Abstract
Loss of zooxanthellae (dinoflagellate Symbiodinium) from corals will sometimes lead to mass mortality of corals. To detect and quantify Symbiodinium released from corals, we developed a zooxanthellae “trap” and a quantitative PCR (qPCR) system with Symbiodinium clades A–F-specific primer sets. The trap was attached to a branch or the surface of several wild stony corals, and the water samples within the traps, including released Symbiodinium, were subjected to qPCR. All tested corals released clade C Symbiodinium at estimates of ~5,900 cells h−1 cm−2 of coral surface. Although all tested Pocillopora eydouxi harboured both clades C and D, some of these colonies released only clade C or released a lesser amount of clade D than that in the tissues. Our Symbiodinium quantification system revealed that wild hermatypic corals constantly release Symbiodinium to the environment. Our result suggests that some corals may discharge certain clades of Symbiodinium alternatively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many marine invertebrates, including stony corals, harbour symbiotic microalgae commonly referred to as zooxanthellae within their tissues. The zooxanthellae, specifically the dinoflagellate genus Symbiodinium, translocate their photosynthetic products to the host corals and contribute to the vigour of corals and the coral reef ecosystem. “Coral bleaching,” which results in mass mortality of corals, has occurred in many tropical and subtropical seas with increasing frequency over the past two decades (Hoegh-Guldberg 1999). As an example of the catastrophic effect of coral bleaching in coral reef ecosystems, coral reefs have decreased by 30–50% in the Caribbean Sea over the last three decades (Gardner et al. 2003) and more than 70% of corals died between 2003 and 2008 in Sekisei Lagoon (Okinawa, Japan) (National Institute for Environmental Study; http://www.nies.go.jp/topics/2008/20080910/20080910.html).
In a strict sense, the term coral bleaching refers to the loss of Symbiodinium and/or reduction of their pigments (Brown 1997). Thus, release of Symbiodinium may be directly correlated with coral bleaching. Causes of Symbiodinium discharge include stresses such as elevated temperature (Gates et al. 1992) or metal exposure (Jones 1997). Symbiodinium discharge has been investigated in a variety of aquarium experiments. Bhagooli and Hidaka (2004) showed that photosynthetically active Symbiodinium could be released from a coral, and Stimson and Kinzie (1991) reported that a Pocillopora damicornis colony in nutrient-enriched water released more Symbiodinium than did the control colony in ambient seawater. Perez et al. (2001) found that Symbiodinium expulsion increased significantly with increasing temperature in the sea anemone Aiptasia pallida. Although these aquarium experiments have provided useful information, data from the field are necessary, as little is known about Symbiodinium release from corals in natural environments. Such information is essential to understanding coral bleaching in the field.
Because Symbiodinium cells are relatively small and have a featureless spherical morphology, it is difficult to enumerate them once they are released into the sea. To detect microorganisms that cannot be feasibly recognized and counted under a microscope, researchers often use quantitative real-time PCR (qPCR) (e.g., Bowers et al. 2000; Dyhrman et al. 2006; Galluzzi et al. 2004; Hosoi-Tanabe and Sako 2005; Kamikawa et al. 2006). qPCR has been applied in a number of studies involving coral-Symbiodinium systems (e.g., Correa et al. 2009; Loram et al. 2007; Mieog et al. 2007). Our previous attempt to quantify Symbiodinium by means of qPCR showed that (1) more than 2 × 106 cells h−1 (enumeration based on using a Symbiodinium culture as the quantification standard) were released from two colonies of Acropora digitifera with a combined total skeletal weight of 210 g and (2) more than 30,000 cells l−1 of Symbiodinium occurred in the water column around a reef (Koike et al. 2007). These data are preliminary but suggestive: The Symbiodinium biomass present in the water column could be a sink for cells from the nearby corals, and this might be indicative of regional coral bleaching and/or a source for cells that might be inherited by other corals.
In Symbiodinium, nine phylogenetically distinct clades have been defined based on rRNA genes (clades A–I) (see Coffroth and Santos 2005; Pochon and Gates 2010). Furthermore, Symbiodinium phylogeny is highly diverse and includes many “types” within a clade (LaJeunesse 2002; LaJeunesse et al. 2003). Phylotypes belonging to different clades can exhibit similar patterns of sensitivity to elevated temperatures but their closely related sister phylotypes can exhibit different patterns of sensitivity to that stress (Tchernov et al. 2004). This study demonstrates that rDNA genotyping may not be diagnostic of thermal sensitivity in Symbiodinium. However, long-term field observations demonstrated that the cladal composition of some corals shifted from clade C to clade D after bleaching events (Baker et al. 2004; Jones et al. 2008). Although Symbiodinium has many types within a clade, roughly speaking, some of the corals with clade D symbionts are more resistant to thermal stress than those with clade C symbionts (Glynn et al. 2001; Rowan 2004). Therefore, it is plausible that the assemblage of Symbiodinium clades present in a coral could confer greater environmental tolerance to the host, and indeed, some hosts seem to replace symbiont clades as a result of changing environmental conditions (e.g., Baker 2001).
Buddemeier and Fautin (1993) proposed the “Adaptive Bleaching Hypothesis (ABH),” which states that stressed corals bleach to change the symbiont clade according to environmental conditions. The hypothesis includes two plausible modes for changing clades: “switching” and “shuffling” (Baker 2003). Switching can be attributed to an uptake of exogenous symbionts, which is a phenomenon found in anemones (Kinzie et al. 2001) and soft corals (Lewis and Coffroth 2004). Shuffling can be a shift from background endogenous symbionts to a major one, which is a phenomenon that is assumed to occur in scleractinian corals (Jones et al. 2008). Recently, qPCR systems revealed that many coral species harbour a low abundance of clade D along with a major clade (Correa et al. 2009; Mieog et al. 2007). These results suggest that in many coral species, symbionts may be shuffled by environmental change, such as an increase in temperature. Regardless of the mode of change, a loss or deterioration of the former dominant symbiont clade must occur as the primary step, perhaps when it becomes unfit in the new environment. However, this phenomenon has not yet been demonstrated in any survey. Based on our previous laboratory experiment in which corals released numerous symbionts to the surrounding water (Koike et al. 2007), we hypothesize that such active release may be attributed to the primary step of the ABH process. To test this hypothesis, we developed and evaluated a new qPCR system that enabled us to enumerate Symbiodinium biomass by clade (A–F) and a “trap” to collect symbionts released from corals in the field. We then attempted to quantify the Symbiodinium released from wild stony corals using the trap and the qPCR system.
Materials and methods
Designing the clade-specific PCR primers
To date, several clade-specific PCR primer sets for qPCR detection of Symbiodinium have been reported. For example, Correa et al. (2009) developed four clade-specific primer sets targeting the ITS region for clade A and 28S rRNA genes for clades B, C, and D. In the present study, we used the intercalator chemistry that employs the SYBR® Green I fluorochrome and we designed the PCR primer sets based on specific regions of the nuclear 28S rRNA gene (28S rDNA) to differentiate clades A–F; as references we used sequences obtained from GenBank for 32 taxa of Symbiodinium (GenBank accession numbers are listed in Table 1). Twelve taxa of another dinoflagellate lineage and one apicomplexan parasite were also referenced as negative targets. The obtained sequences were aligned using Clustal × 2.0 (Larkin et al. 2007) and manually edited by eye. A region was found that was specific to each Symbiodinium clade (A–F) but allowed the elimination of other clades, other dinoflagellate taxa, and the apicomplexan parasite. Each primer set then was designed with Primer Express® v2.0 (Applied Biosystems, Foster City, CA, USA) to amplify about 100 bp of the region and to have almost the same Tm value.
Testing for primer specificity to each Symbiodinium clade using PCR and qPCR
To determine expected conformity with the target clade and unexpected conformity with non-target clades of the established primer sets, a Web site (Primer-BLAST; National Center for Biotechnology Information) search for DNA sequences was conducted with an input setting of 80–200 bp for the PCR product size and including sets of “all GenBank + RefSeq Nucleotides + EMBL + DDBJ + PDB sequences.” Along with these database searches, we conducted experimental confirmations with PCR using the designed primers against cultures of each Symbiodinium clade and other microalgae kept in our laboratory (Table 2). Samples (10 ml) of algae from the exponentially grown cultures were harvested by centrifugation (2,000g, 10 min), and the resultant cell pellets were subjected to total DNA extraction using a SepaGene® kit (Sanko Junyaku, Tokyo, Japan). Because Symbiodinium cells possess a rigid cell wall that cannot be feasibly extracted by the kit, cell pellets were initially homogenized using a glass-bead beating method (Mini-BeadBeater-8, Biospec Products, Bartlesville, OK, USA; 0.5-mm bead, operated at 3,200 rpm for 3 min). Extracted and purified total DNAs were dried, dissolved in 100 μl of TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0), and quantified by measuring the absorbance at 260/280 nm (Lightwave S2000, WPA, Cambridge, UK). A mixture of 1 ng μl−1 DNA, 1 pmol μl−1 of each forward and reverse primer, and the recommended volume of TaKaRa Ex Taq™ Hot Start Version (Takara Bio Inc., Shiga, Japan) were subjected to conventional PCR to verify the clade-specific amplicon from a designated Symbiodinium clade. A thermal-cycling protocol of 1 cycle at 94°C (5 min), 30 cycles at 94°C (5 s), 63°C (10 s) and 72°C (15 s), and 1 cycle at 72°C (5 min) was run in a 2720 Thermal Cycler (Applied Biosystems). The PCR amplicons were electrophoresed on 2% agarose gel and stained with ethidium bromide.
qPCR against the above-extracted DNAs from the Symbiodinium clades and other aligned microalgae were subsequently performed. Mixtures of 1 ng μl−1 DNA, 0.4 pmol μl−1 of each forward and reverse primer, and the recommended volume of TaKaRa SYBR® Premix Ex Taq™ II (Takara), including a ROX reference dye (as a passive reference), were analysed in an ABI PRISM 7000 (Applied Biosystems) under the PCR cycles of 1 cycle at 95°C (30 s) and 40 cycles at 95°C (5 s) and 60°C (31 s). After the qPCR runs, melting profiles of the PCR product vs. temperature (dissociation curves) were obtained for each sample to check that positive amplifications of target DNA without primer–dimers had occurred.
Quantification of Symbiodinium cells using qPCR
Symbiodinium cell numbers in exponentially grown cultures of CS-161 (clade A; purchased from The Commonwealth Scientific & Industrial Research Organization, Australia), CCMP1633 (clade B; purchased from The Provasoli-Guillard National Center for Culture of Marine Phytoplankton, USA), CCMP2466 (clade C), CCMP2556 (clade D), CCMP421 (clade E), and CS-156 (clade F) were counted under a light microscope, and culture aliquots equivalent to 250,000, 25,000, 2,500, and 250 cells each were filter-trapped onto a polycarbonate filter (0.8-μm pore size, 47-mm Φ, Advantec-Toyo, Tokyo, Japan) under gentle vacuuming. The filters were then stored at −20°C for more than a day, thawed at room temperature for 1 h, and immersed in 500 μl TE buffer in a 1.5-ml microtube. The tubes were then placed in boiling water with intermittent vortexing (30 s, 3-min intervals) for a total of 10 min and then triplicate aliquots of 2 μl (each equivalent to 1,000, 100, 10, and 1 cell/reaction) were retrieved and subjected to qPCR using each clade-specific primer set, with H2O as a non-template control (NTC). At the same time, to determine whether each primer set produces a signal from non-target clades, 1,000 cell-equivalent aliquots derived from other clades were also subjected to qPCR.
qPCR Application: how many Symbiodinium cells were released from corals in the natural environment?
Sampling locations and coral specimens
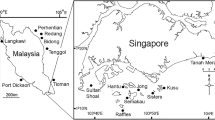
Field sampling was performed in Urasoko Bay, Ishigaki Island, Okinawa, Japan (Fig. 1). Two distinct locations were sampled: Point 1, where substantially healthy corals cover the reef slope, and Point 2, which is shallow (~1 m) and is exposed to turbid and stagnant water, and thus experiences high temperatures. On 27–28 August 2007, two colonies of Acropora muricata and four colonies of Pocillopora eydouxi were selected at Point 1 and two colonies of Porites lutea were selected at Point 2. Additionally, a single colony each of Acropora cf. grandis, Favites abdita, and Cyphastrea serailia was selected at Point 2 on 26 November 2007, and two colonies of Poc. eydouxi at Point 1 were selected on 9 October 2008. Finally, on 29–31 August 2009, three colonies of A. muricata were selected at Point 1, and a single colony of A. muricata and one of Acropora nobilis were selected at Point 2. The subsurface seawater temperatures on these sampling dates were ~28°C (Point 1) and 28°C (Point 2) on 27–28 August 2007; 24°C (Point 2) on 26 November 2007; 26°C (Point 1) on 9 October 2008; and 29°C (Point 1) and 30°C (Point 2) on 29–31 August 2009. At each sampling time point, a coral colour chart (distributed from CoralWatch Project from Project AWARE; http://www.projectaware.org) was used to categorize the bleaching index for the tested corals.
Sampling procedures
SCUBA divers attached a cylindrical trap for collecting released Symbiodinium from corals to a branch or the surface of the selected corals. The trap was composed of transparent PVC material, a cylindrical bottle (97 mm inner diameter, 167 mm depth, 1 l volume), a screw-top cap with a circular opening (73 mm inner diameter), and a textile pouch (with the bottom cut). The bottom-cut pouch was fitted to the circular opening of the cap by pinching it between the cap and the bottle mouth. The trap mouth was closed by squeezing a cord of the pouch after a branch of coral was inserted into the bottle (Fig. 2). To attain neutral buoyancy, a small lead sinker and a floater were tied to the trap. To set the trap on a non-branching encrusting coral (i.e., Por. lutea, F. adbita, or C. serailia), the textile part of the trap was folded up and the bottle opening was placed directly onto the coral’s surface. The trap itself was strung to a coral with a rubber band. At the time of trap setting, the divers were careful not to rub the coral surface with the textile pouch or bottle. Our previous aquarium experiment (Koike et al. 2007) using Acropora digitifera indicated that the release of Symbiodinium from the coral could exhibit daily rhythmicity, with a peak at noon. For this reason, we set the traps for ~3 h between 10:00 and 15:00. At the end of the experiment, the trap was carefully removed from the coral and the mouth was instantly closed by squeezing the pouch cord. The coral branches that had been covered by the trap were marked by twisting a PVC-coated wire next to the trap mouth, and then the branch was snapped off from the colony. For the encrusting corals, it was difficult to retrieve the exact coral area over which the trap mouth had been placed; thus, another easily removable part near the trap was broken off. Each coral sample was placed immediately into an individual plastic bag without seawater, and within 1 h, these samples were placed in a −20°C freezer.
The trap used to collect Symbiodinium released from corals. a The trap consists of a cylindrical bottle and a textile pouch, and it maintains neutral buoyancy with a small lead sinker and floater. b Deployment of the trap around a coral branch. The branch is inserted into the bottle, and the trap mouth is closed by squeezing a cord of the pouch. c Deployment of the trap around an encrusting coral. The trap is strung to the coral surface with a rubber band
On the boat, water samples from the traps were subjected to dissolved oxygen (DO) and temperature measurements using a portable DO meter (LDO-HQ10, HACH, Loveland, CO, USA); these measurements were used to detect possible stresses (e.g., DO decrease due to respiration, photosynthetic deterioration of Symbiodinium). The samples then were transferred to another bottle and kept in an ice-chilled cooler box. They were subjected to filtrations of coral-released Symbiodinium within 6 h, as described below. For the blank of Symbiodinium in seawater, ambient waters were also collected.
To make sure that the trapping of the coral did not cause stress-induced Symbiodinium release, at point 2 the traps were set on each of two branches of A. muricata (total two colonies) and A. nobilis (one colony). Five hours later, the trap-covered branches and adjacent free branches (three from each colony) were snapped off, and the Symbiodinium densities per surface area were determined and compared according to the method described below.
DNA extraction from the Symbiodinium in coral tissue
Frozen corals were thawed at room temperature, Symbiodinium cells were removed from the coral surface by spraying with 0.2-μm-filtered seawater containing 5 mM EDTA with an airbrush, and the subsequent cell suspension was collected in a plastic bag. The suspension was diluted to 50 ml with filtered seawater containing 5 mM EDTA, and the Symbiodinium cell number was counted under a light microscope. To remove animal tissue or cells, Triton X-100 was added to 10 ml of the suspension to give a final concentration of 0.01%. This sample then was vortexed for 30 s and allowed stand for 5 min at room temperature. After centrifugation at 1,450g for 3 min, the supernatant was removed and the filtered seawater containing 5 mM EDTA was added to the cell pellet. These washing steps were repeated three times (LaJeunesse 2002). Finally, each Symbiodinium pellet was subjected to bead beating homogenization and DNA extraction with a kit according to the aforementioned method, and the resultant DNA was dissolved in TE buffer.
Quantification of Symbiodinium in the coral tissues
To quantify Symbiodinium in the coral tissue using qPCR, the DNA extracts could not be used with precision because there was an expected loss of the cells during the Triton X-100 treatment, centrifugation, and/or the extraction steps. Therefore, we estimated cell numbers by classic microscopic counts, as described above, but we estimated relative clade abundances (%) among the total Symbiodinium cell numbers using qPCR. Prior to qPCR, the first screening of the existent clades was performed by conventional PCR. This was necessary to reduce the effort and cost in qPCR and to confirm the presence or absence of minor background clades. Therefore, the minimum detection level (both absolute value and relative % among the major clades) of the minor Symbiodinium clades was determined by conventional PCR. In our primary estimation using each clade of Symbiodinium cultures, amplicons from the DNA extract at a minimum of 0.1 cell (clades B and E) or 1 cell (clades A, C, D, and F) in one PCR volume could be detected reliably on an agarose gel by ethidium bromide staining. These detection levels were confirmed to be invariable, even when the sample was mixed with a 1,000 cell-equivalent of non-target clade cells. Therefore, the presence of minor background clades in the DNA extract obtained from coral tissues can be detected at a level of 0.01% for clades B and E and 0.1% for clades A, C, D, and F. In other words, clade presence below these levels was excluded in the remaining discussion.
The conventional PCR-based screening revealed that all coral specimens possessed only clade C and/or D. Therefore, qPCR for these two clades was performed next. Mixtures of 1 ng μl−1 DNA extract, 0.4 pmol μl−1 of each forward and reverse primer, and the recommended volume of TaKaRa SYBR® Premix Ex Taq™ II, with the included ROX reference dye, were analysed with the ABI PRISM 7000 as described above. For use as quantification standards, DNA extracts from Symbiodinium cultures CCMP2466 (clade C) and CCMP2556 (clade D), each corresponding to 1, 10, 100, and 1,000 cells/reaction, were loaded at every run (all performed in triplicate). Based on these culture-based standardizations, the relative percentages of each clade were estimated. To determine whether these standard cultures were representative of wild Symbiodinium in coral tissues, 106 cell-equivalent DNA extracts (without Triton X-100 treatment) comprising only wild clade C obtained from two colonies of Porites lutea were diluted to a final concentration of 100 cells/reaction (three times replicate of DNA extractions from each colony) and subjected to qPCR together with the 100 cell-equivalent DNA extract from CCMP2466 (clade C). The same comparison was performed for CCMP2556 (clade D) with wild clade D obtained from Favites adbita (four times replicate of DNA extractions from single colony). We did not encounter cases of clades A, B, E, and F in the field (see Results).
Quantification of Symbiodinium released into the trap
The trapped seawater was passed through a 20-μm sieve, and 500 ml of the filtrate was filtered again with a 0.8-μm polycarbonate filter (Advantec-Toyo). Particles smaller than 20 μm were thus trapped onto the filter, and DNA from the particles was extracted using the TE boiling method described above (Koike et al. 2007). This method has advantages over other DNA extraction protocols because even a single cell of target specimen on the filter is never lost. The extracted DNAs were initially subjected to conventional PCR using all of the clade-specific primers, and Symbiodinium clade compositions in the traps were qualitatively estimated. However, if the trap water contained a relatively small number of Symbiodinium cells, coexisting background clades occurring in small numbers might escape detection. Therefore, for the samples obtained from multi-cladal colonies (in our case, clades C and D), nested-PCR was employed. Here, a partial 28S rRNA gene region covering the clade-specific regions was primarily amplified using a clade-universal primer set of Sym-28Sf (5′-TAGCATGAAGTCARACAAG-3′) and ls1.3 M (5′-ACCGATTTGCACGTCAGTA-3′; modified from Wilcox 1998), and then the primary PCR amplicons were subjected to secondary amplification following the above-mentioned clade-specific runs. The primary PCR cycle was 1 cycle at 94°C (5 min), 10–12 cycles at 94°C (30 s), 55°C (30 s), 72°C (1 min), and 1 cycle at 72°C (7 min). qPCR was then employed using the primers for the clades detected with nested and/or conventional PCR, using culture strain-based standards. To test for possible interference due to coral exudates or other extracts (e.g., organic acids or polysaccharides in coral mucus or in other materials) from filter-trapped particles in the qPCR, 10 or 100 cell-equivalent DNAs from the culture strains were initially added to every sample as internal standards, and the extra signal increases in qPCR corresponding to these additional DNAs were monitored. When interference was suspected, the samples were diluted sufficiently to diminish interference. All quantifications from qPCR were measured in triplicate.
Estimation of surface area of the corals
To express Symbiodinium cell density on a coral surface, estimations of coral surface areas were needed. The detached coral skeletons were soaked in melted 1.5% agarose three times, which caused the surface to become coated with a thin layer of agarose. Next, 0.2% methylene blue with ethanol was poured onto the coral and allowed to drip off. The coated layer was then re-melted in 500 ml of hot water, and the resultant methylene blue solution was measured for 660 nm absorbance (maximum absorbance of methylene blue at neutral pH) using a spectrophotometer (Lightwave S2000, WPA, Cambridge, UK). As a standard linear regression, correlations between the 660 nm absorbance and the surface area were estimated using several pieces of equally treated polystyrene foam with known surface areas. In the case of encrusting corals, we did not collect the exact portions where the traps were laid; instead, the area of the trap opening (41.8 cm2) was used.
Results
PCR specificity and quantification of six clades of Symbiodinium
Table 3 lists sequences and Tm values of the qPCR primer sets designed to amplify each 28S rRNA gene of Symbiodinium clades A–F. The desired amplifications against each Symbiodinium clade were simulated on the Primer-BLAST Web site (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) using the settings described in the Materials and Methods. Mismatched regions were sometimes found in all of the searches. For example, clade A Symbiodinium in Anemonia rustica (AY074976) had a four-base difference (3 in the forward primer and 1 in the reverse) with our clade A primers, clade B in Montastraea annularis (DQ200701) had a five-base difference (all 5 in the forward primer) with our clade B primers, clade C in Amphisorus hemprichii (AJ308894) had a three-base difference (all 3 in the forward primer) with our clade C primers, clade D in Haliclona koremella (AJ308899) had a 10-base difference (6 in the forward primer and 4 in the reverse primer) with our clade D primers, and clade F of the strain Z3 culture (DQ174742) had a one-base difference (in the reverse primer) with our clade F primers. In addition, our clade B primer may inadvertently amplify Symbiodinium clades H or F, since they have only five-base substitutions at the priming site. While our primers were not perfect in all instances, they matched the designated clades in the database. Moreover, experimental confirmations were rigorously undertaken for positive PCR amplifications of a designated Symbiodinium clade and negative amplification of other clades using Symbiodinium from established cultures, and other non-Symbiodinium algae were also checked using conventional PCR and qPCR.
Figure 3 summarizes the quantification of the delta Rn curves of clades A–F with the different primer sets. In the graphs for clades A-, C-, D-, E-, and F-specific primers, the fluorescence equivalent to even one cell/reaction of the designated clade increased significantly in comparison with other 1,000 cell-equivalents of non-target clades or NTC. However, for the clade B primer, the signals from one cell/reaction were not easily differentiated from other 1,000 cell clades. Based on these results, the determination limits were ~1 cell/reaction for clades A, C, D, E, and F and ~10 cells/reaction for clade B. The relationship between Ct values (PCR cycles at which a certain delta Rn value is obtained) and a logarithmic plot of cell numbers of each Symbiodinium clade (1, 10, 100, 1,000 cells/reaction) yielded good linear correlations (Fig. 4). In these runs, linear regression fits between cell numbers of standard clades (X) and the corresponding Ct values (Y) were calculated as follows: Y = −0.777 ln X + 29.503 (clade A, r 2 = 0.961); Y = −1.038 ln X + 32.006 (clade B, r 2 = 0.999); Y = −1.249 ln X + 27.811 (clade C, r 2 = 0.995); Y = –1.111 ln X + 27.108 (clade D, r 2 = 0.999); Y = −1.310 ln X + 27.382 (clade E, r 2 = 0.992); and Y = −0.958 ln X + 31.049 (clade F, r 2 = 0.981).
Delta Rn curves versus PCR cycle in qPCR using clade-specific primer sets (clades A–F) against target clades equivalent to 1, 10, 100, and 1,000 cells/PCR and non-target clades of 1,000 cells/PCR. NTC represents a non-template control in which H2O was used. For clade A-, C-, D-, E-, and F-specific primers, the fluorescence equivalent to one cell/reaction of the target clade increased significantly in comparison with other 1,000 cell-equivalents of non-target clades. For the clade B-specific primer, the signals from one cell/reaction were not reliably differentiated from the other 1,000 cells of non-target clades. a–f Represent data for specific primers for clades A–F, respectively
Linear regression curves of Ct values (PCR cycle at which a certain delta Rn value was obtained) versus logarithmic cell numbers of each Symbiodinium clade. The standard DNA solutions equivalent to 1, 10, 100, and 1,000 cells/reaction were analysed by qPCR using clade-specific primer sets. Data are shown as the mean (circle) and standard deviations (bars) for triplicate measurements. Each Symbiodinium clade-specific primer set showed strong linear correlations. a CS-161 clade A, r 2 = 0.961, b CCMP1633 clade B, r 2 = 0.999, c CCMP2466 clade C, r 2 = 0.995, d CCMP2556 clade D, r 2 = 0.999, e CCMP421 clade E, r 2 = 0.992, f CS-156 clade F, r 2 = 0.957
qPCR application: how many Symbiodinium cells were released from corals in the natural environment?
Trap setting
Using our qPCR system, we enumerated Symbiodinium cells released from the corals. For this purpose, the original trap (Fig. 2) was set on the branches of A. muricata, A. cf. grandis, A. nobilis, and Poc. eydouxi or to the surfaces of Por. lutea, F. abdita, and C. serailia. To be sure that the trap would not lead to stress-induced Symbiodinium escape from the corals, the water inside the traps was monitored for DO concentrations and NH4-N concentrations because DO is expected to decrease and NH4-N is expected to increase when corals are under stress. We found significantly higher DO or lower NH4-N in all test cases compared to ambient water. For example, during the survey conducted on 27–28 August 2007, the trap waters from one colony of A. muricata, Poc. eydouxi, and Por. lutea contained 7.9 mg l−1, 8.7 mg l−1, and 9.4 mg l−1 of DO, respectively, all of which were above ambient DO concentrations (6.5 mg l−1 where A. muricata and Poc. eydouxi were collected and 8.5 mg l−1 for Por. lutea). The NH4-N concentrations in the trap waters around A. muricata, Poc. eydouxi, and Por. lutea were 0.62 μmol l−1, 0.08 μmol l−1, and under the detection level (0.05 μmol l−1), respectively; these values were lower than the concentration in ambient water (0.8 μmol l−1). Furthermore, comparison of the Symbiodinium densities between the trap-covered and the trap-free branches (Fig. 5) illustrated that the trap-covered branches did not show any conspicuous decrease in Symbiodinium densities (P > 0.05, Student’s t-test). From these results, we concluded that a maximum of 3 h of trap setting would not result in stress-induced Symbiodinium release from the corals, which is consistent with our previous study (Koike et al. 2007), in which we found that aquarium corals that seemingly did not suffer from stress released Symbiodinium periodically. It should also be noted that water temperature inside the traps was always equal to ambient temperature.
Comparisons of Symbiodinium cell densities between the trap-covered and the trap-free branches in Acropora muricata and Acropora nobilis colonies. None of the tested corals showed significant differences in cell density between trap-covered and trap-free branches (P > 0.05, Student’s t-test). The bars indicate standard error among the branches
Quantification of released and retained Symbiodinium from/in corals
In our field studies, corals bearing only clades C and D were found. To determine whether these “wild” clades C and D were quantifiable by qPCR based on the culture-based standardizations, we compared Ct values (PCR cycles at which a certain delta Rn value is obtained) of both wild cells extracted from corals and cultures of the same clade. The mean Ct values (± SD) from 100 cells of CCMP2466 (clade C) and wild cells from Por. lutea (harbouring only clade C) were 22.8 ± 0.3 (n = 3 extracts × 3 measurements) and 22.5 ± 0.7 (n = 2 colonies × 3 extracts × 3 measurements), respectively. These were significantly congruent with each other (P > 0.05, Student’s t-test). In the same manner, mean Ct values from 100 cells of CCMP2556 (clade D) and wild cells from F. adbita (composed only of clade D) were 22.0 ± 0.3 (n = 3 extracts × 3 measurements) and 21.7 ± 0.3 (n = 4 extracts × 3 measurements), respectively. These results suggested that culture-based quantification could be applied to wild Symbiodinium cells, at least for clades C and D. Because we encountered several instances wherein small numbers of clade D coexisted with clade C, a sample containing virtually one cell from the clade D culture (CCMP2556) and 1,000 cells from the clade C culture (CCMP2466) was prepared and subjected to clade D quantification. As a result, a Ct value (mean ± SD) of 22.7 ± 0.1 (n = 5) was obtained for the single clade D cell, and the value was congruent with that obtained from independent clade D analyses (Ct = 22.6 ± 0.3, n = 5). Based on these independent quantifications of clades C and D using culture-based standardizations, each co-existing percentage could be determined.
Table 4 lists the released cell numbers and clade compositions of Symbiodinium released to the traps and the retained cell density and clade compositions in the coral tissues. The former are represented as released cell numbers/cm2 of coral surface covered by the trap divided by the number of hours of trap setting. For the Symbiodinium densities retained in the corals, a minimum value of 1.0 × 105 cells cm−2 in Poc. eydouxi sample No. 3 (August 2007) and a maximum value of 1.3 × 106 cells cm−2 in Por. lutea sample No. 1 (August 2007) were recorded. Roughly speaking, indexes estimated from the coral colour chart corresponded to the cell densities of Symbiodinium. It should be noted that corals suffering from bleaching (as in the cases shown by low colour indexes of 2 or 3) still retained significant numbers of Symbiodinium.
In the analyses of clade compositions retained within the corals and estimated by qPCR, every colony of Poc. eydouxi (Nos. 1–6) and one colony of F. adbita showed multi-cladal (C and D) presence of Symbiodinium. However, the relative abundance (%) of clade D was low and ranged from traces (under the determination level of qPCR but feasibly detectable at ~0.1% by conventional PCR) to 2.2% in Poc. eydouxi and 1.1% in F. abdita. In these multi-cladal cases, Poc. eydouxi No. 4 exhibited trace levels of clade D release just below the confidence level using qPCR (estimated as 0.5 cell/reaction, which was below the minimum standard of 1 cell/reaction) and roughly calculated to be 1.4 cells cm−2 h−1. It was then estimated at 0.02% of the total release (the remaining 99.98% was clade C), making it much lower than that present in the tissues (clade D = 0.1%). Quantifications by qPCR failed for other multi-cladal colonies, although Poc. eydouxi Nos. 3, 5, and 6 and F. adbita showed cryptic occurrences of clade D in the trap waters after nested-PCR amplifications, whereas Poc. eydouxi Nos. 1 and 2 did not. The estimated releases reached a maximum of ~5,900 cells cm−2 h−1 from Poc. eydouxi No. 4 (August 2007) and a minimum of ~4 cells cm−2 h−1 from A. muricata No. 4 (August 2009). The ratios of released Symbiodinium to retained Symbiodinium within the corals were estimated at 0.0006–1.2%. We also collected ambient water to estimate background Symbiodinium occurrence in the water column. However, we could not detect any signals from ambient sea water. Thus, background Symbiodinium in the water column did not have deleterious effects on the estimation of Symbiodinium cell numbers in the traps.
Discussion
Properties of the qPCR developed in the present study
In the present study, we developed PCR primers to amplify and differentiate DNAs from Symbiodinium clades A–F and examined their specificities and applications using database searches and corroboration with conventional PCR and qPCR. The database searches revealed some Symbiodinium types that had sequence differences from our designated clade primers. Although most of these cases involved symbionts in sea anemones or foraminiferans and thus may be immaterial for coral applications, we did find unconformities such as clade B in the coral Montastraea annularis having five bases different from our clade B primers. Moreover, our clade D primer may not work on a peculiar type of clade D symbiont (type D1: Pochon et al. 2006; Takabayashi et al. 2004) (e.g., in Haliclona koremella) because of the 10-base difference between primers. Nevertheless, at least in our field study, coral-borne clades C and D were reliably quantified using our primers.
Another particular concern about our qPCR system is the possibility that some non-specific signal increases might have been recognized in later PCR cycles (usually above 33 cycles) due to properties of the SYBR® Green chemistry used in qPCR. Such signal increases likely would result from the trace formation of primer–dimers, which cannot be eliminated. However, in the cases of clades A, C, D, E, and F, even 1 cell/reaction was enough to be discriminated from 1,000 times higher contaminants of other clades, and we concluded that minimum detection levels for these clades were above 1 cell/reaction and at least 0.1% relative abundance among other clades. For clade B, SYBR® Green signals around such small amounts of DNA (one cell/reaction) could not be discriminated from high contaminants of other non-target DNAs, probably due to the formation of primer–dimers or the non-specific amplification of DNA. Therefore, a minimum of 10 cells/reaction and at least 1% relative abundance among other clades would be the limit for quantification. Above such ranges, and with appropriate dilutions covering the standard ranges, the quantitative results obtained by our qPCR analyses were adequately rigorous, as linear relationships between a logarithmic plot of cell numbers and Ct values were obtained for all clades (Fig. 4). It should be repeated here that prior to the qPCR analyses, we performed conventional PCR to determine the presence or absence of the clades. This was necessary to reduce the effort and cost of qPCR and to confirm the quantification result in qPCR for background clades. In this conventional PCR analysis, detection levels of background clades were set to be equal to or much more sensitive than qPCR; this means that the first screening did not exclude the qPCR-detectable clades.
qPCR trial to enumerate Symbiodinium cells released from corals in the field
To determine how many and what clades of Symbiodinium are released from corals, we needed to develop novel methodologies beyond the qPCR systems. Thus, we first developed a trap to collect Symbiodinium discharged from corals. Trapped waters were monitored for concentrations of DO and ammonium ions after collection: these samples had higher DO and lower NH4-N than ambient levels. Moreover, there were no significant differences in the Symbiodinium densities between the branches with and without the trap. From these results, we assumed that the setting of the trap did not negatively affect or promote Symbiodinium discharge.
The second methodology we developed was a technique to estimate coral surface area. This was necessary to determine the Symbiodinium density per area. A method using aluminium foil (Marsh 1970) has often been used to determine coral surface area. Hoegh-Guldberg (1988) proposed an alternative method that is more applicable for pitted surfaces and that involves coating surfaces with water-based polyurethane and methylene blue dye. In our newly developed method, we coated the coral skeleton with melted agarose gel instead of polyurethane and then stained it with methylene blue dye. Subsequent analysis for methylene blue adhesion is quite easy and involves re-melting the agarose coating in hot water. The primer coating using agar can be removed from any unwanted area for surface determination of trap-covered branch (e.g., broken parts). The amount of dye on the agarose layer of the coral skeleton then can be determined easily by spectrophotometry after liquefying the agar. Using this approach, we obtained a rather constant value of Symbiodinium density of 105 cells cm−2 in all corals except Porites. Although Symbiodinium cell density can fluctuate with season (e.g., Fagoonee et al. 1999; Fitt et al. 2000), our result was apparently lower than the previous observations made by several authors. This may be because our test corals had undergone intensive bleaching events.
The next issue that we needed to address was how to determine absolute cell numbers or relative percentages of cell presence for each clade, all of which were based on the qPCR results. Expressing this parameter as a “copy number of rRNA gene” would be more accurate but less attractive for ecological purposes. Thus, we calculated cell numbers using the formula obtained from Ct value versus cell numbers of “standard cells.” For standard cells, we used a culture strain for each clade; however, we are not certain that such strains were good representatives of wild Symbiodinium occurring in the field or of the intragenomic variations of copies of the rRNA gene. Comparisons of Ct values between the clade C standard (CCMP2466) or clade D standard (CCMP2556) and the equivalent cell numbers of wild clades C or D from corals were not significantly different. Moreover, in our qPCR system, each qPCR-based cell number was determined based on each independent cladal standard. Therefore, our culture-based standardizations should give independent levels of detection and should not be affected by differences in the rRNA gene copy numbers among the clades. Mieog et al. (2007) reported the cell average for ITS1 copies as 984 ± 109 copy cell−1 (clade C, mean ± SE from two colonies of Acropora millepora, each single colony of Acropora tenuis and Pocillopora damicornis) and 3181 ± 69 copy cell−1 (clade D, mean ± SE from two colonies of A. millepora) for wild specimens. This maximum 10% error does not seem to present an obstacle to the determination of cell number based on rRNA gene copies.
We next tried to estimate how many cells and what clades are released from corals in the field. The tested species (A. muricata, Poc. eydouxi Nos. 1, 2, Por. lutea, A. cf. grandis, C. serailia, and A. nobilis) discharged only clade C Symbiodinium at an estimated 4–655 cells h−1 cm−2 of coral surface. In the case of Poc. eydouxi No. 4, clade D in the trap water was preliminarily quantified and calculated to be 0.02% of the total discharged biomass. This value is still lower than that observed in the tissues (0.1%). Although quantifications of clade D cell numbers were not successful, Poc. eydouxi Nos. 3, 5, and 6 and F. adbita seemed to release clade D. Nonetheless, it is noteworthy that Poc. eydouxi Nos. 1, 2, and 4 apparently released clade C prior to clade D. The highest discharge was recorded in a coral specimen of Poc. eydouxi (No. 4) at ~5,900 cells cm−2 h−1; the rates of hourly discharges were calculated to be 0.0006–1.2%. During the sampling in August 2007, at which time an enormous bleaching event occurred due to extraordinarily high temperature, a high discharge rate, such as in the case of Poc. eydouxi No. 4, might possibly have led to bleaching. Titlyanov et al. (1996) reported that most of discharged Symbiodinium cells from aquarium corals were degraded. We still do not know whether the released Symbiodinium cells from corals in the field were damaged or not.
In August 2007, the low-Symbiodinium bearing colonies of Poc. eydouxi (Nos. 2 and 3; 1.0–1.2 × 105 cells cm−2) had higher clade D ratios to the Symbiodinium cells (2.1 and 2.2%) in their tissues, whereas other colonies (Nos. 1 and 4; 4.3–5.0 × 105 cells cm−2) contained only 0.2 and 0.1% of clade D, respectively. The results were not obtained from long-term observation of a single colony, but the relative increase in clade D retention would be likely if the preferential discharge of clade C was prolonged. This is consistent with the findings of other studies (e.g., that Symbiodinium clade D content can increase after a coral bleaching event) (Baker et al. 2004; Jones et al. 2008).
Symbiodinium clades or types often differ among individuals of the same species and in hosts occurring in different habitats. For example, coral specimens of Montastraea annularis and Montastraea faveolata from shallow water harboured clade A or B, whereas specimens from deep water harboured clade C (Rowan and Knowlton 1995). Furthermore, the cladal composition in these corals differed even though they occurred within a single colony (Rowan et al. 1997). In addition, multi-clades in certain corals have been reported to shift in some instances (e.g., Berkelmans and van Oppen 2006; Thornhill et al. 2006). Two possible mechanistic explanations of this phenomenon exist: symbiont switching and symbiont shuffling (Baker 2003). Symbiont switching is attributed to uptake of exogenous Symbiodinium and has been reported in sea anemones and soft corals (Kinzie et al. 2001; Lewis and Coffroth 2004). Symbiont shuffling is the propagation of “background symbionts” that occur at levels lower than the detection level. Symbiont shuffling is considered to incur a risk of death during the period of shuffling (Mieog et al. 2007). Jones et al. (2008) reported that a thermally sensitive subclade, C2, initially predominated in corals of the Great Barrier Reef, but after bleaching events, including a massive case in 2006, cladal composition shifted to clade D by symbiont shuffling. Although some of the corals with clade D symbionts are thought to be more resistant to thermal stress than those with clade C symbionts (Glynn et al. 2001; Rowan 2004), clade D Symbiodinium can be found in high latitude coral communities (Lien et al. 2007), and the deep water colonies of Montastraea franksi harboured clade D symbionts (Toller et al. 2001). Recent studies have indicated that there are several types that exist within clade D (LaJeunesse et al. 2004; Pochon et al. 2006), and when exposed to elevated temperatures, most closely related sister “types” exhibited significantly different responses (Tchernov et al. 2004). Therefore, we cannot confidently generalize that all clade D Symbiodinium show a high-light high-temperature tolerance. However, recent studies that used highly sensitive qPCR to examine coral-Symbiodinium systems have revealed that many coral hosts harbour background clade D Symbiodinium in low numbers side by side with a major clade (Correa et al. 2009; Mieog et al. 2007). These corals may therefore have the potential for symbiont shuffling. In any case, swapping of Symbiodinium clades seems to be one strategy for corals to survive environmental stress. In this study, we observed a trend of selective retention of clade D and release of clade C in Poc. eydouxi colonies. This might be interpreted as an indicator of ongoing symbiont shifting.
References
Baker AC (2001) Reef corals bleach to survive change. Nature 411:765–766
Baker AC (2003) Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst 34:661–689
Baker AC, Starger CJ, McClanahan TR, Glynn PW (2004) Corals’ adaptive response to climate change. Nature 430:741
Berkelmans R, van Oppen MJH (2006) The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc R Soc B 273:2305–2312
Bhagooli R, Hidaka M (2004) Release of zooxanthellae with intact photosynthetic activity by the coral Galaxea fascicularis in response to high temperature stress. Mar Biol 145:329–337
Bowers HA, Tengs T, Glasgow HB, Burkholder JM, Rublee PA, Oldach DW (2000) Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl Environ Microbiol 66:4641–4648
Brown BE (1997) Coral bleaching: causes and consequences. Coral Reefs 16:S129–S138
Buddemeier RW, Fautin DG (1993) Coral bleaching as an adaptive mechanism. Bioscience 43:320–326
Coffroth MA, Santos SR (2005) Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156:19–34
Correa AMS, McDonald MD, Baker AC (2009) Development of clade-specific Symbiodinium primers for quantitative PCR (qPCR) and their application to detecting clade D symbionts in Caribbean corals. Mar Biol 156:2403–2411
Dyhrman ST, Erdner D, La Du J, Galac M, Anderson DM (2006) Molecular quantification of toxic Alexandrium fundyense in the Gulf of Maine using real-time PCR. Harmful Algae 5:242–250
Fagoonee I, Wilson HB, Hassell MP, Turner JR (1999) The dynamics of zooxanthellae populations: a long-term study in the field. Science 283:843–845
Fitt WK, McFarland FK, Warner ME, Chilcoat GC (2000) Seasonal patterns of tissue biomass and densities of symbiotic dinoflagellates in reef corals and relation to coral bleaching. Limnol Oceanogr 45:677–685
Galluzzi L, Penna A, Bertozzini E, Vila M, Garcés E, Magnani M (2004) Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl Environ Microbiol 70:1199–1206
Gardner TA, Côté IM, Gill JA, Grant A, Watkinson AR (2003) Long-term region-wide declines in Caribbean corals. Science 301:958–960
Gates RD, Baghdasarian G, Muscatine L (1992) Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull 182:324–332
Glynn PW, Maté JL, Baker AC, Calderón MO (2001) Coral bleaching and mortality in Panama and Ecuador during the 1997–1998 El Niño-Southern Oscillation event: spatial/temporal patterns and comparisons with the 1982–1983 event. Bull Mar Sci 69:79–109
Hoegh-Guldberg O (1988) A method for determining the surface area of corals. Coral Reefs 7:113–116
Hoegh-Guldberg O (1999) Climate change, coral bleaching and the future of the world’s coral reefs. Mar Freshw Res 50:839–866
Hosoi-Tanabe S, Sako Y (2005) Species-specific detection and quantification of toxic marine dinoflagellates Alexandrium tamarense and A. catenella by real-time PCR assay. Mar Biotechnol 7:506–514
Jones RJ (1997) Zooxanthellae loss as a bioassay for assessing stress in corals. Mar Ecol Prog Ser 149:163–171
Jones AM, Berkelmans R, van Oppen MJH, Mieog JC, Sinclair W (2008) A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc R Soc B 275:1359–1365
Kamikawa R, Asai J, Miyahara T, Murata K, Oyama K, Yoshimatsu S, Yoshida T, Sako Y (2006) Application of a real-time PCR assay to a comprehensive method of monitoring harmful algae. Microbes Environ 21:163–173
Kinzie RA III, Takayama M, Santos SR, Coffroth MA (2001) The adaptive bleaching hypothesis: experimental tests of critical assumptions. Biol Bull 200:51–58
Koike K, Yamashita H, Oh-Uchi A, Tamaki M, Hayashibara T (2007) A quantitative real-time PCR method for monitoring Symbiodinium in the water column. Galaxea 9:1–12
LaJeunesse TC (2002) Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar Biol 141:387–400
LaJeunesse TC, Loh WKW, van Woesik R, Hoegh-Guldberg O, Schmidt GW, Fitt WK (2003) Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol Oceanogr 48:2046–2054
LaJeunesse TC, Bhagooli R, Hidaka M, deVantier L, Done T, Schmidt GW, Fitt WK, Hoegh-Guldberg O (2004) Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar Ecol Prog Ser 284:147–161
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lewis CL, Coffroth MA (2004) The acquisition of exogenous algal symbionts by an octocoral after bleaching. Science 304:1490–1492
Lien Yi-T, Nakano Y, Plathong S, Fukami H, Wang Jih-T, Chen CA (2007) Occurrence of the putatively heat-tolerant Symbiodinium phylotype D in high-latitudinal outlying coral communities. Coral Reefs 26:35–44
Loram JE, Boonham N, O’Toole P, Trapido-Rosenthal HG, Douglas AE (2007) Molecular quantification of symbiotic dinoflagellate algae of the genus Symbiodinium. Biol Bull 212:259–268
Marsh JA (1970) Primary productivity of reef-building calcareous red algae. Ecology 51:255–263
Mieog JC, van Oppen MJH, Cantin NE, Stam WT, Olsen JL (2007) Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs 26:449–457
Perez SF, Cook CB, Brooks WR (2001) The role of symbiotic dinoflagellates in the temperature-induced bleaching response of the subtropical sea anemone Aiptasia pallida. J Exp Mar Biol Ecol 256:1–14
Pochon X, Gates RD (2010) A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol Phylogent Evol 56:492–497
Pochon X, Montoya-Burgos JI, Stadelmann B, Pawlowski J (2006) Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol Phylogenet Evol 38:20–30
Rowan R (2004) Thermal adaptation in reef coral symbionts. Nature 430:742
Rowan R, Knowlton N (1995) Intraspecific diversity and ecological zonation in coral-algal symbiosis. Proc Natl Acad Sci USA 92:2850–2853
Rowan R, Knowlton N, Baker A, Jara J (1997) Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388:265–269
Stimson J, Kinzie RA III (1991) The temporal pattern and rate of release of zooxanthellae from the reef coral Pocillopora damicornis (Linnaeus) under nitrogen-enrichment and control conditions. J Exp Mar Biol Ecol 153:63–74
Takabayashi M, Santos SR, Cook CB (2004) Mitochondrial DNA phylogeny of the symbiotic dinoflagellates (Symbiodinium, Dinophyta). J Phycol 40:160–164
Tchernov D, Gorbunov MY, de Vargas C, Yadav SN, Milligan AJ, Häggblom M, Falkowski PG (2004) Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc Natl Acad Sci USA 101:13531–13535
Thornhill DJ, LaJeunesse TC, Kemp DW, Fitt WK, Schmidt GW (2006) Multi-year, seasonal genotypic surveys of coral-algal symbioses reveal prevalent stability or post-bleaching reversion. Mar Biol 148:711–722
Titlyanov EA, Titlyanova TV, Leletkin VA, Tsukahara J, van Woesik R, Yamazato K (1996) Degradation of zooxanthellae and regulation of their density in hermatypic corals. Mar Ecol Prog Ser 139:167–178
Toller WW, Rowan R, Knowlton N (2001) Zooxanthellae of the Montastraea annularis species complex: patterns of distribution of four taxa of Symbiodinium on different reefs and across depths. Biol Bull 201:348–359
Wilcox TP (1998) Large-subunit ribosomal RNA systematics of symbiotic Dinoflagellates: morphology does not recapitulate phylogeny. Mol Phylogenet Evol 10:436–448
Acknowledgments
This work was supported by a Grant-in-Aid (No. 21310011) for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to KK. The authors express their sincere thanks to Akiko Yura, Tomo Matsuoka, and Yohei Takaya, Faculty of Applied Biological Science, Hiroshima University, for their support during this investigation. Thanks are also given to Dr. Lawrence M. Liao, Hiroshima University, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Riginos.
Rights and permissions
About this article
Cite this article
Yamashita, H., Suzuki, G., Hayashibara, T. et al. Do corals select zooxanthellae by alternative discharge?. Mar Biol 158, 87–100 (2011). https://doi.org/10.1007/s00227-010-1544-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-010-1544-z