 km distance; GL2: 9 locations between ~33°S and 42°S, 1550 km distance; Table 1; Fig. 2). On the other hand, the two most frequent COI haplotypes of G. rex presented disjunct geographic distributions, with GR4 exclusively found at the three northernmost locations (~28°S to 30°S) and GR1 only at the seven southernmost locations (~31°S to 34°S; Fig. 3). In between, the location FUAD presented a singular pattern, as all sampled individuals (i.e., 14) shared the GR2 haplotype, which is private from this location. In the case of the rbcL marker, the three haplotypes in G. lingulatum were distributed from ~33°S to 40°S, co-occurring in some locations (CUR and QICO, Fig. 2, see Online Resource 2), whereas for G. rex the single haplotype was observed from ~28°S to 34°S (Fig. 3; see Online Resource 2).
Geographic distribution of haplotypes and haplotype networks of Gelidium lingulatum for mitochondrial COI and chloroplastic rbcL markers. Sampling locations where no individuals of the species were found from the northern part of the study area are also indicated. Photographs of a specimen and intertidal patches of species are shown in the lower right. The within-location diversity and the geographical extent of each haplotype are shown. On the map each circle represents a location and the proportion of pie chart indicates the frequency of individuals for each haplotype. The pie chart color-code corresponds to the one used in haplotype networks of each marker. In the networks, each circle represents a haplotype and its size is proportional to the frequency in which the haplotype was encountered (correspondence between circle sizes and numbers of individuals is indicated). Perpendicular bars between each haplotype pair correspond to the number of mutational steps among them. Abbreviations for location codes are as in Table 1
Geographic distribution of haplotypes and haplotype networks of Gelidium rex for mitochondrial COI and chloroplastic rbcL markers. Sampling locations where no individuals of the species were found from the northern and southern sites of the study area are also indicated. Photographs of a specimen and intertidal patches of species are shown in the lower right. See legend of Fig. 2 for details
For the G. lingulatum COI dataset, average values per sampled location of standardized haplotype diversity (H) and nucleotide diversity (%π) were 0.348 ± 0.203 and 0.150 ± 0.106, respectively (Table 1). Of the 20 sampled locations, 17 were polymorphic with up to 5 haplotypes per location (Fig. 2; Table 1). The haplotype network showed that private haplotypes differed from one of the three most frequent haplotypes mostly by 1 (and up to 3) mutational steps and the maximum pairwise difference among G. lingulatum haplotypes is 7 steps (Fig. 2).
A lower diversity was observed for the G. rex COI dataset, with average values per sampled location of standardized haplotype diversity (H) and nucleotide diversity (%π) of 0.165 ± 0.225 and 0.029 ± 0.039, respectively (Table 1). Most of the locations were monomorphic (7 of 11) and polymorphic locations showed up to 4 haplotypes (Table 1; Fig. 3). The haplotypes differed by 1–3 mutational steps in the haplotype network, with all private haplotypes found at 1 single step from one of the two most frequent haplotypes (Fig. 3).
Within-species genetic structure (COI datasets)
The SAMOVA revealed eight different groups for G. lingulatum, with only two groups formed by several locations which are distributed interspersed along the latitudinal gradient covered by the study (i.e., group 1: PAM, LOT, CRNC, CUCA, 1770 km distant between the most extreme sites; group 8: LBO, PCH, CON, MBRA, SBA, 1550 km distant between the most extreme sites; see Fig. 2 and Online Resource 3). In contrast, in G. rex the three detected groups coincide completely with the disjunct distribution of the three most frequent haplotypes, as described above (Online Resource 3). The groups corresponded to (1) northern locations (BURR, APOL and SAUC), (2) the single site FUAD, and (3) southern locations (from CHLO to BUCA).
According to the Mantel test, in G. lingulatum the correlation between genetic distance and geographic distance was not significant (r 2 = 9.658 e−5, F 1,103 = 0.009, P = 0.514; Fig. 4a), while in G. rex the correlation was significant (r 2 = 0.103, F 1,43 = 4.939, P = 0.047), indicating an isolation-by-distance pattern for the latter species (Fig. 4b). However, no isolation-by-distance was observed when the three geographic groups were considered separately, G. rex north (r 2 = 0.199, F 1,6 = 2.339, P = 0.177), G. rex north-f (r 2 = 0.835, F 1,4 = 2.861, P = 0.166), and G. rex south (r 2 = 1.441 e−4, F 1,37 = 0.283, P = 0.598).
Scatter plot of genetic differentiation and geographic distance of pairwise locations for COI marker. a Gelidium lingulatum and b G. rex. Pairwise genetic distances, represented as D, are plotted against pairwise geographic distances (km). Each point corresponds to a pairwise comparison of locations. The results of the statistical analyses and the regression line for significant relationship (G. rex) are also shown. Locations with N < 14 were excluded from analyses
The overall φ ST (at species-level) for G. lingulatum was 0.629. Most pairwise φ ST-values were significant, indicating differentiation among locations. Interestingly, similarities between geographically distant (over 1000 km) locations were observed (e.g., LBO, PCH, CON, MBRA and SBA) (Fig. 2, Online Resource 4). On the other hand, the overall φ ST for G. rex was 0.859. The φ ST-values were significant for all pairwise comparisons among locations from distinct groups. Likewise, genetic differentiation was evidenced among all location pairs from the group G. rex north (i.e., BURR, APOL, SAUC and FUAD), whereas within the group G. rex south, only the location BUCA was significantly different from the other sampled locations of that zone (Fig. 3; Online Resource 5).
Historical demography (COI datasets)
The mismatch distribution for the G. lingulatum COI dataset was fitted to the sudden demographic expansion model (SSD = 0.017, P = 0.398), and the spatial expansion model (SSD = 0.015, P = 0.496; Fig. 5a). Neutrality tests on overall G. lingulatum COI data also supported partially a demographic expansion, with a negative Tajima’s D index (although not significant: D = −0.853, P = 0.223) and a negative and significant Fu’s F s index (F s = −6.287, P = 0.039; Table 1).
Mismatch distribution for COI datasets for Gelidium lingulatum (a), G. rex north (b), G. rex north-f (c) and G. rex south (d), according to spatial expansion models. The observed distributions of the number of pairwise differences (bars) are contrasted to their expected distributions (solid lines) under a model of spatial expansion
For G. rex north and G. rex north-f, a demographic population expansion was not or only poorly supported by both tested models (G. rex north: sudden demographic expansion, SSD = 0.013, P = 0.045 and spatial expansion, SSD = 0.013, P = 0.001; G. rex north-f: sudden demographic expansion, SSD = 0.002, P = 0.025 and spatial expansion, SSD = 0.002, P = 0.089; Fig. 5b, c). Tajima’s D was not significant for G. rex north, while Fu’s F s indicated a demographic expansion (D = −1.179, P = 0.119; F s = −3.295, P = 0.048). In the case of G. rex north-f, both indices showed a demographic expansion (D = −1.509, P = 0.037; F s = −4.090, P = 0.005). In contrast, for G. rex south stronger evidence was found for population expansion both from mismatch analysis (with a higher support for the sudden demographic expansion: SSD = 0.014, P = 0.309; compared to the spatial expansion model: SSD = 0.014, P = 0.162; Fig. 5d), and from neutrality tests, both significant (D = −1.368, P = 0.047; F s = −3.514, P = 0.004; Table 1).
Discussion
The two seaweed species presented contrasting genetic diversity and structure. Gelidium lingulatum had higher genetic diversity, but genetic structure did not follow a clear geographic pattern, while G. rex had low genetic diversity, a phylogeographic break, but shallow genetic structure. In particular, the phylogeographic pattern of G. lingulatum is not consistent with that observed for other intertidal red seaweeds described for the coast of Chile, using the same mitochondrial marker and partially sharing the same geographic area of study (Montecinos et al. 2012; Guillemin et al. 2016b). On the other hand, the G. rex pattern has similarities to the shallow genetic structure of M. pyrifera (Macaya and Zuccarello 2010a).
Contrasts in genetic diversity and structure
Our results for genetic diversity in G. lingulatum and G. rex are within the observed range of previous studies done for other Gelidium species, using the COI marker (e.g., G. elegans: h = 34, H = 0.711, %π = 0.734, Kim et al. 2012; G. vagum: h = 17, S = 16, H = 0.844, %π = 0.173, Yoon et al. 2014; H values not standardized by rarefaction), revealing a high genetic diversity at species-level, particularly in the case of G. lingulatum, and after standardizing the H values to the smaller sample size of G. rex. Another red seaweed (Mazzaella laminarioides) from the coast of Chile showed higher genetic diversity indices (h = 24, S = 62, H = 0.871, %π = 3.42, H value not standardized) compared with G. lingulatum and G. rex, although that study covered a larger geographic area (29°S–54°S) (Montecinos et al. 2012). These three red seaweed species (G. lingulatum, G. rex, M. laminarioides) share some characteristics of their habitat (cohabiting in the rocky intertidal shore, partially co-distributed along the Chilean coast) and of their life history (low autonomous dispersal capacity, triphasic isomorphic life cycle). Nevertheless, they present contrasting genetic diversity; these differences could be associated with the distribution in the intertidal zone (i.e., M. laminarioides lives higher in the intertidal zone than both Gelidium species) or with the type of vegetative reproduction found in the genus Gelidium (i.e., fragmentation and re-attachment to the substratum, Santelices and Varela 1994; Rojas et al. 1996; Perrone et al. 2006). In particular G. lingulatum tends to monopolize the rocky substratum, which could suppress local genetic diversity, in contrast to the individual and unconnected thalli of Mazzaella (Gómez and Westermeier 1991).
The amplitude of the latitudinal range could also have implications in the genetic diversity, because a wider geographic range is related to a larger effective population size and a higher gradient of environmental variability, which can lead to selection and local adaptation (Alberto et al. 2010). This could explain the differences in genetic diversity between the two species studied, considering the wider geographic range of G. lingulatum compared to G. rex. Therefore, our results suggest that the amplitude of the geographic range contribute to the differences in genetic diversity observed for both species, and other red seaweeds from the Chilean coast.
In the case of G. lingulatum, genetic structure was evidenced throughout its range, but without a clear geographical pattern (i.e., haplotypes disappear and reappear repeatedly throughout its geographic range and no phylogeographic break was detected), and there was no genetic isolation-by-distance. On the other hand, G. rex compared to G. lingulatum showed a different pattern with a disjunct haplotype distribution where a separation occurs at ~31°S between the northern, FUAD, and southern populations of its geographic range. This coincides with the biogeographic break at 30°S (Camus 2001) and is also consistent with the phylogeographic breaks described for that region for many intertidal species of invertebrates and macroalgae with limited dispersal abilities (i.e., 29°S–33°S, Tellier et al. 2009; Sánchez et al. 2011; Montecinos et al. 2012; Haye et al. 2014; Guillemin et al. 2016a). However, unlike other intertidal seaweeds (e.g., Lessonia nigrescens complex, Tellier et al. 2011; M. laminarioides, Montecinos et al. 2012), this geographical subdivision is not based on a strong genetic difference, since the separation between populations is only one mutational step, similar to the shallow genetic structure described for M. pyrifera, a kelp species with high dispersal potential via rafting (Macaya and Zuccarello 2010a).
Indeed, the rbcL marker, despite the low sample sizes, revealed no indication for a phylogeographic break due to the complete absence and lower polymorphism for this marker in G. rex and G. lingulatum, respectively. This is consistent with the generally lower mutation rate of this marker, compared to COI (Engel et al. 2008; Grant 2016). Therefore, all these results suggest that G. lingulatum and G. rex (more evident in G. lingulatum) have a long-distance dispersal mechanism, which cannot be explained by intrinsic dispersal abilities alone due to limited autonomous dispersal potential via spores/gametes.
Gelidium rex is a species found in the very low intertidal zone compared to G. lingulatum, which grows closer to the mid-intertidal zone (Santelices 1986). In general, species that are distributed in an area with less environmental variability (e.g., longer immersion times below the tidal gradient as for G. rex) tend to have less genetic structure (Kelly and Palumbi 2010). This might be due to less patchy distributions and larger population sizes in the low intertidal or subtidal zones (i.e., distribution width effect, Robuchon et al. 2014), which would reduce the selection pressure and the action of genetic drift observed in upper intertidal zone (although the distribution of G. rex in the lower intertidal zone tends to be very patchy rather than being a continuous fringe; Santelices and Abbott 1985). For example, for two sister species of laminarian kelps co-distributed along the coast of France, Robuchon et al. (2014) showed that populations of the species inhabiting the shallow subtidal zone (Laminaria hyperborea) had less genetic structure than those of the intertidal species (L. digitata). In addition, this is congruent with the observed pattern of M. laminarioides from the mid-intertidal zone, which has a much stronger genetic structure than both Gelidium species, showing two strongly differentiated haplogroups (separated by 15–45 bp for COI) between 29°S and 37°S, and up to 3 haplogroups considering locations up to 42°S (Montecinos et al. 2012). However, if both species of Gelidium are compared, genetic structure of G. lingulatum did not follow a clear geographic pattern in contrast to G. rex and overall (species-level) φ ST-value is lower for G. lingulatum (0.629) than G. rex (0.859). Therefore, our results do not support the hypothesis that seaweed species from the mid-intertidal zone have more genetic structure compared with organisms from lower zones and it suggests that other factors may be important. A trend of less genetic structure in species from the upper intertidal zone has also been observed in two intertidal barnacles (Jehlius cirratus and Notochthamalus scabrosus) from the Chilean coast (18°S–54°S) (Zakas et al. 2009; Ewers-Saucedo et al. 2016; Guo and Wares 2017). Future studies should also use complementary nuclear markers to improve understanding of the genetic structure of these species within their tidal distribution.
Phylogeographic patterns as a result of rafting dispersal
In G. lingulatum, the lack of a phylogeographic break, patchy distribution of haplotypes within its geographic range and no isolation-by-distance are indications of long-distance dispersal events. Human-mediated transport (Banks et al. 2015) or rafting dispersal (e.g., wood and floating seaweeds, Thiel and Gutow 2005a) can move organisms over long distances. In the case of human-mediated dispersal, transport through maritime traffic (ballast waters and ship hulls) has been shown to affect the phylogeographic patterns of other seaweeds (e.g., Undaria pinnatifida, Voisin et al. 2005; Caulerpa cylindracea, Piazzi et al. 2016). Although G. lingulatum germlings have a high tolerance to total darkness (Santelices et al. 2002), conditions in ballast waters are strongly adverse (i.e., anoxia) and maritime traffic occurs offshore and in ports, not in rocky areas, so that this transport mechanism is much less likely than through rafting dispersal by floating seaweeds. In addition, at least one species from the genus Gelidium has been detected on derelict aquaculture buoys in Coquimbo Bay (30°S) (Astudillo et al. 2009), and so other floating substrata cannot be completely ruled out as dispersal vehicle. Another long-distance dispersal mechanism through drifting fronds has been also reported in G. versicolor on the south coast of England (Dixon and Irvine 1977).
Detached seaweeds are one of the most common floating substrata along the coast of Chile (Hinojosa et al. 2010; 2011; Wichmann et al. 2012). Rafting transport could increase the gene flow between distant populations and thus modify the genetic structure, as had been described for some invertebrates inhabiting holdfasts of D. antarctica (Nikula et al. 2010; Haye et al. 2012). In Chile, both G. lingulatum and G. rex are often found attached to holdfasts of floating and recently stranded bull kelps D. antarctica (higher frequencies in G. lingulatum than G. rex, Macaya et al. 2016), but the continental clade (30°S–44°S) of this bull kelp presents a very different phylogeographic pattern (Fraser et al. 2010) than the two red seaweeds. For example, D. antarctica has a much more genetically structured pattern (i.e., more mutational steps among pairs of haplotypes) compared to G. rex and its geographical haplotype distribution is not similar to the patchy pattern of G. lingulatum. This suggests that other factors during, or after, along-shore rafting journeys could be affecting connectivity among distant populations. Moreover, Macaya et al. (2016) suggested that the physiological capacity to tolerate new environmental conditions at the sea surface during rafting might be directly related to the bathymetric distribution pattern of seaweeds in their benthic habitats. Particularly in turf algae, changes in solar radiation levels during transfer from the benthic to the pelagic environment (rafting at the sea surface) could affect performance and persistence of these algae. Given the intertidal distribution of the two study species, these shifts in light regime should be more critical in G. rex than in G. lingulatum. In addition, the difference in latitudinal distribution between the two species (i.e., wider in G. lingulatum compared to G. rex) could also suggest that there are different tolerance capacities to harsh conditions between them.
Long-distance dispersal could also be consistent with the historical patterns observed. For G. lingulatum and G. rex a recent population expansion was detected, although in the latter this was only observed in southern populations, while in northern populations (particularly, those from BURR to SAUC) this pattern was not so clear. This coincides with the high presence of private haplotypes in both species, which in the case of G. rex were detected only at the northernmost and southernmost sampling sites (i.e., BURR and BUCA). Nikula et al. (2010) reported genetic signatures of population expansion in epifaunal invertebrates (i.e., peracarids) associated with holdfasts of floating bull kelp D. antarctica in subantarctic areas. This suggests that rapid historical population growth might have been favored by frequent rafting events.
Successful immigration after rafting journeys is likely also influenced by other factors such as substratum availability, settlement capacity of immigrant propagules, and the density of the resident population (i.e., density blocking, Waters et al. 2013; Neiva et al. 2014). In dense local populations, new haplotypes that arrive with few immigrant individuals have a high probability of being outcompeted because of their rarity, which leads to rapid elimination of these new haplotypes by genetic drift. For example, this could be happening for G. rex in locations such as FUAD, where a single private haplotype was very frequent among the sampled individuals. This is congruent with records in locations adjacent to FUAD (30°S–31°S), where higher population abundances of this species have been observed in comparison to northern and southern sites (Broitman et al. 2001, Vásquez and Vega 2004). This could be because, as observed in other species of Gelidium (i.e., G. arbuscula, Sosa and García-Reina 1992; Sosa et al. 1998), stoloniferous outgrowths of creeping axes is a common way of propagation; therefore, locally adapted clones could propagate asexually and became predominant through competitive advantage, thereby minimizing the availability of unoccupied substratum and limiting opportunities for recruitment of new genotypes (via sexual reproduction). In addition, the ability of thallus reattachment of these species (Rojas et al. 1996) would favor the monopolization of the substratum. For example, Alberto et al. (1999) suggested that populations of G. sesquipedale from northern France to Morocco maintain the gene flow among populations (<500 km) through occasional transport of detached fronds by local currents during storm events and subsequent reattachment to new substrata.
Conversely, strong disturbances with massive local mortalities (e.g., coastal uplifts after earthquakes) could change this pattern (Castilla et al. 2010; Jaramillo et al. 2012), enhancing the possibility of successful immigration to uncolonized habitats or those with lower population density. Habitat heterogeneity could also be an important factor influencing phylogeography and population connectivity in intertidal seaweeds. For example, the extent of sandy beaches (particularly from 36°S to 40°S on the coast of Chile, Thiel et al. 2007) could reduce the availability of primary substratum for intertidal seaweeds inhabiting rocky shores and thus, genetic drift and small effective population sizes probably contribute strongly to the divergence between their populations (Fraser et al. 2010). Therefore, our results suggest that the phylogeographic patterns of these intertidal algae are affected by rafting dispersal via floating seaweeds, although there may be differential functional capabilities during rafting journeys and/or differential efficiency of recruitment after long-distance dispersal that could explain the divergent patterns between both species. Future studies should also focus on phenology and the relationships between different phases of the life cycle in these species.
Conclusions and outlook
Our phylogeographic study confirms the presence of G. lingulatum along the Chilean coast at least from 29°S to 42°S (no recent records are available for the south, 42°S–56°S, John et al. 2003; Soto et al. 2012), but our surveys suggest that this species does not occur north of 29°S. Similarly, we only found individuals of G. rex between 28°S and 34°S, despite a reported distribution ranging from 16°S to 39°S (Santelices and Abbott 1985), thus suggesting a previous overestimation of the geographical range in both study species (Fig. 1).
Gelidium lingulatum had some genetic structure (i.e., φ ST values are highly significant among several locations), but did not follow a clear geographic pattern (i.e., no phylogeographic break, and haplotypes disappear and reappear repeatedly along its geographical range), contrasting with findings for other red seaweeds with similar life histories and distribution ranges (e.g., M. laminarioides, Montecinos et al. 2012; Nothogenia chilensis, Lindstrom et al. 2015). A shallow genetic structure was observed in G. rex, with a phylogeographic break coinciding with the phylogeographic discontinuity described for other species between 29°S and 33°S (Tellier et al. 2009; Sánchez et al. 2011; Montecinos et al. 2012). We propose that these contrasting patterns of G. lingulatum and G. rex might be due to (1) differences in tidal level and species-specific adaptations in physiology and reproductive biology (e.g., temperatures below 10 °C are limiting for growth of these species, Oliger and Santelices 1981) and (2) differences in extrinsic dispersal capacities, with more effective rafting dispersal for G. lingulatum than for G. rex.
As previously suggested (Macaya et al. 2016), while our study provides support for efficient rafting dispersal, it also indicates that the relative contribution of rafting to contemporaneous population connectivity may vary, depending on seaweed biology (e.g., functional and reproductive characteristics of these epiphytic non-buoyant seaweeds) and population ecology (e.g., density blocking). Further studies should in particular focus on rafting routes, via genetic characterization of the source populations of stranded D. antarctica, particularly those holdfasts carrying G. rex or G. lingulatum individuals. Recent studies indicate that supplies of bull kelp rafts to the shore vary strongly along the coast of Chile (López et al. 2017, Fig. 1), which could affect connectivity among the populations of D. antarctica and of associated epibionts.
References
Alberto F, Santos R, Leitão JM (1999) Assessing patterns of geographic dispersal of Gelidium sesquipedale (Rhodophyta) through RAPD differentiation of populations. Mar Ecol Prog Ser 191:101–108. doi:10.3354/Meps191101
Alberto F, Raimondi PT, Reed DC, Coelho NC, Leblois R, Whitmer A, Serrão EA (2010) Habitat continuity and geographic distance predict population genetic differentiation in giant kelp. Ecology 91:49–56. doi:10.1890/09-0050.1
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Aravena G, Broitman B, Stenseth NC (2014) Twelve years of change in coastal upwelling along the central-northern coast of Chile: spatially heterogeneous responses to climatic variability. PLoS One 9:e90276. doi:10.1371/journal.pone.0090276
Astudillo JC, Bravo M, Dumont CP, Thiel M (2009) Detached aquaculture buoys in the SE Pacific: potential dispersal vehicles for associated organisms. Aquat Biol 5:219–231. doi:10.3354/ab00151
Ballard JW, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13:729–744. doi:10.1046/j.1365-294X.2003.02063.x
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Banks NC, Paini DR, Bayliss KL, Hodda M (2015) The role of global trade and transport network topology in the human-mediated dispersal of alien species. Ecol Lett 18:188–199. doi:10.1111/ele.12397
Billard E, Daguin C, Pearson G, Serrão E, Engel C, Valero M (2005) Genetic isolation between three closely related taxa: Fucus vesiculosus, F. spiralis, and F. ceranoides (Phaeophyceae). J Phycol 41:900–905. doi:10.1111/j.0022-3646.2005.04221.x
Bobadilla M, Santelices B (2005) Variations in the dispersal curves of macroalgal propagules from a source. J Exp Mar Biol Ecol 327:47–57. doi:10.1016/j.jembe.2005.06.006
Boo GH, Park JK, Boo SM (2013) Gelidiophycus (Rhodophyta: Gelidiales): a new genus of marine algae from East Asia. Taxon 62:1105–1116. doi:10.12705/626.7
Boo GH, Mansilla A, Nelson W, Bellgrove A, Boo SM (2014a) Genetic connectivity between trans-oceanic populations of Capreolia implexa (Gelidiales, Rhodophyta) in cool temperate waters of Australasia and Chile. Aquat Bot 119:73–79. doi:10.1016/j.aquabot.2014.08.004
Boo GH, Kim KM, Nelson WA, Riosmena-Rodríguez R, Yoon KJ, Boo SM (2014b) Taxonomy and distribution of selected species of the agarophyte genus Gelidium (Gelidiales, Rhodophyta). J Appl Phycol 26:1243–1251. doi:10.1007/s10811-013-0111-7
Boo GH, Le Gall L, Miller KA, Freshwater DW, Wernberg T, Terada R, Yoon KJ, Boo SM (2016) A novel phylogeny of the Gelidiales (Rhodophyta) based on five genes including the nuclear CesA, with descriptions of Orthogonacladia gen. nov and Orthogonacladiaceae fam. nov. Mol Phylogenet Evol 101:359–372. doi:10.1016/j.ympev.2016.05.018
Bouza N, Caujape-Castells J, González-Pérez MA, Sosa PA (2006) Genetic structure of natural populations in the red algae Gelidium canariense (Gelidiales, Rhodophyta) investigated by random amplified polymorphic DNA (RAPD) markers. J Phycol 42:304–311. doi:10.1111/j.1529-8817.2006.00201.x
Broitman BR, Navarrete SA, Smith F, Gaines SD (2001) Geographic variation of southeastern Pacific intertidal communities. Mar Ecol Prog Ser 224:21–34. doi:10.3354/Meps224021
Camus PA (2001) Biogeografía marina de Chile continental. Rev Chil Hist Nat 74:587–617
Castilla JC, Manríquez PH, Camaño A (2010) Effects of rocky shore coseismic uplift and the 2010 Chilean mega-earthquake on intertidal biomarker species. Mar Ecol Prog Ser 418:17–23. doi:10.3354/meps08830
Coyer JA, Hoarau G, Costa JF, Hogerdijk B, Serrão EA, Billard E, Valero M, Pearson GA, Olsen JL (2011a) Evolution and diversification within the intertidal brown macroalgae Fucus spiralis/F. vesiculosus species complex in the North Atlantic. Mol Phylogenet Evol 58:283–296. doi:10.1016/j.ympev.2010.11.015
Coyer JA, Hoarau G, Van Schaik J, Luijckx P, Olsen JL (2011b) Trans-Pacific and trans-Arctic pathways of the intertidal macroalga Fucus distichus L. reveal multiple glacial refugia and colonizations from the North Pacific to the North Atlantic. J Biogeogr 38:756–771. doi:10.1111/j.1365-2699.2010.02437.x
Cumming RA, Nikula R, Spencer HG, Waters JM (2014) Transoceanic genetic similarities of kelp-associated sea slug populations: long-distance dispersal via rafting? J Biogeogr 41:2357–2370. doi:10.1111/jbi.12376
Dawson MN, Hays CG, Grosberg RK, Raimondi PT (2014) Dispersal potential and population genetic structure in the marine intertidal of the eastern North Pacific. Ecol Monogr 84:435–456. doi:10.1890/13-0871.1
Destombe C, Godin J, Lefebvre C, Dehorter O, Vernet P (1992) Differences in dispersal abilities of haploid and diploid spores of Gracilaria verrucosa (Gracilariales, Rhodophyta). Bot Mar 35:93–98. doi:10.1515/botm.1992.35.2.93
Dixon PS, Irvine LM (1977) Seaweeds of the British Isles. Volume 1. Rhodophyta. Part 1. Introduction, Nemaliales, Gigartinales. The Natural History Museum, London, p 252
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581. doi:10.1046/j.1365-294X.2002.01650.x
Engel CR, Destombe C, Valero M (2004) Mating system and gene flow in the red seaweed Gracilaria gracilis: effect of haploid-diploid life history and intertidal rocky shore landscape on fine-scale genetic structure. Heredity 92:289–298. doi:10.1038/sj.hdy.6800407
Engel CR, Billard E, Voisin M, Viard F (2008) Conservation and polymorphism of mitochondrial intergenic sequences in brown algae (Phaeophyceae). Eur J Phycol 43:195–205. doi:10.1080/09670260701823437
Ewers-Saucedo C, Pringle JM, Sepúlveda HH, Byers JE, Navarrete SA, Wares JP (2016) The oceanic concordance of phylogeography and biogeography: a case study in Notochthamalus. Ecol Evol 6:4403–4420. doi:10.1002/ece3.2205
Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13:853–864. doi:10.1046/j.1365-294X.2003.02004.x
Excoffier L, Lischer HEL (2010) Arlequin (version 3.5): arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi:10.1111/j.1755-0998.2010.02847.x
Försterra G (2009) Ecological and biogeographical aspects of the Chilean Fjord region. In: Häussermann V, Försterra G (eds) Marine benthic fauna of chilean patagonia. Nature in Focus, Puerto Montt, pp 61–76
Fraser CI, Nikula R, Spencer HG, Waters JM (2009a) Kelp genes reveal effects of subantarctic sea ice during the Last Glacial Maximum. Proc Natl Acad Sci USA 106:3249–3253. doi:10.1073/pnas.0810635106
Fraser CI, Hay CH, Spencer HG, Waters JM (2009b) Genetic and morphological analyses of the southern bull kelp Durvillaea antarctica (Phaeophyceae: Durvillaeales) in New Zealand reveal cryptic species. J Phycol 45:436–443. doi:10.1111/j.1529-8817.2009.00658.x
Fraser CI, Thiel M, Spencer HG, Waters JM (2010) Contemporary habitat discontinuity and historic glacial ice drive genetic divergence in Chilean kelp. BMC Evol Biol 10:203. doi:10.1186/1471-2148-10-203
Freshwater DW, Rueness J (1994) Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, based on rbcL nucleotide sequence analysis. Phycologia 33:187–194. doi:10.2216/i0031-8884-33-3-187.1
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gavio B, Fredericq S (2002) Grateloupia turuturu (Halymeniaceae, Rhodophyta) is the correct name of the non-native species in the Atlantic known as Grateloupia doryphora. Eur J Phycol 37:349–359. doi:10.1017/s0967026202003839
Gómez IM, Westermeier RC (1991) Frond regrowth from basal disc in Iridaea laminarioides (Rhodophyta, Gigartinales) at Mehuín, southern Chile. Mar Ecol Prog Ser 73:83–91. doi:10.3354/meps073083
González S, Stotz W, Toledo P, Jorquera M, Romero M (1991) Utilización de diferentes microambientes del intermareal como lugares de asentamiento por Fissurella spp (Gastropoda: Prosobranchia) (Palo Colorado, Los Vilos, Chile). Rev Biol Mar Oceanogr 26: 325–338. http://www.revbiolmar.cl/escaneados/262-325.pdf. Accessed 29 Aug 2016
González A, Beltrán J, Hiriart-Bertrand L, Flores V, de Reviers B, Correa JA, Santelices B (2012) Identification of cryptic species in the Lessonia nigrescens complex (Phaeophyceae, Laminariales). J Phycol 48:1153–1165. doi:10.1111/j.1529-8817.2012.01200.x
Grant SW (2016) Paradigm shifts in the phylogeographic analysis of seaweeds. In: Hu ZM, Fraser CI (eds) Seaweed phylogeography. Springer, Dordrecht, pp 23–62. doi:10.1007/978-94-017-7534-2_2
Guillemin ML, Valero M, Faugeron S, Nelson W, Destombe C (2014) Tracing the trans-pacific evolutionary history of a domesticated seaweed (Gracilaria chilensis) with archaeological and genetic data. PLoS One 9:e114039. doi:10.1371/journal.pone.0114039
Guillemin ML, Valero M, Tellier F, Macaya EC, Destombe C, Faugeron S (2016a) Phylogeography of seaweeds in the South East Pacific: complex evolutionary processes along a latitudinal gradient In: Hu ZM, Fraser CI (eds) Seaweed Phylogeography. Springer, Dordrecht, pp 251–278. doi:10.1007/978-94-017-7534-2_10
Guillemin ML, Contreras-Porcia L, Ramírez ME, Macaya EC, Bulboa-Contador C, Woods H, Wyatt C, Brodie J (2016b) The bladed Bangiales (Rhodophyta) of the South Eastern Pacific: molecular species delimitation reveals extensive diversity. Mol Phylogenet Evol 94:814–826. doi:10.1016/j.ympev.2015.09.027
Guo B, Wares JP (2017) Large-scale gene flow in the barnacle Jehlius cirratus and contrasts with other broadly-distributed taxa along the Chilean coast. PeerJ 5:e2971. doi:10.7717/peerj.2971
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Haye PA, Varela AI, Thiel M (2012) Genetic signatures of rafting dispersal in algal-dwelling brooders Limnoria spp. (Isopoda) along the SE Pacific (Chile). Mar Ecol Prog Ser 455:111–122. doi:10.3354/meps09673
Haye PA, Segovia NI, Muñoz-Herrera NC, Gálvez FE, Martínez A, Meynard A, Pardo-Gandarillas MC, Poulin E, Faugeron S (2014) Phylogeographic structure in benthic marine invertebrates of the southeast Pacific coast of Chile with differing dispersal potential. PLoS One 9:e88613. doi:10.1371/journal.pone.0088613
Hernández CJ (1997) Análisis de la variación estacional e interanual de la cosecha de Gelidium robustum en Baja California Sur, México. Master thesis, Centro Interdisciplinario de Ciencias Marinas, La Paz, Baja California Sur, México
Hinojosa IA, Pizarro M, Ramos M, Thiel M (2010) Spatial and temporal distribution of floating kelp in the channels and fjords of southern Chile. Estuar Coast Shelf Sci 87:367–377. doi:10.1016/j.ecss.2009.12.010
Hinojosa IA, Rivadeneira MM, Thiel M (2011) Temporal and spatial distribution of floating objects in coastal waters of central-southern Chile and Patagonian fjords. Cont Shelf Res 31:172–186. doi:10.1016/j.csr.2010.04.013
Hoffmann A, Santelices B (1997) Flora marina de Chile Central. Ediciones Universidad Católica de Chile, Santiago de Chile
Hu ZM, De-Lin D, López-Baptista J (2016) Seaweed phylogeography from 1994 to 2014: an overview. In: Hu ZM, Fraser CI (eds) Seaweed Phylogeography. Springer, Dordrecht, pp 3–22. doi:10.1007/978-94-017-7534-2_1
Jaramillo E, Dugan JE, Hubbard DM, Melnick D, Manzano M, Duarte C, Campos C, Sánchez R (2012) Ecological implications of extreme events: footprints of the 2010 earthquake along the Chilean coast. PLoS One 7:8. doi:10.1371/journal.pone.0035348
John D, Paterson G, Evans N, Ramírez M, Spencer J, Báez P, Ferrero T, Valentine C, Reid D (2003) Manual de biotopos marinos de la región de Aysén, Sur de Chile (A manual of marine biotopes of Region Aysén, Southern Chile. The Laguna San Raphael National Park, Estero Elefantes, Chonos Archipelago and Katalalixar). The Natural History Museum, London
Kelly RP, Palumbi SR (2010) Genetic structure among 50 species of the Northeastern Pacific rocky intertidal community. PLoS One 5:13. doi:10.1371/journal.pone.0008594
Kiessling T, Gutow L, Thiel M (2015) Marine litter as habitat and dispersal vector. In: Bergmann M, Gutow L, Klages K (eds) Marine anthropogenic litter, Springer, Berlin, pp 141–184. doi:10.1007/978-3-319-16510-3_6
Kim KM, Boo SM (2012) Phylogenetic relationships and distribution of Gelidium crinale and G. pusillum (Gelidiales, Rhodophyta) using cox1 and rbcL sequences. Algae 27:83–94. doi:10.4490/algae.2012.27.2.083
Kim KM, Hwang IK, Park JK, Boo SM (2011) A new agarophyte species, Gelidium eucorneum sp. nov. (Gelidiales, Rhodophyta), based on molecular and morphological data. J Phycol 47:904–910. doi:10.1111/j.1529-8817.2011.01005.x
Kim KM, Hoarau GG, Boo SM (2012) Genetic structure and distribution of Gelidium elegans (Gelidiales, Rhodophyta) in Korea based on mitochondrial cox1 sequence data. Aquat Bot 98:27–33. doi:10.1016/j.aquabot.2011.12.005
Krueger-Hadfield SA, Hoban SM (2016) The importance of effective sampling for exploring the population dynamics of haploid–diploid seaweeds. J Phycol 52:1–9. doi:10.1111/jpy.12366
Krueger-Hadfield SA, Roze D, Mauger S, Valero M (2013) Intergametophytic selfing and microgeographic genetic structure shape populations of the intertidal red seaweed Chondrus crispus. Mol Ecol 22:3242–3260. doi:10.1111/mec.12191
Kützing FT (1868) Tabulae phycologicae; oder, Abbildungen der Tange. Vol. XVIII pp. [i–iii], 1–35, 100 pls. Nordhausen: Gedruckt auf kosten des Verfassers (in commission bei W. Köhne)
Lachkar Z, Gruber N (2012) A comparative study of biological production in eastern boundary upwelling systems using an artificial neural network. Biogeosciences 9:293–308. doi:10.5194/bg-9-293-2012
Lin SM, Fredericq S, Hommersand MH (2001) Systematics of the Delesserlaceae (Ceramiales, Rhodophyta) based on large subunit rDNA and rbcL sequences, including the Phycodryoideae, subfam. nov. J Phycol 37:881–899. doi:10.1046/j.1529-8817.2001.01012.x
Lindstrom SC, Gabrielson PW, Hughey JR, Macaya EC, Nelson WA (2015) Sequencing of historic and modern specimens reveals cryptic diversity in Nothogenia (Scinaiaceae, Rhodophyta). Phycologia 54:97–108. doi:10.2216/14-077.1
López B, Macaya EC, Tellier F, Tala F, Thiel M (2017) The variable routes of rafting: stranding dynamics of floating bull-kelp Durvillaea antarctica (Fucales, Phaeophyceae) on beaches in the SE Pacific. J Phycol 54:70–84. doi:10.1111/jpy.12479
Macaya EC, Zuccarello GC (2010a) Genetic structure of the giant kelp Macrocystis pyrifera along the southeastern Pacific. Mar Ecol Prog Ser 420:103–112. doi:10.3354/meps08893
Macaya EC, Zuccarello GC (2010b) DNA barcoding and genetic divergence in the giant kelp Macrocystis (Laminariales). J Phycol 46:736–742. doi:10.1111/j.1529-8817.2010.00845.x
Macaya EC, López B, Tala F, Tellier F, Thiel M (2016) Float and raft: role of buoyant seaweeds in the phylogeography and genetic structure of non-buoyant associated flora. In: Hu ZM, Fraser CI (eds) Seaweed phylogeography, Springer, Dordrecht, pp 97–130. doi:10.1007/978-94-017-7534-2_4
Matsuhiro B, Urzúa CC (1990) Agars from Gelidium rex (Gelidiales, Rhodophyta). Hydrobiologia 204:545–549. doi:10.1007/bf00040284
Matsuhiro B, Urzúa CC (1991) Agars from Chilean Gelidiaceae. Hydrobiologia 221:149–156. doi:10.1007/bf00028371
Melo RA (1998) Gelidium commercial exploitation: natural resources and cultivation. J Appl Phycol 10:303–314. doi:10.1023/A:1008070419158
Montecinos A, Broitman BR, Faugeron S, Haye PA, Tellier F, Guillemin ML (2012) Species replacement along a linear coastal habitat: phylogeography and speciation in the red alga Mazzaella laminarioides along the south east Pacific. BMC Evol Biol 12:97. doi:10.1186/1471-2148-12-97
Muhlin JF, Engel CR, Stessel R, Weatherbee RA, Brawley SH (2008) The influence of coastal topography, circulation patterns, and rafting in structuring populations of an intertidal alga. Mol Ecol 17:1198–1210. doi:10.1111/j.1365-294X.2007.03624.x
Neiva J, Assis J, Fernandes F, Pearson GA, Serrão EA, Maggs C (2014) Species distribution models and mitochondrial DNA phylogeography suggest an extensive biogeographical shift in the high-intertidal seaweed Pelvetia canaliculata. J Biogeogr 41:1137–1148. doi:10.1111/jbi.12278
Nelson WA, Farr TJ, Broom JES (2006) Phylogenetic diversity of New Zealand gelidiales as revealed by rbcL sequence data. J Appl Phycol 18:653–661. doi:10.1007/s10811-006-9068-0
Nikula R, Fraser CI, Spencer HG, Waters JM (2010) Circumpolar dispersal by rafting in two subantarctic kelp-dwelling crustaceans. Mar Ecol Prog Ser 405:221–230. doi:10.3354/meps08523
Nikula R, Spencer HG, Waters JM (2011a) Evolutionary consequences of microhabitat: population-genetic structuring in kelp- vs. rock-associated chitons. Mol Ecol 20:4915–4924. doi:10.1111/j.1365-294X.2011.05332.x
Nikula R, Spencer HG, Waters JM (2011b) Comparison of population-genetic structuring in congeneric kelp- versus rock-associated snails: a test of a dispersal-by-rafting hypothesis. Ecol Evol 1:169–180. doi:10.1002/ece3.16
Nikula R, Spencer HG, Waters JM (2013) Passive rafting is a powerful driver of transoceanic gene flow. Biol Lett 9:20120821. doi:10.1098/rsbl.2012.0821
Oliger P, Santelices B (1981) Physiological ecology studies on Chilean Gelidiales. J Exp Mar Biol Ecol 53:65–75. doi:10.1016/0022-0981(81)90084-8
Ortega M, Godínez-Ortega J, Garduño G (2001) Catálogo de algas bénticas de las costas mexicanas del Golfo de México y Mar Caribe. Instituto de Biología, Universidad Nacional Autónoma de México, México
Palumbi SR (1994) Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst 25:547–572. doi:10.1146/annurev.ecolsys.25.1.547
Perrone C, Felicini GP, Bottalico A (2006) The prostrate system of the Gelidiales: diagnostic and taxonomic importance. Bot Mar 49:23–33. doi:10.1515/Bot2006.003
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855. doi:10.1046/j.1523-1739.1998.96489.x
Piazzi L, Balata D, Bulleri F, Gennaro P, Ceccherelli G (2016) The invasion of Caulerpa cylindracea in the Mediterranean: the known, the unknown and the knowable. Mar Biol 163:161–174. doi:10.1007/S00227-016-2937-4
Polzin T, Daneshmand SV (2003) On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett 31:12–20. doi:10.1016/s0167-6377(02)00185-2
Ramírez M, Santelices B (1981) Análisis biogeográfico de la flora algológica de Antofagasta (norte de Chile). Bol Mus Nac Hist Nat Santiago de Chile 38:5–20
Ramírez ME, Santelices B (1991) Catálogo de las algas marinas bentónicas de la costa temperada del Pacífico de Sudamérica. Monografías Biológicas 5, Ediciones Universidad Católica de Chile, Santiago de Chile
Robuchon M, Le Gall L, Mauger S, Valero M (2014) Contrasting genetic diversity patterns in two sister kelp species co-distributed along the coast of Brittany, France. Mol Ecol 23:2669–2685. doi:10.1111/mec.12774
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Rojas R, León N, Rojas R (1996) Practical and descriptive techniques for Gelidium rex (Gelidiales, Rhodophyta) culture. Hydrobiologia 327:367–370
Sánchez R, Sepúlveda RD, Brante A, Cárdenas L (2011) Spatial pattern of genetic and morphological diversity in the direct developer Acanthina monodon (Gastropoda: Mollusca). Mar Ecol Prog Ser 434:121–131. doi:10.3354/meps09184
Santelices B (1980) Phytogeographic characterization of the temperate coast of Pacific South-America. Phycologia 19:1–12. doi:10.2216/i0031-8884-19-1-1.1
Santelices B (1986) The wild harvest and culture of the economically important species of Gelidium in Chile. In: Doty MS, Caddy JS, Santelices B (eds) Case of studies of seven commercial seaweed resources. FAO Fisheries Technical Paper, vol 281, FAO, Rome, pp 165–192
Santelices B (1990) New and old problems in the taxonomy of the Gelidiales (Rhodophyta). Hydrobiologia 204:125–135. doi:10.1007/bf00040224
Santelices B, Abbott IA (1985) Gelidium rex sp. nov. (Gelidiales. Rhodophyta) from central Chile. In: Abbott IA, Norris JN (eds) Taxonomy of economic seaweeds with reference to some Pacific and Caribbean species. California Sea Grant College Program, La Jolla, pp 33–36
Santelices B, Montalva S (1983) Taxonomic studies on Gelidiaceae (Rhodophyta) from central Chile. Phycologia 22:185–196. doi:10.2216/i0031-8884-22-2-185.1
Santelices B, Stewart JG (1985) Pacific species of Gelidium Lamouroux and other Gelidiales (Rhodophyta), with keys and descriptions to the common or economically important species. In: Abbott IA, Norris JN (eds) Taxonomy of economic seaweeds with reference to some Pacific and Caribbean species. California Sea Grant College Program, California, La Jolla, pp 17–31
Santelices B, Varela D (1994) Abiotic control of reattachment in Gelidium chilense (Montagne) Santelices and Montalva (Gelidiales, Rhodophyta). J Exp Mar Biol Ecol 177:145–155. doi:10.1016/0022-0981(94)90233-x
Santelices B, Aedo D, Hoffmann A (2002) Banks of microscopic forms and survival to darkness of propagules and microscopic stages of macroalgae. Rev Chil Hist Nat 75:547–555. doi:10.4067/S0716-078X2002000300006
Saunders GW (2005) Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos Trans R Soc Lond B Biol Sci 360:1879–1888. doi:10.1098/rstb.2005.1719
Selkoe KA, Toonen RJ (2011) Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar Ecol Prog Ser 436:291–305. doi:10.3354/meps09238
Slatkin M (1993) Isolation by distance in equilibrium and nonequilibrium populations. Evolution 47:264–279. doi:10.2307/2410134
Sosa P, García-Reina G (1992) Genetic variability and differentiation of sporophytes and gametophytes in populations of Gelidium arbuscula (Gelidiaceae: Rhodophyta) determined by isozyme electrophoresis. Mar Biol 113:679–688
Sosa PA, Valero M, Batista F, González-Pérez MA (1998) Genetic structure of natural populations of Gelidium species: a re-evaluation of results. J Appl Phycol 10:279–284. doi:10.1023/A:1008092023549
Soto EH, Báez P, Ramírez ME, Letelier S, Naretto J, Rebolledo A (2012) Biotopos marinos intermareales entre Canal Trinidad y Canal Smyth, Sur de Chile. Rev Biol Mar Oceanogr 47:177–191
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tapia FJ, Largier JL, Castillo M, Wieters EA, Navarrete SA (2014) Latitudinal discontinuity in thermal conditions along the nearshore of central-northern Chile. PLoS One 9:e110841. doi:10.1371/journal.pone.0110841
Technelysium Pty Ltd (2016) CHROMAS LITE. Available at: http://www.technelysium.com.au/chromas_lite.html. Accessed 12 April 2016
Tellier F, Meynard AP, Correa JA, Faugeron S, Valero M (2009) Phylogeographic analyses of the 30°S south-east Pacific biogeographic transition zone establish the occurrence of a sharp genetic discontinuity in the kelp Lessonia nigrescens: vicariance or parapatry? Mol Phylogenet Evol 53:679–693. doi:10.1016/j.ympev.2009.07.030
Tellier F, Vega JMA, Broitman BR, Vásquez JA, Valero M, Faugeron S (2011) The importance of having two species instead of one in kelp management: the Lessonia nigrescens species complex. Cah Biol Mar 52:455–465
Thiel M, Gutow L (2005a) The ecology of rafting in the marine environment. I. The floating substrata. Oceanogr Mar Biol Ann Rev 42:181–263. doi:10.1201/9780203507810
Thiel M, Gutow L (2005b) The ecology of rafting in the marine environment. II. The rafting organisms and community. Oceanogr Mar Biol Ann Rev 43:279–418. doi:10.1201/9781420037449
Thiel M, Haye PA (2006) The ecology of rafting in the marine environment. III. Biogeographical and evolutionary consequences. Oceanogr Mar Biol Ann Rev 44:323–429. doi:10.1201/9781420006391.ch7
Thiel M, Macaya EC, Acuña E, Arntz WE, Bastias H, Brokordt K, Camus PA, Castilla JC, Castro LR, Cortés M, Dumont CP, Escribano R, Fernández M, Gajardo JA, Gaymer CF, Gómez I, González AE, González HE, Haye PA, Illanes JE, Iriarte JL, Lancellotti DA, Luna-Jorquera G, Luxoroi C, Manríquez PH, Marín V, Muñoz P, Navarrete SA, Pérez E, Poulin E, Sellanes J, Sepúlveda HH, Stotz W, Tala F, Thomas A, Vargas CA, Vásquez JA, Vega JMA (2007) The Humboldt Current system of northern and central Chile. Oceanogr Mar Biol Ann Rev 45:195–344. doi:10.1201/9781420050943
Thomas DT, Freshwater DW (2001) Studies of Costa Rican Gelidiales (Rhodophyta): four Caribbean taxa including Pterocladiella beachii sp nov. Phycologia 40:340–350. doi:10.2216/i0031-8884-40-4-340.1
Valero M, Destombe C, Mauger S, Ribout C, Engel CR, Daguin-Thiebaut C, Tellier F (2011) Using genetic tools for sustainable management of kelps: a literature review and the example of Laminaria digitata. Cah Biol Mar 52:467–483
Vargas DR, Collado-Vides L (1996) Architectural models for apical patterns in Gelidium (Gelidiales, Rhodophyta): hypothesis of growth. Phycol Res 44:95–100
Vásquez JA, Vega JMA (2004) Ecosistemas marinos costeros del Parque Nacional Bosque Fray Jorge. In: Squeo F, Gutiérrez J, Hernández I (eds) Historia Natural del Parque Nacional Bosque Fray Jorge. Ediciones Universidad de La Serena, La Serena, pp 235–252
Voisin M, Engel CR, Viard F (2005) Differential shuffling of native genetic diversity across introduced regions in a brown alga: aquaculture vs. maritime traffic effects. Proc Natl Acad Sci USA 102:5432–5437. doi:10.1073/pnas.0501754102
Waters JM, Fraser CI, Hewitt GM (2013) Founder takes all: density-dependent processes structure biodiversity. Trends Ecol Evol 28:78–85. doi:10.1016/j.tree.2012.08.024
Weersing K, Toonen RJ (2009) Population genetics, larval dispersal, and connectivity in marine systems. Mar Ecol Prog Ser 393:1–12. doi:10.3354/meps08287
Wichmann CS, Hinojosa IA, Thiel M (2012) Floating kelps in Patagonian Fjords: an important vehicle for rafting invertebrates and its relevance for biogeography. Mar Biol 159:2035–2049. doi:10.1007/s00227-012-1990-x
Yoon KJ, Kim KM, Boo GH, Miller KA, Boo SM (2014) Mitochondrial cox1 and cob sequence diversities in Gelidium vagum (Gelidiales, Rhodophyta) in Korea. Algae 29:15–25. doi:10.4490/algae.2014.29.1.015
Zakas C, Binford J, Navarrete SA, Wares JP (2009) Restricted gene flow in Chilean barnacles reflects an oceanographic and biogeographic transition zone. Mar Ecol Prog Ser 394:165–177. doi:10.3354/meps08265
Acknowledgements
This study was financed by the following grants: CONICYT/FONDECYT 1131082 to MT, F. Tala and F. Tellier, CONICYT/FONDECYT 1110437 to EM, and CONICYT/FONDECYT 11121504 to F. Tellier. BL received financial support by PhD-fellowship Beca CONICYT-PCHA/Doctorado Nacional/2014-21140010. Additional support came from International Research Network “Diversity, Evolution and Biotechnology of Marine Algae” (GDRI N ̊ 0803). The collaboration of Óscar Pino, José Pantoja, Alvaro Gallardo, Solange Pacheco, Ricardo Jeldres, María Fabiola Monsalvez, Ariel Cáceres, Ulyces Urtubia, Vieia Villalobos and Tim Kiessling in field activities is gratefully acknowledged. The valuable comments from two anonymous referees were very helpful in improving the original manuscript. We are grateful to Lucas Eastman for checking the language of the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interests.
Human and animals rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Responsibile Editor: O. Puebla.
Reviewed by Undisclosed experts.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
López, B.A., Tellier, F., Retamal-Alarcón, J.C. et al. Phylogeography of two intertidal seaweeds, Gelidium lingulatum and G. rex (Rhodophyta: Gelidiales), along the South East Pacific: patterns explained by rafting dispersal?. Mar Biol 164, 188 (2017). https://doi.org/10.1007/s00227-017-3219-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-017-3219-5