Abstract
Panulirus argus (Latreille in Ann Mus Hist Nat Paris 3:388–395, 1804) is the lobster of greatest economic importance throughout its distribution. In this study, mitochondrial (Cytochrome Oxidase I and 16S ribosomal genes) and nuclear (Adenine Nucleotide Transporter gene) sequences were used to evaluate the taxonomic status of P. argus sampled from five sites in the Caribbean Sea and nine sites in the Southwest Atlantic. Phylogenetic analyses indicate that lobsters from the two regions form two monophyletic groups with a molecular divergence similar to that observed between distinct congeneric lobster species and much larger than that found between conspecific lobster populations. Therefore, the Caribbean and the Southwest Atlantic lobster populations originally attributed to P. argus belong to different species, with an estimated time of isolation of around 16 Million years. An important consequence of these findings is that the fisheries of spiny lobsters from the Caribbean and the Southwest Atlantic species must be managed separately.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spiny lobster P. argus (Latreille 1804) occurs from North Carolina (USA) to Rio de Janeiro (Brazil) and supports valuable fisheries throughout its distribution, where it is the lobster of greatest economic importance (Holthuis 1991). Many studies have focused on the ecology, behaviour, physiology and fisheries of P. argus (McWilliam and Phillips 2007; Phillips and Melville-Smith 2006), with few papers regarding the genetic structure at specific sites throughout its distribution (Glaholt and Seeb 1992; Hateley and Sleeter 1993; Silberman et al. 1994; Sarver et al. 1998; Diniz et al. 2005; Naro-Maciel et al. 2011).
Large levels of genetic differentiation, based on few partial sequences of the mitochondrial 16S and cytochrome oxidase I (COI) genes, have been reported between individuals P. argus from five locations in the Caribbean and one location in Brazil (Sarver et al. 1998). This led the authors to suggest the provisional recognition of two subspecies: P. argus argus, representing populations from Venezuela to Bermuda and P. argus westonii, representing populations from Brazil. A similarly high divergence between the Caribbean and the Southwest Atlantic lobster populations was observed by Diniz et al. (2005), who used sequences from the control region of the mitochondrial DNA of P. argus, but no taxonomic conclusions were ventured by the authors. Those phylogenetic works were based only on parts of the mitochondrial genome so that, to date, no study has used nuclear markers to analyse the taxonomic status of Panulirus species from the Atlantic.
The absence of a definitive conclusion about the taxonomic status of the Caribbean and the Atlantic lobsters is harmful to the development of new research and to the understanding of lobster population dynamics, ecology and physiology. Until now, populations of P. aff. argus from the two areas have been treated as a single species by all governments and international fisheries organisations. This could result, among other problems, in an overestimation of fishing stock size. The correct identification of fisheries species is fundamental for their management and conservation. This is particularly important for fisheries of spiny lobsters where a high market value and limited management practices have resulted in its overexploitation in many producer countries, including Brazil, Colombia and Nicaragua (FAO 2011).
In this study, we use sequences of a nuclear gene (Adenine Nucleotide Transporter—ANT) and of two mitochondrial regions (cytochrome oxidase I—COI and large ribosomal subunit—16S) to demonstrate that P. aff. argus from the Caribbean and from the Southwest Atlantic belong to two different species, which diverged around 16 Million years (My) ago.
Materials and methods
Sampling
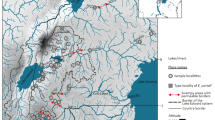
Individuals of P. aff. argus were obtained from local artisanal fisheries or bought fresh from local markets, where the collection site could be attested. Pereopod samples were collected from lobsters from five sites in the Caribbean: Caracas (Venezuela), San Andres, Cartagena, La Guajira and Santa Marta (Colombia); and from nine sites along 4,000 km of the Southwest Atlantic coast (Fig. 1; Table 1). One individual of P. echinatus and two individuals of P. laevicauda were collected to be used as outgroups in the phylogenetic analyses.
DNA extraction, amplification and sequencing
DNA was purified using a genomic DNA extraction kit (GE Life Sciences) according to the specifications of the manufacturer. Portions of two mitochondrial regions were amplified using primers: L-CO1490 (GCA ACG ATG ATT TTT CTC) and H-CO2198 (GCC TTT TGG GGC CTT GGG) for cytochrome oxidase I (COI) (Folmer et al. 1994); and 16Sar-L (GAT AAA ATA ACT TAA AAT TA) and 16Sbr-H (GCT TAA TCG AAC ACA CAG) for 16S (Palumbi et al. 1991). There are few nuclear markers available for decapods in the literature. The markers used so far are usually evolutionarily much conserved and, hence, only useful for phylogenetic studies of higher taxa. We tried to amplify different nuclear regions (like the ITS-1 and ITS-2 ribosomal spacers, introns of the extension factors EF1 and EF2 and of the ANT and GAPDH genes). The primers for the adenine nucleotide transporter (ANT; also known as ATP/ADP translocase) produced the best results in terms of amplification reliability and inter/intraspecific genetic differentiation ratios. The ANT gene is conserved enough to have regions for which primers that amplify across several families of decapods can be developed and presents an intron with variation levels compatible with intra-familial molecular systematic studies (Teske and Beheregaray 2009). A portion of the ANT nuclear gene was amplified using primers DecapANT-F (CCT CTT GAY TTC GCK CGA AC) and DecapANT-R (TCA TCA TGC GCC TAC GCA C) (Teske and Beheregaray 2009). PCR reactions were set up using 1 unit of Taq polymerase (GE Life Sciences), 0.2 mM of each dNTP, 0.3 μM of each primer, 2.5 mM of MgCl2, 2.5 μL of PCR buffer (10X) and 1 μL of DNA (10–100 ng) as a template in a final volume of 25 μL. The thermocycling conditions for the mitochondrial regions were one initial cycle of 4 min at 94 °C, followed by 30 cycles of 50 s each at 94, 50 and 72 °C, and one final extension step of 5 min at 72 °C. For the nuclear gene, the annealing temperature was 57 °C. Negative controls (without DNA template) were used in all PCR reactions. Both strands of PCR products were purified with an ExoSAP-IT purification kit (USB) and were sequenced in an ABI 3500 automatic sequencer with the same sets of primers used for the PCR.
Phylogenetic analyses
The DNA sequences obtained were edited with SEQMAN 7.0 (DNASTAR Inc.) and aligned with the CLUSTALW (Thompson et al. 1994) algorithm present on MEGA 5.0 (Tamura et al. 2011). The Kimura 2-parameter (K2P; Kimura 1980) distance model was used on MEGA 5.0 to estimate nucleotide divergences. All sequences obtained were deposited in GenBank (Accession Numbers ANT: sequences 1–6, JQ412160 to JQ412165; 16S: Haplotypes 1–14, GU475989 to GU475995, JQ412152 to JQ412158; COI: Haplotypes 1–26, GU476034 to GU476055, JQ412167 to JQ412170; P. echinatus, 16S: JQ412159; COI: JQ412171; P. laevicauda, ANT: JQ412166). The program PHASE 2.1 (Stephens et al. 2001; Stephens and Scheet 2005) was used to identify alleles of heterozygote sequences found in the ANT gene.
Estimates of sequence polymorphism were obtained through DNASP 5 (Librado and Rozas 2009). For Maximum Likelihood and Bayesian analyses, the COI and 16S sequences were combined as one dataset with two different matrices using the program MESQUITE 2.6 (Maddison and Maddison 2009). The Maximum Likelihood algorithm of MEGA 5.0 was used to obtain the best nucleotide substitution model through the estimation of the lowest Bayesian Information Criterion (BIC) score. All analyses were done using the nucleotide evolution model available that closely resembled the one indicated by MEGA 5.0. For the COI-16S combined dataset, the HKY model (Hasegawa et al. 1985) with estimates of invariable sites (I) and gamma distribution (G) was used, and for the ANT gene dataset, the F81 model was used (Felsenstein 1981).
The Maximum Likelihood analysis was performed with the program PHYML (Guindon and Gascuel 2003) using default parameters and 1,000 Bootstrap branch support replicates. The Bayesian analysis was carried out using MR. BAYES (Huelsenbeck and Ronquist 2001), allowing the program to estimate parameters for each gene partition. Bayesian posterior probabilities (BPP) were obtained by performing two separate runs with four Markov chains. Each run was conducted for 10 million generations, sampled every 1,000 generations. A consensus tree was calculated after excluding the first 25 % of the iterations as burn-in.
Divergence time estimation
To infer the divergence time between the individuals of P. aff. argus from the Caribbean and from the Southwest Atlantic, we used sequences of the COI gene obtained in this work together with sequences from other species of the Palinuridae and Synaxidae family retrieved from GenBank (Table 2). This analysis was performed using BEAST 1.6.1 (Drummond and Rambaut 2007), with a Bayesian relaxed-clock uncorrelated lognormal approach with estimated rate, the HKY model of sequence evolution and a Yule process for the tree prior. Each genus was determined as a monophyletic group, and a calibration was set on the Palinurus, Panulirus, Puerulus and Palibythus group using a lognormal distribution based on fossil evidence of Palinurus palaciosi (110–125 My; Vega et al. 2006) and Panulirus destombesi (99–112 My; Garassino and Breton 2010). The genus Palibythus was included in the Palinuridae family as indicated by a recent phylogenetic study (Tsang et al. 2009). A mean substitution rate was set using a normal distribution with 0.024 mean and standard deviation of 0.002. Markov-Chain Monte Carlo (MCMC) simulations were run for 80 million generations, with the first 10 % discarded as burn-in and the lower and upper bounds of the 95 % highest posterior density (HPD) interval obtained for every node.
Results
DNA polymorphism
Aligned lengths of sequences of 46 individuals were 658 bp of cytochrome oxidase I and 524 bp of 16S. For the ANT gene, 490 bp were sequenced in 25 individuals (Table 1). The average transition/transversion ratio was 2.4 for COI and 1.7 for 16S sequences. The COI gene was the most variable, and the ANT gene was the least variable in the individuals analysed (Table 3).
Phylogenetic analyses
Indels were found only on 16S sequence alignment and were removed before analyses. Sequences from two individuals of P. laevicauda were used as outgroups, together with sequences from two species of the family Scyllaridae, Thenus orientalis and Ibacus orientalis, retrieved from GenBank.
With the COI-16S dataset, trees with similar topologies were retrieved for both phylogenetic approaches, and hence, only the Maximum Likelihood tree is shown with both bootstrap and BPP values included (Fig. 2). P. aff. argus from the Southwest Atlantic and from the Caribbean formed reciprocally monophyletic groups that diverged (distance corrected with Kimura 2-parameter model) between 10.2 and 11.4 % for the combined mitochondrial set (14.4–17.8 % for COI and 5.0–5.8 % for 16S). The COI genetic divergence values were similar to those observed between other species of Panulirus (13.7–36.1 %) and congeneric species of other genera of the Palinuridae (4.3–24.3 %).
Maximum Likelihood phylogenetic tree based on the COI-16S concatenated sequences of P. aff. argus. The bootstrap and posterior probability values are indicated only for the nodes with over 70 % support for the analysis of Maximum Likelihood (1,000 replicates) and the Bayesian inference (100 replicates), respectively. Tree was rooted using P. echinatus as an outgroup
The ANT gene presented only seven polymorphic sites, of which three were diagnostic between the Caribbean and the Southwest Atlantic regions, two were polymorphic within the Caribbean population and two were mutations of the same allele present only in the Southwest Atlantic in seven homozygote and four heterozygote individuals. The resulting phylogenetic trees retrieved from Maximum Likelihood and Bayesian inference had very few topological differences in the Caribbean group, and the individuals from Southwest Atlantic formed a monophyletic group in both analyses (Fig. 3). The few differences disappeared when only P. laevicauda was used as an outgroup, probably because the two other outgroup species were too divergent, which led to a very large number of indels in the alignments. The phylogeny retrieved from analyses with the nuclear gene agreed with the mitochondrial results (Figs. 2 and 3). The divergence estimated between individuals of P. aff. argus from the Caribbean and from the Southwest Atlantic was similar to that observed between other species of Panulirus (Fig. 4) and much higher than the variation found within each region.
Maximum Likelihood phylogenetic tree based on ANT sequences of P. aff. argus. The Maximum Likelihood analysis (1,000 replicates) and the Bayesian inference (100 replicates) bootstrap and posterior probability values, respectively, are indicated only for the nodes with over 50 % support. P. laevicauda, Ibacus peronii and Thenus orientalis were used as outgroups for the ML analysis. Bayesian inference used only P. laevicauda as an outgroup (see text)
Divergence time
The phylogeny analyses show P. aff. argus as a monophyletic group with a clear separation between the Caribbean and the Southwest Atlantic clades (Fig. 4), which diverged around 16 Million Years ago (My) (95 % HPD 8.4–26.1 My). The divergence time between species of the genus Panulirus varied from 4.7 My (95 % HPD 0.6–11.1 My), between P. echinatus and P. penicillatus to 32.3 My, which is the age of the most recent common ancestor of the Panulirus genus (95 % HPD 16.8–54.4 My).
Discussion
The analyses of both nuclear and mitochondrial sequences clearly show that the Caribbean and the Southwest Atlantic populations originally attributed to Panulirus argus belong to different species. The two groups formed reciprocally monophyletic clades irrespective of geographical distance. Furthermore, the high divergence found with mitochondrial genes between the Caribbean and the Southwest Atlantic clades (K2P around 16 % for COI and 5.5 % for 16S) was similar to that observed between distinct congeneric species and much larger than that found between conspecific lobster populations. Within the genus Panulirus, the interspecific divergence levels vary between 10 and 32 % for COI (Ptacek et al. 2001; Cannas et al. 2006; Naro-Maciel et al. 2011) and between 3 and 19 % for the 16S sequences (Sarver et al. 1998; Ptacek et al. 2001). Mean interspecific levels of gene divergence within the genus in combined analysis of the 16S and COI genes were of about 8 % (Groeneveld et al. 2007), which is similar to the 10 % observed between concatenated sequences of the Caribbean and the Southwest Atlantic P. aff. argus.
The levels of genetic divergence observed for the ANT gene were smaller between the Caribbean and the Southwest Atlantic Panulirus (~ 1 % K2P) than those observed between congeneric species of Halicarcinus and Hymenosoma crabs (Teske and Beheregaray 2009). Using similar areas of the sequence, which included introns and exons, the divergence between Halicarcinus cookie and H. varius varied between 1.2 and 5 %, and between Hymenosoma geometricum and Hymenosoma sp., it varied between 2.1 and 7 %. However, even though the calculated divergence between the Caribbean and the Southwest Atlantic was not very high, the polymorphisms found were specific to each region, once again indicating absence of gene flow between the two areas.
In addition to the fixed genetic differences, the Caribbean and the Southwest Atlantic P. aff. argus also present consistent morphological differences, recognised by workers of the fishing industry (Sarver et al. 1998) but discarded as intraspecific variation by most taxonomists to date. The Caribbean species presents a darker red colour with deeper grooves in the cephalothorax region and a pattern of dorso-abdominal spots more characteristic of the species description, whereas the Southwest Atlantic species has a lighter colour and shallow grooves in the carapace, with a higher concentration of smaller abdominal spots. The Southwest Atlantic species also has a distinct colouration pattern of the pereopods and the pleopods. The former have segments striped longitudinally in the Caribbean species, whereas, in the Southwest Atlantic, the segments have purple stains. The pleopods of the Caribbean species are yellow and black, with the black colour of the three first segments forming a falcate discontinued shape, while in the Southwest species the black colour of all pleopods follows their ovate shape. Although colouration is usually considered of limited use in taxonomic studies, among other things because it is easily lost by traditional methods of preservation, it is a character used for the identification of other Panulirus species (Holthuis 1991) and has been successfully used to distinguish cryptic decapod species (Berry 1974; Bruce 1975; Knowlton 1986).
A high level of genetic divergence has already been reported by Sarver et al. (1998) between individuals of P. argus from the Caribbean and from the Southwest Atlantic (20 % for COI and 8 % for 16S), but their analyses were based on a reduced sample of the Southwest Atlantic coast (in the case of COI, only one sequence from each region was used). The authors suggested that lobsters from the two regions be given subspecific status (P. argus argus for the Caribbean individuals and P. argus westonii for the Southwest Atlantic ones), but no formal description was made. The same happened in the analysis of Diniz et al. (2005), who found a large divergence (control region; K2P up to 38 %) between the Caribbean and the Southwest Atlantic spiny lobsters, but who did not make any taxonomic inference from their results, again because few individuals (N = 5, from two locations only) from Southwest Atlantic had been sampled.
All factors discussed here contribute to a re-evaluation of the taxonomic status of P. argus and the acknowledgement of a new species. According to Latreille (1804), the type locality could be the Antilles (“Je la soupçonne des Grandes Indes”), but Lamarck (1818) stated that Latreille really intended Brazil as the type locality. Based on that confusion, the type locality of P. argus has been classified as “unknown” (Holthuis 1991). Although a holotype was not designated for the species, possible syntypes from the “Antilles” were deposited at the Paris Museum of Natural History, which suggests that the binomial P. argus should be used for the Caribbean species (Holthuis 1991). It is important therefore that analyses of all syntypes, of samples from both species from multiple localities and of the holotypes of other species now in synonymy (e.g. Palinurus americanus, P. ricordi), be performed to verify if any of the given names can be reassigned. Since Sarver et al. (1998) did not describe the two subspecies they named nor did they present drawings or assigned types for either subspecies (Chan 2010), the names suggested by them (P. argus argus and P. argus westonii) are not formalised and must be considered nomina nuda (Chapter 4, Article 13 of the International Code for Zoological Nomenclature; ICZN 1999; Chan 2010). Hence, we suggest that the binomial P. argus be used for the Caribbean species, whereas the Southwest Atlantic species should be called Panulirus sp., until it is formally described. A detailed morphological taxonomic analysis is being done and will be published elsewhere.
In the present work, the two species were not found co-occurring in any of the sampling sites. However, there are anecdotal indications of the occasional capture of lobsters with the diagnostic colouration pattern of P. argus by industrial fishing boats off the coast of Ceará State, in Northeast Brazil. The southern limit of the distribution of Panulirus sp. on the Brazilian coast is Rio de Janeiro (20°S), and the northern limit of P. argus is North Carolina (35°N). We found Panulirus sp. along the coast up to 2°S (Pará State) and P. argus down to 8°N (Venezuela). P. argus is not cited in the area between parallels 8°N and 2°S (for a fisheries faunal list of the French Guyana, see Guéguen 2000; for Suriname, see FAO 2008; Holthuis 1959). This does not mean that the species is absent, but indicates that, if anything, it is rare in the area. It would be interesting to analyse samples from the Guyanas and Suriname if they are found, to verify where the limit of the two species may be and whether they occur in sympatry in those areas. Silberman et al. (1994), analysed restriction fragment length polymorphisms (RFLPs) of mitochondrial genomes of 259 Caribbean lobsters, and registered the occurrence of three more divergent RFLP patterns, two from Florida and one from Venezuela. Those three individuals were genetically identified by Sarver et al. (2000) as belonging to the Southwest Atlantic Panulirus sp. (“P. argus westonii” in their paper). The presence of individuals bearing Panulirus sp. mitochondrial DNA in Florida may be related to introgression or to the presence of recruits resulting from long-range natural or anthropogenic dispersal. Occasional individuals of P. aff. argus have been found in West Africa (Holthuis 1991), and Freitas and Castro (2005) reported the presence of P. cf. argus in Cape Verde. In both cases, intentional or accidental (possibly through ballast water) anthropogenic transport was suggested as the source of the introductions, but none of the introductions resulted in the establishment of local populations.
The high genetic differentiation between the Caribbean and the Southwest Atlantic lobsters has been explained as the result of the outflow of the Amazon River in the Atlantic Ocean (Sarver et al. 1998; George 2005). According to geological records, the deposition of sediment and freshwater by the Amazon River started at the end of the uplift of the Andes mountains, which inverted the river flow towards the Atlantic Ocean at the beginning of late Miocene, around 10 My (Hoorn 1993, 1996; Hoorn et al. 1995; Potter 1997; Campbell Jr et al. 2006). Currently, the Amazon River discharges about one-fifth of the world’s freshwater runoff into the Atlantic (Curtin 1986a, b), which causes an alteration of salinity and sediment discharge up to 500-km seaward and 30-m depth (Rocha 2003). The divergence time estimated here between the mitochondrial DNA sequences of the Caribbean P. argus and the Southwest Atlantic Panulirus sp. was of 16 My, which is earlier, but not incompatible considering confidence intervals (8.4 to 26.1 My), with the beginning of the Amazon outflow. During the Miocene (23 to 5 My), the Atlantic Ocean suffered many oceanographic changes, including the formation of a thermocline as a consequence of the ice build-up on Antarctica (Berger 2011). These changes may have helped initiate the speciation process that would later be completed with the formation of a barrier of low salinity water by the outflow of the Amazon River.
The obvious implication of the results of this paper is that spiny lobster fisheries will have to be managed separately for the Caribbean and the Southwest Atlantic species. The possible presence of Southwest Atlantic Panulirus sp. in Florida should be further investigated, including analyses of mitochondrial and nuclear genes to verify if the samples result from current migration or past introgression. The occasional presence of P. aff. argus in Africa is also puzzling, and it would be interesting to genetically verify whether those lobsters belonged to the Caribbean or the Southwest Atlantic species to help formulate hypotheses on their origin.
References
Berger WH (2011) Geologist at sea: aspects of ocean history. Ann Rev Mar Sci 3:1–34
Berry PF (1974) A revision of the Panulirus homarus-group of spiny lobsters (Decapoda, Palinuridae). Crustaceana 27:31–42
Boisselier-Dubayle MC, Bonillo C, Cruaud C, Couloux A, Richer de Forges B, Vidal N (2010) The phylogenetic position of the “living fossils” Neoglyphea and Laurentaeglyphea (Decapoda: Glypheidea). C R Biol 333:755–759
Bruce AJ (1975) Coral reef shrimps and their color patterns. Endeavour 34:23–27
Campbell KE Jr, Frailey CD, Romero-Pittman L (2006) The Pan-Amazonian Ucayali Peneplain, late Neogene sedimentation in Amazonia, and the birth of the modern Amazon River system. Palaeogeogr Palaeoclimatol Palaeoecol 239:166–219
Cannas R, Cau A, Deiana AM, Salvadori S, Tagliavini J (2006) Discrimination between the Mediterranean spiny lobsters Palinurus elephas and P. mauritanicus (Crustacea: Decapoda) by mitochondrial sequence analysis. In: Thessalou-Legaki M (ed) Issues of Decapod Crustacean Biology. Hydrobiologia 557:1–4
Chan TY (2010) Annotated checklist of the world’s marine lobsters (Crustacea: Decapoda: Astacidea, Glypheidea, Achelata, Polychelida). Raffles Bull Zool 23:153–181
Curtin TB (1986a) Physical observations in the plume region of the Amazon River during peak discharge. 2. Water masses. Cont Shelf Res 6:53–71
Curtin TB (1986b) Physical observations in the plume region of the Amazon River during peak discharge. 3. Currents. Cont Shelf Res 6:73–86
Diniz FM, Maclean N, Ogawa M, Cintra IHA, Bentzen P (2005) The hypervariable domain of the mitochondrial control region in Atlantic spiny lobsters and its potential as a marker for investigating phylogeographic structuring. Mar Biotechnol 7:462–473
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214–221
FAO (2008) Fisheries Country Profile. The Republic of Suriname, FAO, Rome
FAO (2011) FISHSTAT plus Version 2.3: Universal software for fishery statistical time series. Capture Production 1950–2009. FAO, Rome
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–373
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Freitas R, Castro M (2005) Occurrence of Panulirus argus (Latreille, 1804) (Decapoda, Palinuridae) in the Northwest islands of the Cape Verde Archipelago (Central-East Atlantic). Crustaceana 78:1191–1201
Garassino A, Breton G (2010) Panulirus destombesi n. sp. (Crustacea, Decapoda, Palinuridae) de l’Albien (Crétacé inférieur) de Wissant (Pas-de-Calais, France). Geodiversitas 32:391–397
George RW (2005) Tethys Sea fragmentation and speciation of Panulirus spiny lobsters. Crustaceana 78:1281–1309
Glaholt RD, Seeb J (1992) Preliminary investigation into the origin of the spiny lobster, Panulirus argus (Latreille, 1804), population of Belize, Central America (Decapoda, Palinuridea). Crustac Int J Crustac Res 62:159–165
Groeneveld JC, Gopal K, George RW, Matthee CA (2007) Molecular phylogeny of the spiny lobster genus Palinurus (Decapoda: Palinuridae) with hypotheses on the speciation in the NE Atlantic/Mediterranean and SW Indian Ocean. Mol Phylogenet Evol 45:102–110
Guéguen F (2000) Distribution et abondance des poissons démersaux et de quelques autres organismes benthiques marins du plateau continental (0–60 m) de Guyane Française. Comptes Rendus de la Académie des Sciences—Series III—Sciences de la Vie 323:775–791
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hasegawa M, Kishino H, Yano TA (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Hateley JG, Sleeter TD (1993) A biochemical genetic investigation of spiny lobster (Panulirus argus) stock replenishment in Bermuda. Bull Mar Sci 52:993–1006
Holthuis LB (1959) The Crustacea Decapoda of Suriname (Dutch Guiana). Zool Verh 43:1–296
Holthuis LB (1991) FAO species catalogue. Vol. 13. Marine lobsters of the world. An annotated and illustrated catalogue of species of interest to fisheries known to date. FAO Fisheries Synopsis 125. FAO, Rome
Hoorn C (1993) Marine incursions and the influence of Andean tectonics on the depositional history of Northwestern Amazonia—results of a palynostratigraphic study. Palaeogeogr Palaeoclimatol Palaeoecol 105:267–309
Hoorn C (1996) Miocene deposits in the Amazonian foreland basin. Science 273:122–123
Hoorn C, Guerrero J, Sarmiento GA, Lorente MA (1995) Andean tectonics as a cause for changing drainage patterns in Miocene Northern South-America. Geology 23:237–240
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
ICZN (1999) International Code of Zoological Nomenclature, 4th edn. International Trust for Zoological Nomenclature, London
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Knowlton N (1986) Cryptic and sibling species among the decapod Crustacea. J Crustac Biol 6:356–363
Lamarck JBPA (1818) Histoire naturelle des animaux sans vertèbres. Vol. 5:1–612. Imprimerie d’Abel Lanoe, Paris
Latreille PA (1804) Des langoustes du Muséum National d’Histoire Naturelle. Ann Mus Hist nat Paris 3:388–395
Librado P, Rozas J (2009) DnaSP v. 5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Maddison WP, Maddison DR (2009) Mesquite: a modular system for evolutionary analysis. Version 2.6. http://mesquiteproject.org
McWilliam PS, Phillips BF (2007) Spiny lobster development: mechanisms inducing metamorphosis to the puerulus: a review. Rev Fish Biol Fish 17:615–632
Naro-Maciel E, Reid B, Holmes KE, Brumbaugh DR, Martin M, DeSalle R (2011) Mitochondrial DNA sequence variation in spiny lobsters: population expansion, panmixia, and divergence. Mar Biol 158:2027–2041
Ovenden JR, Booth J, Smolenski A (1997) Mitochondrial DNA phylogeny of red and green rock lobsters (genus Jasus). Mar Freshw Res 48:1131–1136
Palero F, Crandall KA, Abelló P, Macpherson E, Pascual M (2009) Phylogenetic relationships between spiny, slipper and coral lobsters (Crustacea, Decapoda, Achelata). Mol Phylogenet Evol 50:152–162
Palumbi SR, Martin A, Romano S, Mcmillan WO, Stice L, Grabowski G (1991) The simple fool’s guide to PCR, version 2.0. University of Hawaii, Honolulu
Phillips BF, Melville-Smith R (2006) Panulirus species. In: Phillips BF (ed) Lobsters: biology, management, aquaculture and fisheries. Blackwell Publishing, Singapore
Potter PE (1997) The Mesozoic and Cenozoic paleodrainage of South-America: a natural history. J South Am Earth Sci 10:331–344
Ptacek MB, Sarver SK, Childress MJ, Herrnkind WF (2001) Molecular phylogeny of the spiny lobster genus Panulirus (Decapoda: Palinuridae). Mar Freshw Res 52:1037–1047
Rocha LA (2003) Patterns of distribution and processes of speciation in Brazilian reef fishes. J Biogeogr 30:1161–1171
Sarver SK, Silberman JD, Walsh PJ (1998) Mitochondrial DNA sequence evidence supporting the recognition of two subspecies or species of the Florida spiny lobster Panulirus argus. J Crustac Biol 18:177–186
Sarver SK, Freshwater DW, Walsh PJ (2000) The occurrence of the provisional Brazilian subspecies of spiny lobster (Panulirus argus westonii) in Florida waters. Fish Bull 98:870–873
Silberman JD, Sarver SK, Walsh PJ (1994) Mitochondrial DNA variation and population structure in the spiny lobster Panulirus argus. Mar Biol 120:601–608
Stephens M, Scheet P (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing data imputation. Am J Hum Genet 76:449–462
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Teske PR, Beheregaray LB (2009) Intron-spanning primers for the amplification of the nuclear ANT gene in decapod crustaceans. Mol Ecol Resour 9:774–776
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Tsang LM, Chan T-Y, Cheung MK, Chu KH (2009) Molecular evidence for the Southern Hemisphere origin and deep-sea diversification of spiny lobsters (Crustacea: Decapoda: Palinuridae). Mol Phylogenet Evol 51:304–311
Vega FJ, García-Barrera P, Perrilliat MC, Coutiño MA, Mariño-Pérez R (2006) El Espinal, a new plattenkalk facies locality from the Lower Cretaceous Sierra Madre Formation, Chiapas, southeastern Mexico. Rev Mex Cien Geológ 23:323–333
Acknowledgments
We thank D. Almeida, E. Araujo, L. Barbosa, P. Coelho Filho, H. Cunha, M.C.O. Matos, N. Leite Jr., F. Magalhães, R. Monteiro, M.C. Ostrowski, P. Paiva, A. Reis, G. Rey, E. Rios, E. Routledge, and S. Ribeiro for help with sample collections. We also thank S.K. Sarver and J. Gusmão for suggestions on an early version of the manuscript and C. Schrago for helping with the Bayesian divergence time analysis. This paper was made possible by grants from the Brazilian Ministries for Fisheries and Aquaculture (MPA) and Science, Technology and Inovation (MCTI), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). This work is part of the MSc thesis of JLT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. I. Taylor.
Rights and permissions
About this article
Cite this article
Tourinho, J.L., Solé-Cava, A.M. & Lazoski, C. Cryptic species within the commercially most important lobster in the tropical Atlantic, the spiny lobster Panulirus argus . Mar Biol 159, 1897–1906 (2012). https://doi.org/10.1007/s00227-012-1977-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-012-1977-7