Abstract
The life cycle of the fish ectoparasitic isopod Gnathia trimaculata is described based on both field samplings and laboratory observations. Species identification of the larvae was based on morphological observation and supported by molecular analysis. As the results of field samplings in several sites of southwestern and central Japan (24–34°N, 124–139°E) from 2005–2011, approximately 900 third-stage larvae of G. trimaculata were found on 25 elasmobranch species, and 220 first- and second-stage larvae were found on three teleost species. No third-stage larvae were found on the teleosts, and the larvae of younger stages never infested elasmobranchs. Therefore, G. trimaculata is supposed to shift its host from teleosts to elasmobranchs as it develops. We discuss the developmental periods, life span, distribution, and predation risk of the present species compared with other gnathiid species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are more than 1,500 parasitic species of the approximately 10,000 isopod crustacean species worldwide (Schotte et al. 2008 onward). For example, cymothoids are well-known ectoparasites of the skin, mouth, and gills of marine and freshwater teleost fishes. Some isopod groups such as the family Bopyridae specialize in parasitism on other crustaceans and often exhibit reduced body plans. Most reproduce on the definitive hosts. However, adult isopods belonging to the family Gnathiidae reproduce in benthic substrata after leaving their hosts (Lester 2005).
Gnathiid larvae are temporal ectoparasites of teleosts and elasmobranchs. They suck their host’s body fluid with their needle-like mouthparts until their abdomens are remarkably swollen. Then, they leave their hosts and settle in the benthic substrata to molt into the next stage. Gnathiid larvae, traditionally called as zuphea (unfed phase) or praniza (fed phase), completely differ in morphology from their adult forms, whereas most non-parasitic isopod larvae resemble miniature adults. In contrast, gnathiid adults differ from the larvae in morphology and exhibit sexual dimorphism; males have well-developed mandibles, which females lack. Females have swollen pereonites to brood their eggs. Adults are always non-parasitic and only reproduce in benthic substrata such as sponges, mud burrows, or similar structures (reviewed in Tanaka 2007).
Among the more than 190 species included in 12 genera in the gnathiid family, the life cycles of nine species belonging to four genera have been described (reviewed in Smit and Davies 2004; Hadfield et al. 2009; Tanaka and Nishi 2011). They usually have three larval stages and the adults do not molt. Remarkably, metamorphosis into the male adult occurs through the pre-male stage after the third stage in Caecognathia calva Vanhöffen, 1914 (Wägele 1988). In Elaphognathia discolor (Nunomura 1988), the small male needs only two molts after eclosion, whereas the larger male and female complete three larval stages (Tanaka and Nishi 2011).
Among the life cycles of nine gnathiid species that have been described, the larval host fish have been identified in four: Paragnathia formica (Hesse, 1864) by Monod (1926); Gnathia africana Barnard, 1914 by Smit et al. (2003); Gnathia pilosus Hadfield, Smit, and Avenant-Oldewage, 2008 by Hadfield et al. (2009); Elaphognathia cornigera (Nunomura, 1992) by Tanaka and Aoki (1998). The larvae of these species infest teleost fishes; however, the life cycle of gnathiids from elasmobranch fishes has never been reported. Recently, several gnathiid species have been described based on larvae collected from host elasmobranchs (Smit and Basson 2002; Nunomura and Honma 2004; Coetzee et al. 2008, 2009; Ota and Hirose 2009a, 2009b; Ota 2011). Some authors have recognized that the third-stage larvae of these species infest elasmobranchs, although younger stages have never been found (McKiernan et al. 2005; Coetzee et al. 2009). This may indicate that the younger larvae feed on different hosts. However, the younger larvae of these gnathiid species and their hosts have not been discovered to date in the natural environment.
The third-stage larva of Gnathia trimaculata Coetzee, Smit, Grutter, & Davies, 2009 infests elasmobranch fishes. This species was originally described from two shark species, Carcharinus melanopterus (Quoy & Gaimard, 1824) and Carcharhinus amblyrhynchos (Bleeker 1856), found in the Great Barrier Reef, Australia (Coetzee et al. 2009), and subsequently reported from one shark species Carcharinus falciformis (Bibron, 1839), and three rays Rhinoptera javanica Müller & Henle, 1841; Taeniura meyeni Müller & Henle, 1841, and Himantura sp. in Okinawa-jima Island, southwestern Japan (Ota and Hirose 2009a).
Recently, we found third-stage larvae of G. trimaculata swimming near the sea bottom in Aki-no-hama, northern coast of Izu-Oshima I., central Japan. They were always in the unfed phase, that is, zuphea. Furthermore, we collected G. trimaculata larvae infesting three benthic teleost species of the family Tripterygiidae in Akino-hama and Nabeta Bay (Shimoda, southern end of Izu Peninsula, central Japan). The G. trimaculata larvae on the teleosts were recognized as large and small types of unidentified gnathiid larvae. The small type molted into the large type, and the large type molted into the third-stage larvae of G. trimaculata. Therefore, the large and small types of gnathiid larvae were the first- and second-stage larvae of G. trimaculata.
Gnathia trimaculata was originally reported from elasmobranchs from tropical (Coetzee et al. 2009) and subtropical areas (Ota and Hirose 2009a), while the present collection sites, Akino-hama and Nabeta Bay, are situated in the warm-temperate area of central Japan. In this paper, we verified the species identification of the larvae based on the larval morphologies and DNA-barcode, referring to the G. trimaculata specimens from the subtropical Japan. Second, we report the exclusive occurrence of third-stage larvae on elasmobranchs in our field survey. Finally, we discuss the life cycle of G. trimaculata and the host shift from teleosts to elasmobranchs, based on records of the laboratory culture.
Materials and methods
Field sampling and laboratory rearing of the gnathiid from teleosts (Fig. 1)
We collected gnathiid larvae from teleosts at two sites in central Japan: Aki-no-hama (4–15 m depth), the northeast coast of Izu-Oshima Island (34°47′15′′N, 139°24′44′′E) during June, July, and August 2010 (Summer) and March and April 2011 (Spring); and Nabeta Bay (0.1–1 m depth), south Izu Peninsula (34°39′57′′N, 138°56′13′′E) on July 2010 and November 2011, central Japan. These sampling sites are rocky shores located in the temperate zone (see electronic supplementary material); water temperature was approximately 20 °C from June to August 2010 and approximately 16 °C from March to April 2011. At Aki-no-hama, the host teleosts were collected by a scuba diver with hand net (0.5 mm mesh), polyethylene bag, and polyester mesh net (0.5 mm mesh). We also captured zuphea larvae freely swimming at the sea bottom and fixed them in 70 % ethanol for morphological observation or 99 % ethanol for DNA analysis. At Nabeta Bay, the host teleosts were captured with a hand net (0.5 mm mesh) by snorkeling. We identified the fishes following by Nakabo (1993) and Froese and Pauly (2011) and measured the total length (TL).
We kept each of the host teleosts caught from Izu-Oshima I. in a 1.3-l seawater tank with a filtration system and a cooling (26 °C from June to August) or heating system (22 °C from March to April). To estimate the feeding periods of gnathiids, they were observed every day until the larvae left the host. After this, we kept each gnathiid larva individually in a 70-ml glass vial or plastic case filled with seawater and recorded when it molted. The gnathiid larvae that successfully molted or died were fixed in 70 or 99 % ethanol.
After molting, we measured the maximum head width and the total length (from anterior margin of the clypeus to posterior margin of the pleotelson) of each specimen under the microscope. Then, we removed the appendages from the bodies of each type with sharpened tungsten needles and observed them under the microscope. From the results of the morphological observations and laboratory culture, we classified each type of larva into the developmental stages as zuphea 1 (Z1), 2 (Z2), and 3 (Z3), and praniza 1 (P1), 2 (P2), and 3 (P3), following previous studies of gnathiid life cycles (Smit et al. 2003; Tanaka 2003; Hadfield et al. 2009). When we combined Z1 and P1, Z2 and P2, and Z3 and P3, we simply classified them as the first-, second-, and third-stage larvae.
Field sampling and laboratory rearing of the gnathiid from elasmobranchs (Fig. 1)
To obtain third-stage larvae of G. trimaculata, we collected elasmobranchs in subtropical and warm-temperate Japan: Nakagusuku Bay (26°12′N, 127°49′E), Yomitan Village (26°21′N, 127°42′E), Ishigaki-jima I. (24°28′N, 124°18′E), Kume-jima I. (26°19′N, 129°48′E), Otsuki town (32°47′N, 132°42′E), and Nabeta Bay (34°39′57′′N, 138°56′13′′E) from 2005 to 2011. The elasmobranchs were caught using a set net (Yomitan and Otsuki town), gill net (Nakagusuku Bay and Kume-jima), line fishing (Ishigaki-jima and Nabeta Bay), and longline (Nabeta Bay). Some of the gnathiid specimens from Mobula tarapacana (Philippi, 1892) captured in a set net off Tanegashima I. and Rhincodon typus Smith, 1828, captured in a set net off Okinawa-jima I. were kindly donated by the staff members of Okinawa Churaumi Aquarium and Kagoshima City Aquarium.
We identified the elasmobranchs and measured total lengths (TL) or disk widths (DW). We could not record the host sizes when host bodies were damaged or incomplete. The mouth and gill chambers of elasmobranchs were dissected, and the gnathiid larvae were collected. The host’s mucus was removed from gnathiids by using a paper towel. Larvae were fixed in 70 % ethanol for morphological observation or 99 % ethanol for molecular analysis. We kept 2–20 individual larvae from each host separately in 0.5-mm mesh containers in a 160-l seawater tank with a filtration system at 22–27 °C. We observed the larvae twice or three times a week until they metamorphosed into adults. After metamorphosis, the adults were photographed and fixed in 70 or 99 % ethanol.
DNA extraction and amplification
We preserved tissue samples (juveniles and/or adults) of gnathiids in ethanol at −30 °C and suspended 1–4 specimens in 400 μl CTAB (hexadecyltrimethyl ammonium bromide) buffer [2 % CTAB, 1.0 M NaCl, 75 mM EDTA (pH 8.0), 35 mM Tris–HCl (pH 8.0)] containing 0.1 % sodium dodecyl sulfate (SDS) and 0.2 % beta-mercaptoethanol, followed by incubation at 65 °C for 1 h, according to Hirose et al. (2009). We added proteinase K to the samples to get a final concentration of 0.1 mg ml−1 and incubated the samples overnight at 37 °C. We extracted DNA with phenol–chloroform as described by Sambrook et al. (1989). We performed PCR amplification of the partial cytochrome c oxidase subunit I (COI) using Ex Taq DNA polymerase (Takara) and a combination of crust-cox1f and crust-cox1r primers (Podsiadlowski and Bartolomaeus 2005). We performed PCR amplification under the following conditions: 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min 30 s, with a final extension at 72 °C for 7 min. PCR-amplified DNA fragments (approximately 1,300 bp) were cloned using pMD20-Tvector (Takara) with a DNA Ligation Kit (Ligation Mighty Mix, Takara). We randomly selected and sequenced at least five clones for each sample and sequenced them. We treated the PCR products with ExoSAP-IT (GE Healthcare) before sequencing using DTCS Quick Start Master Mix (Beckman Coulter), and we analyzed the products using a CEQ8800 (Beckman Coulter) automated DNA sequencing system. Intermediate primers were designed for sequencing (Table 1). The sequences determined in this study can be retrieved from the GenBank/EMBL/DDBJ DNA databases under the accession numbers provided in Table 2.
Phylogenetic analyses
We initially aligned the sequences using MUSCLE (Edgar 2004) and then eliminated poorly aligned positions and divergent regions by Gblocks (Castresana 2000) for phylogenetic analysis. We performed the following analyses on the aligned DNA sequences of the partial COI region: maximum likelihood (ML) using TREEFINDER January 2008 version (Jobb 2008), and maximum parsimony (MP) and neighbor-joining (NJ) using PAUP* 4.0 beta10 (Swofford 2003). To select the appropriate nucleotide substitution model, we used jModeltest ver. 0.1.1 (Posada 2008). Based on the Akaike information criterion, we selected the TVM + G model (transversional model with gamma distribution) as the best model. We searched the unweighted MP trees using a heuristics approach with 100 random initial trees. Statistical support for the ML, MP, and NJ trees was evaluated using a nonparametric bootstrap test with 1,000 re-sampling events.
Results
Species identification
In the laboratory culture of small type and large type of gnathiid larvae that spontaneously left the host teleosts from Izu-Oshima Island (see Laboratory observation below), the small type molted into the large type, and the large type molted into the zuphea larvae that had the same morphology as the third-stage larvae of Gnathia trimaculata, which swim freely near the sea bottom. Therefore, we concluded that the small type and the large type were the first- and the second-stage larvae of G. trimaculata, respectively. This identification was supported by COI gene sequences (see Molecular analysis below). In this study, the small larvae and large larvae from the teleosts were classified into Z1 (unfed) or P1 (fed) larva, and Z2 or P2 larva, respectively, and the third-stage larvae from elasmobranchs were classified into Z3 or P3 larva.
Occurrence of G. trimaculata (Table 3)
We collected 15 individuals of three teleost fish species belonging to the family Tripterygiidae from Izu-Oshima I. and Nabeta Bay: Enneapterygius etheostomus (Jordan and Snyder, 1902) (N = 11), Enneapterygius miyakensis Fricke, 1987 (N = 1), and Springerichthys bapturus (Jordan and Snyder, 1902) (N = 3). A total of 220 G. trimaculata larvae (140 first and 80 second stage) attached to the hosts’ fins and skin (Fig. 2a, b) were collected, and the maximum density was 40 larvae (39 first and 1 second stage) on an individual of S. bapturus. Although we collected 13 individuals of Z3 larvae that were freely swimming near the sea bottom at Izu-Oshima I. (Fig. 2c), we could not find those third-stage larvae on the benthic teleosts.
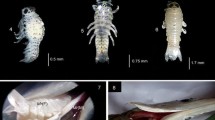
Field observations of G. trimaculata larvae at Aki-no-hama, Izu-Oshima I. a, b first- and second-stage larvae of Gnathia trimaculata on Enneapterygius etheostomus (a) and Springerichthys bapturus (b), c third-stage larvae swimming near the sea bottom, photographed by the second author. Arrow shows the gnathiid larvae attached on the host. Z1 = unfed first-stage larvae (zuphea 1), P1 = satiated first-stage larvae (praniza 1), P2 = satiated second-stage larvae (praniza 2). Scale bar = 10 mm (a, b) and 1 mm (c)
We collected a total 175 elasmobranch fishes of 27 species and unidentified species from 22 genera of 13 families. A total of 893 individual G. trimaculata larvae were collected from 23 species and two unidentified elasmobranchs from Okinawa-jima I., Kume-jima I., Ishigaki-jima I., off Otsuki Town, and Nabeta Bay. All were third stage, and no first and second stages were collected from elasmobranchs (Table 3). Among the third-stage larvae, the unfed phase was rarely found: 14 of 143 gnathiid larvae from Taeniura meyeni Müller and Henle 1841; and 2 of 122 from Himantura sp.
Gnathiid larvae attached mainly to the wall of the interbranchial septum, gill filaments, and the floor of oral cavity of elasmobranchs and rarely attached to the body surface near the gill slits. Among the hosts, large benthic rays (>DW 50 cm) were often infested by a large number of gnathiids except for Himantura undulata Bleeker, 1852. A total of 425 of 893 gnathiids (47.6 %) were collected from eight rays; 13–143 (mean ± SD = 53.1 ± 50.9) from T. meyeni (N = 2), Gymnura japonica Temminck and Schlegel, 1850 (N = 2), and Himantura spp. (N = 4). Himantura undulata (DW 60–93 cm, N = 4) was infested by only 0–10 gnathiids (2.5 ± 4.36) as well as small (<DW 40 cm) benthic ray species. Although sharks were infested by smaller numbers of gnathiids than benthic rays, the benthic shark Stegostoma fasciatum Hermann, 1783, was infested by a mean number of 48.5 gnathiids. The pelagic sharks such as Carcharhinus species were rarely infested by gnathiids.
Laboratory observation (Table 4)
We collected 56 gnathiids (three Z1 individuals, 51 P1, and one P2 larvae) from S. bapturus (Jordan and Snyder 1902) (N = 3) in Aki-no-hama, Izu-Oshima I. in March and April 2011 (spring). We reared 50 P1 larvae in aquaria until molting and preserved other larvae in 70 % ethanol for morphological observation or 99 % for DNA analysis. All P1 larvae molted into Z2 larvae. We reared 33 P2 larvae from E. etheostomus (N = 3) from June to August 2010 (summer), and 23 of 33 P2 larvae (70 %) molted into Z3 larvae. We measured the total length and head width of Z3 larvae (N = 23), Z2 larvae (N = 50), and Z1 larvae (N = 3).
The total length of Z1 larva was 1.16–1.30 mm (mean ± SD = 1.21 ± 0.08 mm; N = 3) and the head width was 0.28–0.32 mm (0.3 ± 0.02 mm; N = 3). Among the specimens of P1 larvae from Aki-no-hama, we recognized brown pigmentation fringing the dorso-lateral margin of their clypeus, head, pereonites, pleonites, uropodal rami, and pleotelson (Fig. 3a). Coloration of the anterior hindgut of P1 larvae caught in nature was red, white, or green. In our laboratory conditions at 22 °C, 29 of 50 P1 larvae left the host fish 1–7 days (4.10 ± 1.32 days) after the capture of their host, and they settled and molted into Z2 larvae after 3–6 days (4.10 ± 0.72 days). The duration from the capture of the host to the molting of each larva was 6–11 days (8.21 ± 1.42 days). We could not record an accurate period for the remaining 21 P1 larvae, but they left the host fish within 5 days and molted into Z2 within 5 days.
Within 1 or 2 days of molting, Z2 larvae started to swim actively. The total length of Z2 larvae was 1.66–2.14 mm (1.83 ± 0.11 mm; N = 50) and head width was 0.44–0.50 mm (0.46 ± 0.01 mm; N = 50). We recognized four distinct brown lines on the dorsal surface of Z2 and P2 larvae (Figs. 2a, b, 3b). The pigmentation often disappeared in P2 larvae before molting. There were morphological differences in antenna 2 and pleopods between Z1 and Z2. The antenna 2 of Z2 larvae presents pectinate scales on the surface of peduncle article 4, and plumose setae on pleopodal rami of Z1 larvae were long for both rami, whereas those of Z2 larvae were relatively short. In our laboratory conditions at 26 °C, 23 of 34 P2 larvae left the host fish after 2–6 days (3.52 ± 1.08 days) from the capture of their host and then molted into Z3 larvae after 2–5 days (3.57 ± 0.73 days). The combined time between the capture of the host and molt of each larva was 6–9 days (7.09 ± 0.85 days).
Z3 larvae started to swim actively within 1 or 2 days after molting. The total length of Z3 larvae was 2.54–3.18 mm (2.90 ± 0.73 mm; N = 23) and the head width was 0.67–0.82 mm (0.73 ± 0.03 mm; N = 23). The whole bodies of Z3 larvae were yellow with four brown lines dorsally but the middle two lines were often unclear or absent (Figs. 2c, 3c). We could not record the feeding period of Z3 larvae on the host elasmobranchs. The swollen pereonites 4–6 of P3 larvae were often pigmented in dark green with four black lines within yellow pigmentation, having 0–3 black spots on the lateral sides.
We chose 215 of 893 P3 larvae from 15 host rays and sharks belonging to 10 species and one unidentified species from Okinawa-jima I., Ishigaki-jima I., Otsuki town, and Nabeta Bay and reared them until they metamorphosed into adults in the laboratory at 22–27 °C. We reared 2–20 individuals separately in each mesh sack. Among them, 116 of 215 larvae (54.0 %) successfully metamorphosed into 82 male adults and 34 female adults, and the other 99 larvae died or disappeared. In our laboratory conditions, the P3 larvae metamorphosed into adults within 5–17 days (7.32 ± 1.79 days; N = 82) for males and 17–29 days (21.4 ± 3.34 days; N = 34) for females (Fig. 3e, f). During metamorphosis, males underwent molting of posterior half and then anterior half within approximately a day. After metamorphosis, the exoskeleton began to harden and turned white. Green pigmentation observed in the previous stage remained on dorsal pereonites 5 and 6, and ventral pereonites 4–6 of the male adults. When several male adults were put in the same mesh sack, we found some mutilated bodies of dead males and some males lost their pereopods, probably because of competition among the male adults. A new male adult was found to remain with some P3 larvae before metamorphosis in a crease in the mesh. The P3 larvae with the male adult were easily recognized as females because of their yellow immature eggs in the pereonites 4–6. Then, they metamorphosed into female adults. However, some P3 females never metamorphosed into adults; they were alive as P3 larvae for over 1 month without molting. When all P3 larvae in the mesh sack were females, they did not metamorphose into adult females, resulting in the low metamorphosis rates from some host elasmobranchs and the occurrence of low numbers of female adults in some P3-larva populations from host elasmobranchs (Table 4). Male adults survived for at least 52 days. Both male and female adults could not swim fast but crawled at the bottom.
Molecular analysis
A total of six haplotypes were obtained from sequencing the partial mitochondrial COI gene of seven gnathiid species (Table 2). The sequences of the partial COI gene were identical among the specimens of G. trimaculata larva (Z2) from two different host fishes and sites, S. bapturus at Nabeta Bay and E. etheostomus at Izu-Oshima I. We confirmed analysis to a specific subset of 858–861 base pairs of the partial COI sequences. The average base frequencies of all three codon positions of five haplotypes from the three Gnathia species were A = 0.205, C = 0.197, G = 0.219, T = 0.379, and the A + T composition frequency was 58.4 %, although the composition of E. rangifer was A = 0.253, C = 0.160, G = 18.4, T = 40.3, and the A + T composition frequency was 41.6 %. The average base frequencies of third codon positions of five haplotypes from three Gnathia species were A = 0.221, C = 0.187, G = 0.187, T = 0.450, and the A + T composition frequency was 62.6 %, although the composition of E. rangifer was A = 0.339, C = 0.180, G = 9.8, T = 0.45, and the A + T composition frequency was 79.4 %. The aligned sequence from E. rangifer and G. limicola had one site of insertion/deletion that consisted of three base pairs compared with sequences from G. trimaculata and G. maculosa. The alignment dataset of partial COI (861 positions) was eliminated to give 585 positions as poorly aligned and divergent regions by Gblocks. The following analyses were performed with this no gaps dataset. The difference between the partial mtCOI DNA sequences within G. maculosa from different collection sites was 16/858 bp (1.86 %). The difference between the partial mtCOI DNA sequences within G. trimaculata from different collection sites and different developmental stages was 0–3/858 bp (0–0.35 %). Within Gnathiidae species, the lowest percentage of sequence divergence, 25.3 % (217/858 bp differences), was between G. maculosa collected from Kume-jima I. and G. trimaculata larvae collected from Nabeta Bay and Izu-Oshima I. The highest divergence of 32.9 % (282/858 bp differences) was between G. limicola and G. trimaculata. The mean sequence divergence among the Gnathiidae species was 29.3 %.
Figure 4 shows the ML tree using the TVM + G substitution model (log-likelihood = − 3,316.9791). For data with all codon positions, the transversional model with gamma distribution (TVM + G) model was selected as best, using the Akaike information criterion (AIC). Therefore, we chose TVM + G for ML and NJ analyses. Because the topologies of the phylogenetic trees were nearly identical in the ML, MP, and NJ trees, the strict-consensus MP tree and the NJ tree under the TVM + G substitution model are not shown. The monophyletic group of G. maculosa and G. trimaculata was supported by high bootstrap values (ML = 97 %, MP = 100 %, and NJ = 100 %, and ML = 97 %, MP = 100 %, and NJ = 100 %, respectively). Although the monophyly of G. maculosa + G. trimaculata group was supported by relatively high bootstrap values (ML = 97 %, MP = 85 %, and NJ = 100 %), the monophyly of Gnathia was not supported.
Discussion
Life cycle
In our survey, approximately 900 larvae of Gnathia trimaculata from a variety of elasmobranchs were collected. All of them were the third-stage larvae; the first and the second stages were never found, suggesting that the first- and the second-stage larvae infest other hosts. McKiernan et al. (2005) reported that only the third-stage larvae of two unidentified gnathiid species were parasitic on the epaulette shark Hemiscyllium ocellatum. Coetzee et al. (2009) also noted that all larvae of G. trimaculata from host sharks Carcharinus spp. were third stage. The question remains of where the first- and second- stage larvae live in and what type of hosts the younger larvae parasitize. In this study, we found that the first- and second-stage larvae of G. trimaculata infested teleosts in the natural environment.
Figure 5 summarizes the life cycle of G. trimaculata inferred from our field sampling and laboratory observations: Z1 larvae attach to the skin or fins of teleosts such as E. etheostomus and suck their body fluid for up to 7 days until their thoraxes are swollen (P1). After feeding, the P1 larvae leave the host and rest at the sea bottom for 3–6 days before molting into Z2 stage. Z2 larvae swim and attach to the teleost host to suck the body fluid. The feeding period of Z2 larvae was estimated as up to 6 days. Then, the P2 larvae leave the host and molt into the Z3 stage within 2–5 days. The Z3 larvae actively swim to attach to a variety of elasmobranch hosts such as benthic species (e.g. Dasyatidae spp.), and pelagic sharks (e.g. Carcharinus species) at a lower frequency. As we have never found the third-stage larvae on any teleosts, even though the first- and second-stage larvae infest them, the third-stage larvae selectively infest elasmobranchs. After feeding on the elasmobranchs, the P3 larvae metamorphose into male adults in 5–17 days or females in 17–29 days.
Schematic life cycle of Gnathia trimaculata showing the periods of each developmental stage. The first- and second-stage larvae parasitize benthic teleosts while the third-stage larvae parasitize a variety of elasmobranchs. The benthic habitat used for reproduction and molting is still unknown. Square shows a sectioned drawing of the ray gill chamber infested by third-stage larvae in left horizontal view. OC = oral cavity, GS = gill slit, GF = gill filaments, IS = interbranchial septum
In general, adults of gnathiid species reproduce without feeding in benthic habitats such as the inside of sponges, mud burrows, rock crevices, and terebellid tubes (reviewed in Tanaka 2007). While the habitat of the adult G. trimaculata was not determined, a single male adult was found by chance at the sea bottom at Izu-Oshima I. sampling site (Arima personal observation).
Life span
Some authors have estimated the life span from hatching to metamorphosis into adults in several gnathiid species based on data from the period of attachment to the hosts and the periods of resting and molting into the next stages or adults. The attachment periods of gnathiid larvae on the host fish were reported in three species: Z1, Z2, and Z3 larvae of Paragnathia formica (Hesse, 1864) infest suitable host fishes (teleosts) for 10–36, 13, and 48 h, respectively (Stoll 1962); Z1, Z2, and Z3 larvae of Gnathia africana Barnard, 1914 for 2.3, 2.7, and 10.1 h, respectively (Smit et al. 2003). Z1, Z2, and Z3 larvae of Gnathia pilosus Hadfield, Smit, and Avenant-Oldewage, 2008 for 3.2–3.7, 3.3–4.4, and 3.8–4.8 h, respectively (see Table 2. in Hadfield et al. 2009). In the present study, Z1 and Z2 larvae of G. trimaculata attached to the host fish 4.10 and 3.52 days on average, respectively. Thus, the feeding period of G. trimaculata on teleosts was much longer than those of the other species parasitic on teleost fishes examined to date. We could not estimate the attachment period of the third-stage larvae on elasmobranch hosts in this study. McKiernan et al. (2005) reported that the third-stage larvae of two undescribed species attached to the host shark H. ocellatum for 4–6 days and some praniza larvae still remained on the host for up to 17 days.
After feeding, P1, P2, and P3 larvae leave the hosts to rest and molt into the next stages. The periods between detachment from the host and the molting were recorded in the three species mentioned above: the periods from P1 stage to Z2 stage, P2 to Z3, and P3 to adult of P. formica were 13, 12, and 7–10 weeks, respectively (Stoll 1962); 8, 10, 8–10 days (male adult) or 17 days (female adult), respectively, in G. africana (Smit et al. 2003); 35, 35, and 19–71 days (male adult) or 14–75 days (female adult), respectively, in G. pilosus (Hadfield et al. 2009). Furthermore, Tanaka (2003) reported 8–13 days for praniza phase in Elaphognathia cornigera (Nunomura, 1992). In this study, these periods of G. trimaculata were 3–6 days from P1 to Z2, 2–5 days from P2 to Z3, and 5–17 days from P3 to male adult or 17–29 days from P3 to female adult. The periods of P1–Z2 and P2–Z3 in G. trimaculata were much shorter than those of the other species reported to date, while the feeding periods were longer. The P3 stage of the female is significantly longer than that of male in G. trimaculata as well as G. africana (Smit et al. 2003) and G. pilosus according to the average period data of P3 to adults (see Table 2; Hadfield et al. 2009). The eggs were already observable in the ovaries of female gnathiids of these species at the P3 stage before metamorphosis. The difference in the periods between males and females may be caused by the difference in the duration of spermatogenesis and oogenesis in gnathiid isopods.
We noted the P3 female of G. trimaculata did not metamorphose into an adult if a male was absent. When the P3 female was not with male adult, it survived for more than a month under laboratory conditions. In the natural environment, after leaving its host, the P3 female may spend a long time on the sea bottom before mates. If the P3 female of G. trimaculata metamorphosed into adult before encountering male, it may fail to reproduce. It would therefore be adaptive for G. trimaculata female to survive as P3 larva until it finds a male.
We estimated the periods in each stage: Z1–P1 (up to 7 days), P1–Z2 (3–6 days), Z2–P2 (up to 6 days), P2–Z3 (2–5 days), and P3–adult (5–17 days in males, 17–29 days in females). After molting, Z2 and Z3 larvae started to swim within 1 or 2 days. Assuming that the attachment period of the third-stage larvae takes a week, we can estimate the duration from eclosion to maturation in G. trimaculata takes up to 54 days in males and 66 days in females, without considering the time spent finding the hosts for feeding in nature. The duration from eclosion to maturation was estimated in six gnathiid species of four genera (Stoll 1962; Wägele 1988; Smit et al. 2003; Tanaka 2003; Hadfield et al. 2009; Tanaka and Nishi 2011). Among them, the duration of the present species is similar to that of G. africana, that is, 62 days. In G. africana, the attachment period is shorter and the rest-molt time is longer than in the present species. Thus, the proportion of each period in the life history is quite different between the two species. As the male adults survived for at least 52 days after metamorphosing, the male adults of G. trimaculata may survive for more than 100 days. We noted the life span of G. trimaculata in nature would be longer than that in laboratory conditions. In nature, zuphea larvae would spend more time for seeking their host fishes. Moreover, the female lifespan may be elongated by delayed metamorphosis when females are not able to encounter males soon after leaving the final host.
Utility of partial mtCOI sequence
Although larvae and adult gnathiid isopods have numerous morphological characters that can be used for taxonomy, their multiple life stages make them difficult to identify in the field. DNA sequences can be useful for species identification if the sequences show species-specific variation. Grutter et al. (2000) and Ota et al. (2010) showed that ribosomal DNA ITS2 sequences are useful for distinguishing gnathiid isopods. However, the “Folmer region” at the 5′ end of the cytochrome oxidase subunit I mitochondrial region has emerged as a standard DNA barcoding region (Ratnasingham and Hebert 2007). The sequence variation in the COI barcode region of Crustacea will be effective for species discrimination. For example, in 31 genera of 54 species of Decapoda, the average Kimura-2-parameter distance between species averaged 0.46 %, whereas congeneric divergences averaged 17.16 % (Costa et al. 2007). In 13 Daphnia (Cladocera) species, the divergence of mtCOI sequences was within species 0–4.3 % and 13.18–30.65 % between species, and the divergence of mtCOI sequences was within species 0–3.09 % and 5.58–31.39 % between species in 11 Gammarus (Amphipoda) species (Costa et al. 2007). This is the first report of sequences of the partial mtCOI region in the family Gnathiidae. The divergence of the partial mtCOI sequences was 0–1.86 % within species and 25.3–32.9 % between species. The divergence of the COI gene sequence among seven well-defined Ligidium species (Isopod; Oniscidea; Ligiidae) range of 14.4–23.3 % (Klossa-Kilia et al. 2006), and another terrestrial isopod (Hawaiioscia, Isopod; Oniscidea; Halophilosciidae) showed a range of 14.1–15.3 % (Rivera et al. 2002). These data are not in conflict with our data. The present study confirms that partial mtCOI sequences are useful for distinguishing gnathiid isopods species throughout their life stages. Unfortunately, a small numbers of mtCOI sequences are available from correctly identified species in Gnathiidae at present, and it is desirable to integrate sequence data of more species and more localities in future studies.
Distribution
As many expeditions or surveys have traditionally been conducted along European coasts, the North Atlantic Ocean, and the Southern Ocean (Antarctic Ocean), several gnathiid species are known to have a wide geographic range; for example, Caecognathia elongata (Kröyer, 1847): Kara Sea, Norwegian Sea, and Greenland (Arctic Ocean), and the Georgia Strait (the Pacific coast of Canada) (Monod 1926); P. formica: the estuary from Morocco to Scotland and Mediterranean Sea (Monod 1926); Gnathia maxillaris (Montagu, 1804): the intertidal from Portugal to southern North Sea (Monod 1926); Caecognathia calva (Vanhöffen, 1914): circumpolar in the Antarctic (Wägele 1987). However, most other gnathiid species have been recorded only at each type locality. This is possibly due to the lack of biogeographic surveys on gnathiid species rather than endemism (Smit and Davies 2004). Gnathia trimaculata was previously reported from the tropical–subtropical coral reefs, i.e., Great Barrier Reef, Australia, and Okinawa-jima I. (southwestern Japan) (Coetzee et al. 2009; Ota and Hirose 2009a). The present study showed that G. trimaculata is also distributed in the warm-temperate waters of Izu Peninsula and Izu-Oshima I. (Central Japan). The present species has a potentially wide distribution range in the western Pacific Ocean.
While the dispersal ability of gnathiids is rarely known, Tanaka (2007) pointed out that the passive movement dependent on currents or host movement/migration potentially contributes to the dispersal of gnathiids at larger scales. Ota and Hirose (2009a) previously noted that some large host elasmobranchs having high mobility such as tiger shark Galeocerdo cuvier Péron and Lesueur, 1822, might considerably expand the distribution of G. trimaculata. Gnathiid larvae of some species were known to infest a variety of host fish species, suggesting low host specificity; for example, the larvae of P. formica were found on 16 teleost species of six families (Monod 1926) and G. pilosus on 16 teleost species of nine families (Hadfield et al. 2009). Because the larvae of G. trimaculata shift their host from teleosts to elasmobranchs, both hosts would be necessary to complete the life cycle, and thus, the distribution range of both suitable hosts would overlap with that of G. trimaculata. This study revealed the third-stage larvae of G. trimaculata infested over 23 elasmobranch species from 10 families including both benthic and pelagic species, and both ray and shark species, indicating that the third-stage larva of G. trimaculata may be generalist as an ectoparasite of elasmobranchs. In contrast, we found the first- and second-stage larvae of G. trimaculata from only three host teleost species of one family. Considering the wide distribution range of G. trimaculata, more host teleost species are likely to be found in future studies.
Predation risks
Some authors have demonstrated that gnathiid larvae suffer from predation by fishes. Grutter (2002) listed gnathiid larvae obtained from the diet of 21 fish species belonging to five families: Labridae, Chaetodontidae, Gobiidae, Syngnathidae, and Embiotocidae. When the host fishes are benthic teleosts such as the small goby Caenogobius annularis Gill, 1859 feeding on small crustaceans, the host fishes also feed on ectoparasitic crustaceans (Tanaka 2002). In coral reefs, the cleaner wrasse Labroides dimidiatus (Valenciennes, 1839) is thought to be an important predator of ectoparasites, preying on over 1,200 ectoparasites, mainly gnathiids, each day (Grutter 1996).
The attachment sites of gnathiids on the host fish seem to be important for avoiding predation from cleaner fishes. Among the gnathiids parasitic on elasmobranchs, Heupel and Bennet (1999) reported gnathiids mainly attached to the cloaca and the claspers of benthic sharks Hemiscyllium ocellatum (Bonneaterre, 1788), whereas some authors found them attached to the gills or branchial chamber walls of several benthic ray species (Paperna and Por 1977; Honma and Chiba 1991a, 1991b). In the present study, G. trimaculata mainly infested gill chambers or oral cavities of elasmobranchs. A large number of G. trimaculata infested the large benthic rays, whereas benthic and pelagic sharks were rarely infested. Small benthic rays such as Neotrygon kuhlii (Müller and Henle, 1841) were often infested by another gnathiid species Gnathia maculosa Ota and Hirose, 2009 (Ota unpublished). The gill slits of benthic rays are generally narrow and open ventrally, facing the sea bottom. Each gill chamber is deep, and it would be difficult for cleaning fishes to approach gnathiids inside the gill chamber of benthic rays (see Fig. 5). Therefore, predation risks for gnathiids would be substantially lower on elasmobranchs than those of gnathiids on teleosts’ fins and skin, suggesting that the parasite may prefer to stay on the elasmobranch host.
P3 gnathiid larvae parasitic on elasmobranchs tend to have a larger body size than those parasitic on teleosts. Poulin (1995) showed that gnathiids were generally small in body size compared with other parasitic and free-living isopods. Grutter (1997) also demonstrated that larger gnathiid larvae were selectively preyed on by the cleaner fish L. dimidiatus. Therefore, large gnathiid larvae suffer more serious predation pressure than small larvae. In fact, the P3 larvae of most gnathiid species that are exclusively parasitic on teleosts are less than 5 mm in body length: P. formica (mean 3.8 mm), G. africana (3.8 mm), G. pilosus (2.2 mm), G. aureamaculosa (1.5 mm), and E. cornigera (2.5 mm) (Upton 1987; Smit et al. 2003; Tanaka 2003; Hadfield et al. 2009; Ferreira et al. 2009). Jones et al. (2007) suggested that P3 larvae of G. falcipenis Holdich and Harrison, 1980 and Gnathia sp. “C” did not feed on elasmobranchs based on 16S mtDNA sequence data from the gnathiid diet, and their P3 larvae were less than 2.5 mm in total length (see Fig. 1 in Jones et al. 2007). In contrast, gnathiid species parasitic on elasmobranchs tend to have a larger body length; G. pantherina (mean 5.2 mm), G. capillata (4–8 mm), G. grandilaris (mean 6.1 mm), G. trimaculata (mean 4.0 mm in Australia and 7.0 mm in Japan), G. maculosa (mean 5.1 mm), G. nubila (mean 8.4 mm), and G. teruyukiae (mean 8.0 mm) (Smit and Basson 2002; Nunomura and Honma 2004; Coetzee et al. 2008, 2009; Ota and Hirose 2009a, 2009b; Ota 2011). Therefore, we consider the low predation risks on gnathiid larvae infesting elasmobranchs may allow an increase in body size of gnathiids.
Needless to say, the large body size is thought to be advantageous for reproduction. Non-feeding gnathiid adults do not molt or grow, and female adults are semelparous. Thus, the body size in the third-stage larvae reflects the adult’s size. Adult females show a positive correlation between body size and brood size in E. cornigera within the same months (Tanaka and Aoki 2000) and in Caecognathia robusta Sars 1879 (see Table 12 in Klitgaard 1997).
Ontogenetic shift in host utilization
In the present study, third-stage larvae of G. trimaculata were exclusively found on elasmobranch species, whereas first- and second-stage larvae were found on teleost species and never on elasmobranchs (Table 3). We demonstrated that second-stage praniza larvae on teleost molted to third-stage zuphea larvae that were morphologically identical to those on elasmobranchs. Moreover, the molecular phylogeny based on mtCOI sequences strongly supported that the second-stage larvae on teleost hosts and adults metamorphosed from larvae infesting elasmobranchs are the same species. These evidences indicate the occurrence of host shift from teleost to elasmobranch in G. trimaculata. The body fluid of host is qualitatively quite different between marine teleosts and elasmobranchs. Elasmobranch fishes in seawater regulate concentrations of urea and other body fluid solutes (trimethylamine oxide, Na+, and Cl−) in their body fluid to remain hyper-osmotic to their environment (reviewed in Neil 2006). As gnathiid larvae suck and digest fishes’ body fluid, the digestive system between third- and first/second-stage larvae of G. trimaculata must be physiologically different between the third- and the first/second-stage larvae of G. trimaculata. Although the digestion mechanism of the fish’s body fluid is unclear in gnathiids, specific symbiotic bacteria have been found in the posterior hindgut of some gnathiid species (Juilfs and Wägele 1987; Davies 1995; Davies and Smit 2001). These symbiotic bacteria have also been found in blood-sucking insects and leeches, and they are supposed to serve an important function in hematophagous nutrition; these bacteria have not been found in non-parasitic isopods and other crustaceans (see Juilfs and Wägele 1987; Davies 1995). Davies (1995) also reported that the bacterial flora in Gnathia maxillaris (Montagu, 1804) consisted of Pseudomonas sp., Bacillus sp., and Streptococcus sp. that were tentatively identified from morphological, physiological, and biochemical properties. The intestinal bacterial flora is possibly different between the larvae infested on teleosts and those on elasmobranchs.
The third-stage larvae of other gnathiid species parasitic on elasmobranchs may also shift from unknown hosts to elasmobranchs. The first author has found eight other gnathiid species on elasmobranchs: G. maculosa, G. nubila, G. grandilaris, G. teruyukiae, and four undescribed Gnathia species. Because the larvae collected from the elasmobranch hosts always metamorphosed into adults at the next molting, they were the final stage (probably the third stage) larvae; larvae of younger stages have not been found on elasmobranchs (Ota, unpublished). Smit (personal communication) and McKiernan et al. (2005) also found third-stage larvae of the other Gnathia species on elasmobranch hosts. These gnathiid species on elasmobranchs may also infest teleost hosts in their younger stages, as does G. trimaculata.
References
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Coetzee ML, Smit NJ, Grutter AS, Davies AJ (2008) A new gnathiid (Crustacea: Isopoda) parasitizing two species of requiem sharks from Lizard Island, Great Barrier Reef, Australia. J Parasitol 94(3):608–615
Coetzee ML, Smit NJ, Grutter AS, Davies AJ (2009) Gnathia trimaculata n. sp. (Crustacea: Isopoda: Gnathiidae), an ectoparasite found parasitising requiem sharks from off Lizard Island, Great Barrier Reef, Australia. Syst Parasitol 72:97–112
Costa FO, deWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, Hebert PDN (2007) Biological identifications through DNA barcodes: the case of the Crustacea. Can J Fish Aquat Sci 64:272–295
Davies AJ (1995) Studies on the gut flora of the haematophagous fish parasite Gnathia maxillaris Montagu. Bull Eur Ass Fish Pathol 15(1):32
Davies AJ, Smit NJ (2001) The life cycle of Haemogregarina bigemina (Adeleina: Haemogregarinidae) in South African hosts. Folia Parasitol 48:169–177
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ferreira ML, Smit NJ, Grutter AS, Davies AJ (2009) A new species of gnathiid (Crustacea: Isopoda) parasitizing teleosts from Lizard Island, Great Barrier Reef, Australia. J Parasitol 95(5):1066–1075
Froese R, Pauly D (2011) Fishbase. World wide web electronic publication. www.fishbase.org, version (08/2011). Accessed 19 Dec 2011
Grutter AS (1996) Parasite removal rates by the cleaner wrasse Labroides dimidiatus. Mar Ecol Prog Ser 130:61–70
Grutter AS (1997) Size-selective predation by the cleaner fish Labroides dimidiatus. J Fish Biol 50:1303–1308
Grutter AS (2002) Cleaning symbiosis from the parasites’ perspective. Parasitology 124:S65–S81
Grutter AS, Morgan JAT, Adlard RD (2000) Characterising parasitie gnathiid isopod species and matching life stages with ribosomal DNA ITS2 sequences. Mar Biol 136:201–205
Hadfield KA, Smit NJ, Avenant-Oldewage A (2009) Life cycle of the temporary fish parasite, Gnathia pilosus (Crustacea: Isopoda: Gnathiidae) from the east coast of South Africa. J Mar Biol Assoc UK 89(7):1331–1339
Heupel MR, Bennet MB (1999) The occurrence, distribution and pathology associated with gnathiid isopod larvae infecting the epaulette shark, Hemiscyllium ocellatum. Int J Parasitol 29:321–330
Hirose M, Yokobori S, Hirose E (2009) Potential speciation of morphotypes in the photosymbiotic ascidian Didemnum molle in the Ryukyu Archipelago, Japan. Coral Reefs 28:119–126
Honma Y, Chiba A (1991a) Pathological changes in the branchial camber wall of stingrays, Dasyatis spp., associated with the presence of juvenile gnathiids (Isopoda, Crustacea). Gyobyo Kenkyu 26:9–16
Honma Y, Chiba A (1991b) Histological studies on the juvenile gnathiid (Isopoda, Crustacea) parasitic on the branchial chamber wall of the stingray, Dasyatis akajei, in the Sea of Japan. Rep Sado Mar Biol Stat, Niigata Univ 21:37–47
Jobb G (2008) TREEFINDER ver. Jan 2008. Munich, Germany. www.treefinder.de
Jones CM, Nagel L, Hughes GL, Cribb TH, Grutter AS (2007) Host specificity of two species of Gnathia (Isopoda) determined by DNA sequencing blood meals. Int J Parasitol 37:927–935
Juilfs HB, Wägele JW (1987) Symbiotic bacteria in the gut of the blood-sucking Antarctic fish parasite Gnathia calva (Crustacea: Isopoda). Mar Biol 95:493–499
Klitgaard AB (1997) The distribution and habitats in the North Atlantic of two gnathiid species (Crustacea, Isopoda) and their reproductive biology in the Denmark Strait and north of Iceland. Meddr Grønland, Biosci 47:5–32
Klossa-Kilia E, Kilias G, Tryfonopoulos G, Koukou K, Sfenthourakis S, Parmaklis A (2006) Molecular phylogeny of the Greek populations of the genus Ligidium (Isopoda, Oniscidea) using three mtDNA gene segments. Zool Scr 35:459–472
Lester RJG (2005) Isopoda (isopods). In: Rohde K (ed) Marine parasitology. CSIRO Publishing, Collingwood, pp 138–144
McKiernan JP, Grutter AS, Davies AJ (2005) Reproductive and feeding ecology of parasitic gnathiid isopods of epaulette sharks (Hemiscyllium ocellatum) with consideration of their role in the transmission of a haemogregarine. Int J Parasitol 35:19–27
Monod T (1926) Les Gnathiidae. Essai monographique (morphologie, biologie, systématique). Mém Soc Sci Nat Maroc 13:1–667
Nakabo T (1993) Fishes of Japan with pictorial keys to the species. Tokai University Press, Tokyo. (In Japanese)
Neil H (2006) Osmoregulation in elasmobranchs: a review for fish biologists, behaviourists and ecologists. Mar Freshw Behav Physiol 39(3):209–228
Nunomura N, Honma Y (2004) Gnathia capillata, a new species of the genus Gnathia (Crustacea, Isopoda) from Sado Island, the Sea of Japan. Contr boil Lab Kyoto Univ 29:343–349
Ota Y (2011) A new species of the gnathiid isopod, Gnathia teruyukiae (Crustacea: Malacostraca), from Japan, parasitizing elasmobranch fish. Bull Natl Mus Nat Sci Ser A Suppl 5:41–51
Ota Y, Hirose E (2009a) Description of Gnathia maculosa and a new record of Gnathia trimaculata (Crustacea, Isopoda, Gnathiidae), ectoparasites of elasmobranchs from Okinawan coastal waters. Zootaxa 2114:50–60
Ota Y, Hirose E (2009b) Gnathia nubila n. sp. and a new record of Gnathia grandilaris (Crustacea, Isopoda, Gnathiidae) that parasitizes elasmobranchs from Okinawan coastal waters, Japan. Zootaxa 2238:43–55
Ota Y, Tanaka K, Hirose M, Hirose E (2010) Description of a new species of Elaphognathia (Isopoda: Gantheidae) from Japan based on morphological and molecular traits. J Crust Biol 30:710–720
Paperna I, Por FD (1977) Preliminary data on the Gnathiidae (Isopoda) of the Northern Red Sea, the Bitter Lakes and the Eastern Mediterranean and the biology of the Gnathia piscivora n sp. Rapp Comm Int Mer Medit 24(4):195–197
Podsiadlowski L, Bartolomaeus T (2005) Organization of the mitochondrial genome of Mantis shrimp Peudosquilla ciliata (Crustacea: Stomatopoda). Mar Biotech 7:618–624
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Poulin R (1995) Evolutionary influences on body size in free-living and parasitic isopods. Biol J Linnean Soc 54:231–244
Ratnasingham S, Hebert PD (2007) Bold: the barcode of life data system (http://www.barcodinglife.org). Mol Ecol Notes 7:355–364
Rivera MAJ, Howarth FG, Taiti S, Roderick GK (2002) Evolution in Hawaiian cave-adapted isopods (Oniscidea: Philosciidae): vicariant speciation or adaptive shifts? Mol Phylogenet Evol 25:1–9
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York
Schotte M, Boyko C, Bruce NL, Poore GCB, Taiti S, Wilson GDF (Eds) (2008 onwards) Isopoda statistics. World Marine, Freshwater and Terrestrial Isopod Crustaceans. http://www.marinespecies.org/isopoda. Accessed 22 Feb 2012
Smit NJ, Basson L (2002) Gnathia pantherina sp. n. (Crustacea: Isopoda: Gnathiidae), a temporary ectoparasite of some elasmobranch species from southern Africa. Folia Parasitol 49:137–151
Smit NJ, Davies AJ (2004) The curious life-style of the parasitic stages of gnathiid isopods. Adv Parasitol 58:289–391
Smit NJ, Basson L, Van As JG (2003) Life cycle of the temporary fish parasite, Gnathia africana (Crustacea: Isopoda: Gnathiidae). Folia Parasitol 50:135–142
Stoll C (1962) Cycle évolitif de Paragnathia formica (Hesse) (Isopode—Gnathiidae). Cah Biol Mar 3:401–416
Swofford D (2003) PAUP* 4.0b7a, Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA
Tanaka K (2002) Predation risks involved in the parasitic behaviour of gnathiid isopods (Crustacea). Jpn J Benthol 57:85–89 (In Japanese with English abstract)
Tanaka K (2003) Population dynamics of the sponge-dwelling gnathiid isopod Elaphognathia cornigera. J Mar Biol Assoc UK 83:95–102
Tanaka K (2007) Life histology of gnathiid isopods-current knowledge and future directions. Plankton Benthos Res 2(1):1–11
Tanaka K, Aoki M (1998) Crustacean infauna of the demosponge Halichondria okadai (Kabota) with reference to life cycle of Gnathia sp. (Isopoda: Gnathiidea). In: Watanabe Y, Fusetani N (eds) Sponge science: multidisciplinary perspectives. Springer, Tokyo, pp 259–267
Tanaka K, Aoki M (2000) Seasonal traits of reproduction in a gnathiid isopod Elaphognathia cornigera (Nunomura, 1992). Zool Sci 17:467–475
Tanaka K, Nishi E (2011) Male dimorphism in the harem-forming gnathiid isopod Elaphognathia discolor (Crustacea: Isopoda). Zool Sci 28:587–592
Upton NPD (1987) Asynchronous male and female life cycles in the sexually dimorphic, harem-forming isopod Paragnathia formica (Crustacea: Isopoda). J Zool Lond 212:677–690
Wägele J-W (1987) Description of the postembryonal stages of the Antarctic fish parasite Gnathia calva Vanhöffen (Crustacea: Isopoda) and synonymy with Heterognathia Amar & Roman. Polar Biol 7:77–92
Wägele J-W (1988) Aspects of the life-cycle of the Antarctic fish parasite Gnathia calva Vanhöffen (Crustacea: Isopoda). Polar Biol 8:287–291
Acknowledgments
We thank Makio Yanagisawa (Okinawa Churaumi Aquarium), Hirohito Arima (Izu-Oshima Island) and Mitsuko Chikuchishin (Kagoshima City Aquarium) for providing specimens. Dr. Daisuke Uyeno (University of the Ryukyus) helped us with collecting the specimens. A part of the material examined was collected during the KUMEJIMA 2009 Expedition organized by the Transdisciplinary Research Organization for Subtropical and Island Studies of the University of the Ryukyus, the Center for Marine Bioscience & Biotechnology of the National Taiwan Ocean University, the Raffles Museum of Biodiversity Research of the National University of Singapore, and the Biodiversity Research Center of the Academia Sinica. The expedition operated under a permit granted to Dr. T. Naruse by the Okinawa Prefectural Governor and the Kumejima Fisheries Cooperative. The present study includes the contribution from Shimoda Marine Research Center (No. 758). This study was partly supported by a Grant-in-Aid JSPS Fellows (No. 23-527) (YO), a joint-research in Japanese Association for Marine Biology (YO, KT, and EH), and “International Research Hub Project for Climate Change and Coral Reef/Island Dynamics” from University of the Ryukyus (EH). The authors would like to thank three anonymous reviewers for valuable comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. A. Poulet.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ota, Y., Hoshino, O., Hirose, M. et al. Third-stage larva shifts host fish from teleost to elasmobranch in the temporary parasitic isopod, Gnathia trimaculata (Crustacea; Gnathiidae). Mar Biol 159, 2333–2347 (2012). https://doi.org/10.1007/s00227-012-2018-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-012-2018-2