Abstract
Increases in temperature can shorten planktonic larval durations, so that higher temperatures may reduce dispersal distances for many marine animals. To test this prediction, we first quantified how minimum time to settlement is shortened at higher temperatures for the ascidian Styela plicata. Second, using latitude as a correlate for ocean temperature and spatial genetic structure as a proxy for dispersal, we tested for a negative correlation between latitude and spatial genetic structure within populations, as measured by anonymous DNA markers. Spatial genetic structure was variable among latitudes, with significant structure at low and intermediate latitudes (high and medium temperatures) and there was no genetic structure within high-latitude (low temperature) populations. In addition, we found consistently high genetic diversity across all Australian populations, showing no evidence for recent local bottlenecks associated S. plicata’s history as an invasive species. There was, however, significant genetic differentiation between all populations indicating limited ongoing gene flow.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dispersal plays a fundamental role in population connectivity, colonisation and re-colonisation events, reproductive opportunities, gene flow and species persistence (Underwood and Fairweather 1989; Johnson and Gaines 1990; Hanski 1998; Bilton et al. 2001; Wiens 2001). For many marine species, dispersal occurs during a planktonic larval phase, and the duration of this larval period can range from minutes to months depending on the species (reviewed by Shanks et al. 2003; O’Connor et al. 2007). How exactly larval duration affects dispersal distance (i.e. the geographical distance moved from the natal area) is a topic of considerable interest and debate. Because dispersal distance cannot be directly observed (with the exception of some species with larval periods of minutes to hours), indirect methods of estimating dispersal distance are widely employed (reviewed by Levin 2006; Jones et al. 2009). For example, Shanks et al. (2003) found a rough correlation between larval duration and dispersal distance (estimated using a variety of methods) across many marine species (although this relationship is quite complex, Shanks 2009). Similarly, Siegal et al. (2003) documented a strong correlation between dispersal as estimated from genetic information (using an isolation by distance approach, see Kinlan and Gaines 2003) and larval duration. In contrast, Weersing and Toonen (2009) and Bradbury et al. (2008) concluded that any correlation between dispersal estimated from genetics and larval durations was weak. Weersing and Toonen (2009) further identified that these correlations were strongly affected by species lacking planktonic larvae.
Temperature and dispersal
These aforementioned studies have focused on the relationship between dispersal and larval duration across species; however, larval duration can vary within species as well. For example, individual larvae can be competent to settle for a prolonged period of time (Pechenik 1990; Hadfield et al. 2001), and populations can differ in mean larval durations (as estimated from otoliths, Wellington and Victor 1992; Bay et al. 2006). One factor that strongly influences larval development (within and between species) is temperature; in experimental conditions, higher temperatures increase metabolic rates and reduce larval durations across a wide spectrum of marine invertebrates and fishes (Hoegh-Guldberg and Pearse 1995; O’Connor et al. 2007). In general, increases in temperature shorten development time, and thus higher temperatures are predicted to reduce dispersal (O’Connor et al. 2007). This prediction is topical given the likelihood of climate change in the near future, but there have been few empirical tests in natural populations.
To test whether temperature affects dispersal, two studies have looked at multiple species distributed over a range of latitudes, and then examined patterns of genetic structure by latitude. The assumptions are that genetic structure and gene flow (arising from dispersal) are inversely related and that latitude and temperature are also negatively correlated. Thus, there should be greater population structure at low latitudes if dispersal is reduced at warm temperatures. For example, Bradbury et al. (2008) examined published data for marine fishes and found that higher-latitude species had longer pelagic larval durations than low-latitude fishes. This trend was accompanied by reduced genetic structure at higher latitudes (Bradbury et al. 2008). Similarly, Kelly and Eernisse (2007) found greater genetic differentiation among low-latitude chiton species when compared to high-latitude species. However, within species of chitons, there was no indication of greater population structure at high latitudes. To our knowledge, Kelly and Eernisse (2007) is the only study to date that has tested whether within-species differences in dispersal or genetic structure vary by temperature or latitude.
In this study, we tested for a correlation between temperature or latitude and dispersal both in the lab and the field using the ascidian, Styela plicata. S. plicata is a solitary, hermaphroditic ascidian commonly found at high densities in marinas and harbours. Following spawning, eggs are negatively buoyant and larvae emerge after 8+ h. These non-feeding larvae are free swimming up to a day or more before settling. This short larval duration allowed us to manipulate eggs and larvae in the lab and suggested that dispersal distances in natural populations may be short (perhaps <100 m, certainly <1 km).
Although temperature has been shown to affect larval duration in Hong Kong S. plicata (Thiyagarajan and Qian 2003), we first confirmed that increases in temperature shorten egg and larval duration in Australian S. plicata. Secondly, using latitude as a correlate for ocean temperature (Shea et al. 1992) and spatial genetic structure as a correlate for dispersal, we predicted that spatial genetic structure within populations would decrease with increasing latitude. We tested this hypothesis for populations along the east coast of Australia. The ubiquity and local high densities of S. plicata allowed us to implement a sampling scheme of replicated horizontal transects across the relatively homogeneous linear habitat provided by marinas.
Genetic structure in an introduced species
Like many ascidians in the fouling community, S. plicata has a cosmopolitan distribution and presumably has expanded its range in association with human ships (Lambert 2005; de Barros et al. 2009). The native range of S. plicata may be in southeast Asia (based on historical records and mtDNA diversity, de Barros et al. 2009) and S. plicata has been found in Australia since 1878 (Heller 1878), where it is considered an introduced species (Kott 1985; Hewitt et al. 2004; Wyatt et al. 2005). Because all ascidian larvae develop directly (they do not feed, Cloney 1982), long distance dispersal should only occur at the adult stage where the adults are attached to a floating object, with boats being frequent transport vectors (Lambert 2007). If introductions are rare and established populations largely isolated from each other, then genetic drift (associated with the introduction event or following local establishment) might lead to genetic differentiation among some populations and reduced genetic diversity relative to the native species (Dlugosch and Parker 2008). Alternatively, if introductions are common and populations well connected, then genetic differentiation would not be expected between populations (or not between the sets of populations that have recently exchanged genes) and genetic diversity should be similar across populations, or even highest in introduced populations due to admixture (Roman and Darling 2007).
Genetic surveys of cosmopolitan solitary ascidians have found a mixture of results between these two extremes (for a review of genetic surveys in invasive ascidians, see Dupont et al. 2010). For example, a study of mtDNA diversity in S. plicata found that some haplotypes were globally distributed (indicative of recent shared ancestry), whereas some haplotypes were restricted to one or a few populations (such that ongoing genetic connections are probably rare) (de Barros et al. 2009). Similar results have been reported in another solitary ascidian, Microcosmus squamiger (Rius et al. 2008), and the colonial ascidian Botryllus schlosseri (López-Legentil et al. 2006). In Styela clava, worldwide populations formed multiple genetic clusters based on microsatellite loci, indicative of multiple independent introductions (Dupont et al. 2010). For both M. squamiger and S. clava, the major genetic groupings were geographically incongruous, with genetic similarity between populations in different oceans yet divergence between some proximate populations. For S. clava and B. schlosseri, however, there was also genetic differentiation among nearby marinas, suggesting limited local gene exchange (López-Legentil et al. 2006; Dupont et al. 2009). In the present study, we examine the patterns of genetic diversity in S. plicata along the eastern coast of Australia for evidence of genetic differentiation among marinas and also for evidence of strong genetic disjunctions, such as those caused by independent introduction events.
Within individual sampling locations, we would not expect the invasion history to affect spatial autocorrelation over our sampling transects (60 m); different locations might have different alleles due to idiosyncratic invasion histories, but how these alleles segregate spatially should reflect small-scale movements of individuals within the population. There are only two scenarios in which we can envision the history of introduction impacting spatial genetic structure within populations. The first scenario is one where multiple sympatric gene pools (cryptic species) are inadvertently sampled and these taxa are spatially clumped in some populations. For example, spatial genetic autocorrelation would be increased if cryptic species were sympatric (and aggregated by species) in some locations and compared to locations with a single species or with no aggregation by species. In S. plicata, a previous survey of mtDNA haplotypes has found populations containing two divergent (3% for COI) groups of haplotypes (de Barros et al. 2009), perhaps indicative of two cryptic species and aggregation is well known for solitary ascidians (Svane and Young 1989). To investigate the possibility of cryptic species, we used methods based on patterns of linkage disequilibria (Pritchard et al. 2000) to test for sympatric cryptic taxa. A second scenario where demographic history would affect spatial genetic structure would be if some established populations differ in heritable traits that affect dispersal. If, for instance, our low-latitude individuals disperse less (at the local, within population scale) than our high-latitude populations, we might be measuring inherent genetic differences between populations (driving a spurious correlation of spatial genetic structure with latitude), not an environmentally mediated response to latitude-associated factors such as temperature. If we found strong population structure between northern and southern sampling locations (such as might be caused by introductions from different ancestral source populations), such a scenario would seem more likely than if there was no genetic structure between locations. For these reasons, broad-scale and individual-based patterns of genetic diversity were examined to infer the effect of invasion history on standing genetic variation and to investigate whether this history could bias our results with regards to spatial genetic structure.
Materials and methods
Experimental test for temperature effects on larval duration
We investigated the effects of temperature on the minimum time to settlement, examining both embryonic development and swimming time for Australian S. plicata. Reproductively mature S. plicata adults were collected from floating piers at the Manly marina in Moreton Bay, QLD, Australia, in June 2007. From dissected adult gonad tissue, sperm was collected from eight individuals and eggs from ten other individuals, according to the methods described by Galletly et al. (2007). Five per cent of the combined sperm solution was diluted with 2 L of fresh filtered seawater (FFSW) to which all the eggs were added and left to sit for 5 min at room temperature (~22°), allowing fertilisation to occur. Then, 10 mL of the mixture containing fertilised eggs was distributed among twelve Petri dishes, filled with 20 mL of FFSW and covered. Four Petri dishes were placed into each of the three treatment temperatures.
Three temperature treatments (26, 18 and 14°C) were selected based on the Australian Bureau of Meteorology’s (www.bom.gov.au) long-term average sea surface temperatures from 1961 to 1990 for each latitude class described later in this study. The Petri dishes were removed from their treatment temperature (maintained in dark, constant temperature rooms or cabinets) every thirty minutes for brief observation (for the first 21 h, then hourly thereafter). The time of the first 36 individuals to reach hatching per temperature treatment was recorded. At hatching, the 36 focal larvae were placed in individual Petri dishes for the remainder of the experiment, and the time of a settlement-response observation (Marshall et al. 2003) was recorded. Because we collected the first 36 individuals that hatched in each temperature, the data do not represent the range of larval durations that could be observed within each temperature but, instead, reflects the minimum time to settlement—a more appropriate measure for the goals of our study.
Data were analysed using SYSTAT version 11 (2005). Hatching time, swimming time and settlement time were each examined where temperature was treated as a fixed, categorical factor and its effects were tested with ANOVA.
Field collections and molecular markers
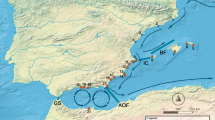
The spatial genetic structure within linear transects of S. plicata populations was estimated at three latitudes with the expectation that within-population genetic structure and latitude would be negatively correlated. S. plicata individuals were sampled in a nested design from nine marina sites within bays, along the eastern coast of Australia in 2007 (Fig. 1). The nine sampling sites were grouped into three latitude classes: low, intermediate, and high, from Queensland (QLD), New South Wales (NSW) and Victoria (VIC), respectively (Fig. 1). This hierarchical sampling scheme was selected to provide replication within the three latitude classes. Within each site, 39–67 individuals were sampled from one 60-m transect along the longest length of a floating pier (except for St. Kilda, where the transect was taken from a long static wall). The location of each individual along the transect was recorded to provide the geographic distance between pairs of individuals within each site.
Map of Australia showing the Styela plicata sampling sites. Insert a shows the east coast of Australia with the location of the three latitude classes. Major cities (grey star) and marina sites (black dot) are indicated for each latitude class: b low latitude, c intermediate latitude and d high latitude
Following collection, clean gonad tissue was dissected from each individual and stored in 100% ethanol or 64% isopropyl for DNA Extraction. DNA was extracted from gonad tissue using a modified rapid salt extraction method (Aljanabi and Martinez 1997). To quantify DNA visually, the DNA samples were electrophoresed on agarose with an H5 DNA ladder (Bioline), and then diluted to gain similar concentrations across all individuals for genetic protocols.
Microsatellite primers previously developed for Styela clava (Dupont et al. 2006) were used on the S. plicata individuals. PCR amplification of microsatellite fragments included M13-tagged primers to enable fluorescent dye labelling (Schuelke 2000). The final PCR products were pooled in pairs (2B12-FAM/1B3-HEX and 3F3-FAM/2G5-HEX), purified using a standard ethanol precipitation, diluted and the fragments separated on Mega BACE 1000 DNA-sequencer capillary system (Amersham Biosciences) with ET400 ladder mix (Amersham Biosciences). Results were viewed with Fragment Profiler software (Amersham Biosciences). There was consistent amplification for a subset of the primers (2B12, 3F3, 1B3 and 2G5) in the S. plicata individuals.
Although amplifications characteristic of microsatellites were not discernable, reliable and repeatable non-specific band amplification occurred, providing us with anonymous DNA (aDNA) markers. Dominant aDNA markers are randomly amplified fragments of nuclear DNA, for which genetic similarity is determined from the presence or absence of same-sized fragments (McDonald et al. 1996). Prospective aDNA markers (48 fragment sizes ranging from 52 to 349 bp from four primer sets) were screened for reliability across PCR amplifications. Error rates were calculated based on 8 individuals that were each re-amplified 8 times for all primer sets. The mean error rate per locus was calculated as the number of phenotypic mismatches to a reference phenotype, divided by the number of replicates (Pompanon et al. 2005), then averaged across the 8 individuals. Loci that had a mean error rate greater than 15% were discarded leaving 33 robust loci for the genetic analyses. Although AFLP’s (Vos et al. 1995) are more commonly used dominant markers than aDNA, our criteria for marker inclusion were stringent; indeed, we also screened S. plicata for AFLP’s but the amplified AFLP loci failed to meet the criteria outlined above. Therefore, our final 33 aDNA loci represent high-quality dominant markers. Whilst codominant markers such as microsatellites may be more sensitive for detecting small-scale genetic differentiation, the use of dominant aDNA markers should not bias our overall results and conclusions. For all 483 individuals, loci were scored manually for their band presence or absence.
Latitudinal differences in spatial genetic structure
To determine whether there were latitudinal differences in spatial genetic structure, geographic and genetic distance data were combined in multilocus spatial autocorrelation analyses (Smouse and Peakall 1999; Peakall and Smouse 2006) using GENALEX v6.1 (Peakall and Smouse 2006) and compared across the three latitude classes. These analyses measure the strength of correlation between genetic and geographic distance using pairwise genetic and geographic distance matrices to generate an autocorrelation coefficient (r), plotted as a function of geographic distance. A significant r value indicates a correlation between genetic and geographic distance. Following Peakall et al. (2003), we conduct two series of correlation analyses. The first estimates the correlation within discrete distance classes (5 m intervals in this case) and is representative of traditional autocorrelation analyses (with modifications on how significance is assessed), and where the correlation estimator first crosses the x-axis (i.e. r = 0) is indicative of the scale of discernable spatial population structure. The second method examines the cumulative correlation from zero (i.e. 0–5, 0–10, 0–15 m, etc.), and the first cumulative distance at which r is not significantly different from zero can also indicate the scale of spatial population structure. To test for significance, null distributions of no spatial structure were derived from permutations of the actual data and 95% confidence intervals were determined by bootstrapping (Smouse and Peakall 1999; Peakall et al. 2003).
Testing for cryptic population structure
To test for cryptic population structure within and between populations, we used the methods implemented in the program Structure (ver. 2.3, command line, Pritchard et al. 2000; Falush et al. 2003, 2007). In these methods, a number of groups (K) are assumed and the probability that each individual belongs to a given group is determined using an algorithm that seeks to minimize linkage disequilibria within each group (Pritchard et al. 2000). Models can allow for admixed individuals that have mixed genotypes from multiple groups or only allow genotypes to come from a single group (=no admixture). Our molecular markers were coded following the recommendations of Falush et al.(2007) for dominant markers, where recessive genotypes (=no band) are coded as homozygous recessive and dominant genotype (=band present) are coded as heterozygotes with the second allele as an unknown. Admixture and no admixture models were run with 50,000 steps following a burn-in of 10,000 steps for K = {1,5}, and we ran three replicates for each condition (default options were chosen for all parameters not related to recessive loci). The relative likelihood of K values was evaluated following Evanno et al. (2005), whereby the maximal ΔL is indicative of the most likely value for K.
Regional genetic patterns along the east coast of Australia
Because S. plicata is globally distributed, genetic diversity and geographic differentiation among established non-native populations can provide insights regarding the frequencies of bottlenecks (associated with colonization or within marina dynamics) versus connections (gene flow) between established populations. For each population, we estimated unbiased expected heterozygosity (assuming Hardy–Weinberg equilibrium) and the proportion of polymorphic loci, using GENALEX v6.1 (Peakall and Smouse 2006). For dominant loci, heterozygosity can be estimated at the population level, as the genotype of individuals without bands at a given locus is known; they are homozygous recessive. Genetic structuring was estimated using analyses of molecular variance (AMOVA) as implemented in GENALEX. In our AMOVA results, a significant effect of latitude (ΦRT) or between marina sites within latitude (ΦPR) would imply that genetic structuring has occurred at that level, suggesting limited gene flow (relative to genetic drift). ΦPT (analogous to FST) values were also estimated between all pairwise combinations of sites. For each AMOVA, significance was assessed with 1,000 random permutations.
To evaluate whether there was a positive relationship between geographic distance and genetic differentiation, we used the geographic distance (in kilometres) via the shortest waterway between each pair of populations (marina sites) and pairwise ΦPT, as this is the analogous measure to F-statistics (Excoffier et al. 1992). The strength and significance of the slope were determined using a Mantel test in GENALEX v6.1 (Peakall and Smouse 2006), with 1,000 permutations.
Results
Temperature effects on larval duration
Temperature significantly affected the time until hatching (F 2,105 = 123922, P < 0.0001) and larval swimming time (F 2,76 = 4.337, P = 0.016) (Fig. 2). S. plicata embryos took longer to develop in cooler temperatures, with an average hatching time of 8.0, 15.6 and 25.4 h post-fertilisation at 26°, 18° and 14, respectively. Likewise, larvae spent more time in the swimming phase at cooler temperatures with an average settlement time of 16.16, 22.1 and 28.8 h post hatching, at 26, 18 and 14°C, respectively.
Temperature effects on minimum larval duration in Styela plicata. Graph shows the average larval duration (in hours) of the first 36 S.plicata larvae (from the Manly marina site only) across different temperature treatments: 26° (dark grey), 18° (medium grey) and 14° (light grey). Larval duration is broken into two stages: embryonic development (fertilisation to hatching time) and swimming time (hatching to settlement time), each with 95% confidence error bars (not visible on embryonic development as they are too small)
Latitudinal differences in spatial genetic structure
The final data set for spatial genetic analyses comprised 483 individuals from nine sampling locations. Fifteen out of the 48 loci had a mean error rate greater than 15% and were discarded, leaving a total of 33 loci with a total mean error rate of 6% (0.7% std. error). After discarding the problematic loci, only 1.1% of the data used in the analyses that followed had missing data.
We found differences in spatial autocorrelation between the low-latitude sampling locations (QLD) and the high- latitude sampling location (VIC) with significant spatial genetic structure at the low- and intermediate latitude locations (Figs. 3, 4). For individuals that were 0-5 m apart in the low and intermediate latitude, there was significant spatial genetic structure, as the observed r was greater than the permuted null, and the bootstrapped confidence interval did not include zero. The rank order of r values for individuals up to 5 m apart were as predicted with low > intermediate > high. However, the values for low and intermediate locations were not statistically different from each other (in either standard autocorrelation or cumulative distance classes, Figs. 3, 4), and r at the intermediate latitude exceeded r at low latitude for 5–10 m and the cumulative distance classes greater than 5 m (i.e. 0–10, 0–15 m, etc.). In contrast, the high-latitude class (VIC) had no spatial genetic structure at any distance.
Latitudinal effects on spatial genetic structure in Styela plicata. Correlograms show the strength of the correlation between genetic and geographic distance, measured as a non-cumulative combined spatial correlation, r, for all individuals within each discrete distance class (in metres). Confidence intervals of 95% for the null hypothesis of randomly distributed individuals (dotted lines) and 95% confidence error bars for r as are shown. Asterisks indicate where r values are significantly different from the null distribution. Autocorrelations are displayed by latitude class: a low latitude, b intermediate latitude, and c high latitude
Spatial genetic structure across increasing distance class sizes in Styela plicata. The graph compares the strength of correlation between genetic and geographic distance measured as the cumulative combined spatial correlation, r, for all individuals within increasing distance classes (in metres). The latitude classes shown are: low latitude (dark grey), intermediate latitude (medium grey) and high latitude (light grey). Confidence intervals of 95% for the null hypothesis of a randomly distributed individuals (dotted lines) and 95% confidence error bars for r are shown. Asterisks indicate where r values are significantly different from the null distribution
Testing for cryptic population structure
In Structure analyses, ΔL was greatest for K = 2 (i.e. the difference between L(K = 1) and L(K = 2)) for both admixed and non-admixed models. All sampled populations, however, contained individuals with intermediate posterior probabilities of membership to each group (P < 0.8 to any single group). Thus, there is no compelling case for two cryptic reproductively isolated taxa. There was some evidence for geographic partitioning, with VIC populations having a greater proportion of individuals than NSW or QLD with high probabilities of group one genotypes; the frequency of individuals with posterior probabilities of membership in group 1 greater than 0.85 was 37% in VIC versus 19% in NSW and QLD (combined) for the admixed model, and 79 and 38%, respectively, for the non-admixed model. More details regarding Structure results can be found in Supplementary data.
Regional genetic patterns along the east coast of Australia
Genetic diversity was high across populations. Most loci were polymorphic in most populations with a slight trend for reduced polymorphism at high-latitude locations (Table 1). There were no private alleles observed in any population, and heterozygosity was similar across populations. The hierarchical analysis AMOVA showed that 11% of the genetic diversity was partitioned according to latitude (ΦRT = 0.106, P = 0.001), 3% was attributed to differences among marina sites within latitude (ΦPR = 0.028, P = 0.001), and the remaining 86% was contained within populations. Although there was a positive correlation between population pairwise ΦPT and geographic distance (R 2 = 0.63, P < 0.001: Fig. 5), there were essentially two (non-overlapping) sets of ΦPT values. Populations from within the same latitude class (QLD, NSW, and VIC) had fairly low ΦPT values, whereas comparisons between regions were higher. The geographically more distant QLD-VIC comparisons were not greater than neighbouring state comparisons (QLD-NSW and NSW-VIC).
Discussion
Ambient temperatures affect the rate at which larvae develop and therefore can influence larval development time (O’Connor et al. 2007). Changes in larval duration may, in turn, affect dispersal distances. In Styela plicata, we showed that increased temperatures shortened time to settlement in the lab. Our results with wild populations were also concordant with latitude affecting dispersal, as we found increased spatial genetic structure at low and intermediate (warmer) latitudes but not our high-latitude location (cold). These results demonstrate that dispersal may vary across latitudinally distributed populations within species. Whereas our observed latitudinal differences match predictions, we cannot conclusively demonstrate that differences in temperature are the cause of the cline in spatial genetic structure. Although S. plicata is cosmopolitan and presumably introduced to Australia via human activities, we found that populations are moderately differentiated, suggesting that genetic connections between established populations are infrequent. Within Australian S. plicata populations, there may be admixture between two genetically differentiated groups; however, there is no evidence for these groups being reproductively isolated. All these findings are discussed in detail below.
Larval duration and temperature
We found a significant effect of temperature on larval duration (Fig. 2), consistent with expectations and results from a previous study using S. plicata (Thiyagarajan and Qian 2003). The two studies differ in that Thiyagarajan and Qian (2003) examined the effect of temperature on average larval duration using S. plicata from Hong Kong, whereas we focused on minimal larval duration using Australian S. plicata. The minimal larval duration (in the lab) gives us an estimate of when larvae are competent to settle in the field; early settlers are most likely to contribute to spatial genetic structure. The temperatures tested in this experiment represented the average annual temperature experienced at each of the latitude classes in this study. Assuming that S. plicata breed at these average temperatures, the minimal time to settlement would be approximately 24, 38 and 53 h long at our low-, intermediate and high-latitude sampling sites, respectively. Minimal settling time might even be less than measured, as experimental larvae were provided with sterile scuffed Petri dishes, not natural habitat. Most likely, spawning occurs over a range of temperatures in each population, as the reproductive season in S. plicata may vary in length and timing within at each latitude class (Yamaguchi 1975), resulting in a range of average larval durations over the breeding season. Nonetheless, our results and those of Thiyagarajan and Qian (2003) suggest that minimum and mean larval durations could differ across latitudinally arrayed populations.
Latitudinal differences in spatial genetic structure
Genetic results were consistent with the prediction that spatial genetic structure within populations of S. plicata would differ by latitude (Figs. 3, 4). Spatial genetic structure is indicative of reduced gene flow that could result from short larval durations. The greatest spatial genetic structure and its sharp decline at the low- and intermediate latitude locations are evidence for short dispersal distances, probably resulting in many offspring settling very close to their parents. The spatial autocorrelation for the intermediate population was similar to the low-latitude populations. This may reflect a threshold of critical causative factors rather than a smooth gradient along the east coast of Australia. The absence of any spatial genetic structure in the high-latitude populations suggests that offspring do not settling near their parents, indicating average dispersal distances that exceeded our transect length of 60 m.
Because the minimal larval duration (from spawn to settlement) of S. plicata is ~1–2.5 days (Fig. 2), it is perhaps surprising that spatial genetic structure was detectable within our 60-m transect design; however, realized larval movement may be much less than that predicted from development time or swimming ability (Olson and McPherson 1987; Bingham and Young 1991). In the case of some solitary ascidians, eggs and even larvae can be retained on the benthos by mucous strings (Svane and Havenhand 1993; Marshall 2002). Long larval swimming can impact post-settlement performance for many invertebrates so that individuals that settle quickly may have greater survival (Pechenik 1990), and this post-settlement survival (or mortality) is captured by genetic estimates of dispersal. Moreover, vertical movements of larvae could exceed horizontal movements. S. plicata spawns in the late afternoon (West and Lambert 1976) or night (Yamaguchi 1975) and eggs are negatively buoyant. Once hatched (10+ h; Fig. 2), many ascidian larvae swim to darkness or shadows (reviewed by Svane and Young 1989) and there is some evidence for vertical migration in the congener Styela clava (Bourque et al. 2007). In summary, the larval biology of solitary ascidians suggests that dispersal distance might be very short, and this matches our genetic results. Whereas transects longer than 60 m may have improved our resolution on the exact spatial scale of genetic structure and would be more relevant to the species biology, many of the sampling sites did not have linear structures exceeding 60 m.
Although spatial structure is usually inferred to result from short inter-generational distances, it is also conceivable that larvae from the same spawning event disperse and travel together (i.e. gregarious larvae) or choose neighbours based on genetically controlled traits (as in Grosberg and Quinn 1986). These processes could also create spatial genetic structure and patchy distributions (which are known to occur for many ascidians, Svane and Young 1989). If interactions between larvae cause spatial genetic structure, however, there is no good a priori reason that these patterns should differ by latitude, as we observe for S. plicata.
Similar spatial autocorrelation methods to those used here have been applied to other sessile marine invertebrates and revealed significant genetic structure over short distances for corals (within 10 m: Miller and Ayre 2008; within 100 m: Underwood et al. 2007), sponges (within 10 m: Blanquer et al. 2009; within 0.5 m: Calderon et al. 2007), and the ascidian Botryllus schlosseri (within 5 m: Yund and O’Neil 2000). The frequent observation of spatial genetic structure over short distances highlights the utility of spatial autocorrelation methods for evaluating dispersal among continuously distributed marine animals. Our results, furthermore, demonstrate that this type of method can be applied to testing specific hypotheses about dispersal.
To our knowledge, our study represents the first demonstration that genetic structure differs by latitude among populations of an animal with planktonic larvae. We can demonstrate statistical differences, however, only between the high-latitude location and the other two locations (medium and low latitudes). Because our sampling design only included three latitude groups, the difference is consistent with expectations, yet other factors affecting dispersal (or gene flow) may vary along the Australian coast. If dispersal-affecting factors covary along a coastline, then any study surveying a single coastline, for example, the present study (and that of Kelly and Eernisse 2007), will not be able to disentangle these potential causative factors. Future studies will be required to verify the generality of these results, particularly including surveys along multiple coastlines. Such investigations will help establish (or reject) a link between dispersal and temperature.
Regional genetic patterns in an introduced species
Our results for Australian S. plicata are intermediate between the extreme scenarios expected for introduced species, namely high differentiation due to multiple bottlenecks versus panmixia due to frequent genetic exchange. We found consistent levels of genetic diversity among populations and no private alleles (i.e. alleles restricted to a single population, Table 1). This widespread homogeneity indicates that all populations had a similar pool of ancestral alleles. Similar levels of genetic diversity are consistent either with a lack of bottleneck at introduction or substantial post-introduction gene flow between multiple populations (a comparison to native populations could be informative, but we do not have such information). Widespread post-introduction gene exchange seems unlikely, however, we did detect substantial population structure at the scale of latitude (over 1,853 km: ΦRT = 0.11; P = 0.001) and among marina sites within latitude (from 3 to 138 km: ΦPR = 0.028; P = 0.001). Although these results suggest that genetic exchange between marina populations has been infrequent, the greater similarity of proximate marinas (Fig. 5) could result from local recreational boat traffic acting as occasional transport vectors. Large commercial harbours were not sampled in our study and thus their role as “transport hubs” (Floerl et al. 2009) cannot be evaluated. Our results are similar to surveys of Styela clava and Botryllus schlosseri, where significant genetic structure was found between populations along a coastline (López-Legentil et al. 2006; Dupont et al. 2009). However, in our results, there is a correlation between geographic distance and genetic differentiation (Fig. 5), whereas there was no correlation for S. clava (Dupont et al. 2009). There was no indication of a substantial genetic disjunction, as would be expected with multiple independent introductions, but without sampling in the native species range, all conclusions must be tentative.
Invasion history is unlikely to have biased our estimates of spatial genetic structure. Although results from the program Structure indicated two groups, these groups were present in all populations and individuals with ambiguous probabilities of membership to each group were common. Thus, there is no indication of two reproductively isolated taxa that could potentially aggregate differently in our sampled populations. Whereas we did find significant geographic differentiation among latitude sites (Fig. 5 and Structure results), alleles were shared among populations so that inherent genetic differences in dispersal ability are unlikely.
The utility of introduced species for studying temperature and dispersal
To what extent can the results from an invasive species sampled from artificial habitats be applied to understanding population dynamics in natural populations? The case for invasive species as models for studying ecological and evolutionary processes has been argued elsewhere (Wares et al. 2005; Sax et al. 2007). For trying to evaluate factors influencing dispersal, cosmopolitan species of fouling communities have additional useful properties. First, artificial substrates (pontoons, piers, pylons, etc.) are also globally distributed so that habitat substrate can be held constant. Second, because of the limited water circulation in marinas and many harbours, larval movements should be better correlated to swimming ability and development time, and less idiosyncratic due to local water circulation patterns. Third, the widespread species distributions potentially allow sampling strategies that reduce confounding environmental factors. For example, sampling sites along multiple coastlines would disentangle latitude and temperature effects from coastline effects. Finally, if non-indigenous species are often genetically homogeneous, then each population would have the capacity to respond in a similar manner (phenotypically or via selection at the population level) to the environmental factor of interest and historical contingency would be less of a confounding factor when compared to native species.
References
Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acid Res 25:4692–4693
Bay LK, Buechler K, Gagliano M, Caley MJ (2006) Intraspecific variation in the pelagic larval duration of tropical reef fishes. J Fish Biol 68:1206–1214
Bilton DT, Freeland JR, Okamura B (2001) Dispersal in freshwater invertebrates. Ann Rev Ecol Syst 32:159–181
Bingham BL, Young CM (1991) Larval behavior of the ascidian Ecteinascidia turbinata Herdman—an in situ experimental study of the effects of swimming on dispersal. J Exp Mar Biol Ecol 145:189–204
Blanquer A, Uriz MJ, Caujape-Castells J (2009) Small-scale spatial genetic structure in Scopalina lophyropoda, an encrusting sponge with philopatric larval dispersal and frequent fission and fusion events. Mar Ecol Prog Ser 380:95–102
Bourque D, Davidson J, MacNair NG, Arsenault G, LeBlanc AR, Landry T, Miron G (2007) Reproduction and early life history of an invasive ascidian Styela clava Herdman in Prince Edward Island, Canada. J Exp Mar Biol Ecol 342:78–84
Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE (2008) Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proc R Soc Biol Sci Ser B 275:1803–1809
Calderon I, Ortega N, Duran S, Becerro M, Pascual M, Turon X (2007) Finding the relevant scale: clonality and genetic structure in a marine invertebrate (Crambe crambe, Porifera). Mol Ecol 16:1799–1810
Cloney RA (1982) Ascidian larvae and the events of metamorphosis. Am Zool 22:817–826
de Barros RC, da Rocha RM, Pie MR (2009) Human-mediated global dispersion of Styela plicata (Tunicata, Ascidiacea). Aquat Inv 4:45–57
Dlugosch KM, Parker IM (2008) Founding event in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Dupont L, Viard F, Bishop JDD (2006) Isolation and characterization of twelve polymorphic microsatellite markers for the invasive ascidian Styela clava (Tunicata). Mol Ecol Notes 6:101–103
Dupont L, Viard F, Dowell MJ, Wood C, Bishop JDD (2009) Fine- and regional-scale genetic structure of the exotic ascidian Styela clava (Tunicata) in southwest England, 50 years after its introduction. Mol Ecol 18:442–453
Dupont L, Viard F, Davis MH, Nishikawa T, Bishop JDD (2010) Pathways of spread of the introduced ascidian Styela clava (Tunicata) in Northern Europe, as revealed by microsatellite markers. Biol Invasions. doi:10.1007/s10530-009-9676-0
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Falush D, Stephens M, Pritchard J (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Floerl O, Inglis GJ, Dey K, Smith A (2009) The importance of transport hubs in stepping-stone invasions. J Appl Ecol 46:37–45
Galletly BC, Blows MW, Marshall DJ (2007) Genetic mechanisms of pollution resistance in a marine invertebrate. Ecol Appl 17:2290–2297
Grosberg RK, Quinn RJ (1986) The genetic control and consequences of kin recognition by the larvae of a colonial marine invertebrate. Nature 322:456–459
Hadfield MG, Carpizo-Ituarte EJ, del Carmen K, Nedved BT (2001) Metamorphic competence, a major adaptive convergence in marine invertebrate larvae. Am Zool 41:1123–1131
Hanski I (1998) Metapopulation dynamics. Nature 396:41–49
Heller C (1878) Beitrage zur nahern Kenntnis der Tunicaten. Sitzber Akad Wiss Wien 77:2–28
Hewitt CL et al (2004) Introduced and cryptogenic species in Port Phillip Bay, Victoria, Australia. Mar Biol 144:183–202
Hoegh-Guldberg O, Pearse JS (1995) Temperature, food availability, and the development of marine invertebrate larvae. Am Zool 35:415–425
Johnson ML, Gaines MS (1990) Evolution of dispersal—theoretical models and empirical tests using birds and mammals. Ann Rev Ecol Syst 21:449–480
Jones GP, Almany GR, Russ GR, Sale PF, Steneck RS, van Oppen MJH, Willis BL (2009) Larval retention and connectivity among populations of corals and reef fishes: history, advances and challenges. Coral Reefs 28:307–325
Kelly RP, Eernisse DJ (2007) Southern hospitality: a latitudinal gradient in gene flow in the marine environment. Evolution 61:700–707
Kinlan B, Gaines SD (2003) Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology 84:2007–2020
Kott P (1985) The Australian Ascidiacea—Phlebobranchia and Stolidobranchia. Mem Queenl Mus 23:1–440
Lambert G (2005) Ecology and natural history of the protochordates. Can J Zool 83:34–50
Lambert G (2007) Invasive sea squirts: a growing global problem. J Exp Mar Biol Ecol 342:3–4
Levin LA (2006) Recent progress in understanding larval dispersal: new directions and digressions. Int Comp Biol 46:282–297
López-Legentil S, Turon X, Planes S (2006) Genetic structure of the star sea squirt, Botryllus schlosseri, introduced in southern European harbours. Mol Ecol 15:3957–3967
Marshall DJ (2002) In situ measures of spawning synchrony and fertilization success in an intertidal, free-spawning invertebrate. Mar Ecol Prog Ser 236:113–119
Marshall DJ, Pechenik JA, Keough MJ (2003) Larval activity levels and delayed metamorphosis affect post-larval performance in the colonial, ascidian Diplosoma listerianum. Mar Ecol Prog Ser 246:153–162
McDonald JH, Verrelli BC, Geyer LB (1996) Lack of geographic variation in anonymous nuclear polymorphisms in the American oyster, Crassostrea virginica. Mol Biol Evol 13:1114–1118
Miller KJ, Ayre DJ (2008) Population structure is not a simple function of reproductive mode and larval type: insights from tropical corals. J Anim Ecol 77:713–724
O’Connor MI, Bruno JF, Gaines SD, Halpern BS, Lester SE, Kinlan BP, Weiss JM (2007) Temperature control of larval dispersal and the implications for marine ecology, evolution and conservation. Proc Natl Acad Sci USA 104:1266–1271
Olson RR, McPherson R (1987) Potential vs. realized larval dispersal: fish predation on larvae of the ascidian Lissoclinum patella (Gottschaldt). J Exp Mar Biol Ecol 110:245–256
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57:1182–1195
Pechenik JA (1990) Delayed metamorphosis by larvae of benthic marine invertebrates: does it occur: is there a price to pay? Ophelia 32:63–94
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Gen 6:847–859
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rius M, Pascual M, Turon X (2008) Phylogeography of the widespread marine invader Microcosmus squamiger (Ascidiacea) reveals high genetic diversity of introduced populations and non-independent colonizations. Divers Distrib 14:818–828
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Tr Ecol Evol 22:454–464
Sax DF, Stachowicz JJ, Brown JH, Bruno JF, Dawson MN, Gaines SD, Grosberg RK, HastingS A, Holt RD, Mayfield MM, O’Connor MI, Rice WR (2007) Ecological and evolutionary insights from species invasions. Tr Ecol Evol 22:465–471
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotech 18:233–234
Shanks AL (2009) Pelagic larval duration and dispersal distance revisited. Biol Bull 216:373–385
Shanks AL, Grantham BA, Carr MH (2003) Propagule dispersal distance and the size and spacing of marine reserves. Ecol App 13:S159–S169
Shea DJ, Trenberth KE, Reynolds RW (1992) A global monthly sea-surface temperature climatology. J Clim 5:987–1001
Siegal D, Kinlan BP, Gaylord B, Gaines SD (2003) Lagrangian descriptions of marine larval dispersion. Mar Ecol Prog Ser 260:83–96
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573
Svane IB, Havenhand JN (1993) Spawning and dispersal in Ciona intestinalis (L.). Mar Ecol 14:53–66
Svane IB, Young CM (1989) The ecology and behaviour of ascidian larvae. Oceangr Mar Biol Ann Rev 27:45–90
Thiyagarajan V, Qian PY (2003) Effect of temperature, salinity and delayed attachment on development of the solitary ascidian Styela plicata (Lesueur). J Exp Mar Biol Ecol 290:133–146
Underwood AJ, Fairweather PG (1989) Supply-side ecology and benthic marine assemblages. Tr Ecol Evol 4:16–20
Underwood JN, Smith LD, Van Oppen MJH, Gilmour JP (2007) Multiple scales of genetic connectivity in a brooding coral on isolated reefs following catastrophic bleaching. Mol Ecol 16:771–784
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acid Res 23:4407–4414
Wares JP, Hughes AR, Grosberg R (2005) Mechanisms that drive evolutionary change: insights from species introductions and invasions. In: Sax D, Stachowicz J, Gaines SD (eds) Species invasions: insights into ecology, evolution and biogeography. Sinauer, Sunderland
Weersing K, Toonen RJ (2009) Population genetics, larval dispersal, and connectivity in marine systems. Mar Ecol Prog Ser 393:1–12
Wellington GM, Victor BC (1992) Regional differences in duration of the planktonic larval stage of reef fishes in the eastern Pacific Ocean. Mar Biol 113:491–498
West AB, Lambert CC (1976) Control of spawning in the tunicate Styela plicata by variations in a natural light regime. J Exp Zool 195:263–270
Wiens JA (2001) The landscape context of dispersal. In: Clobert J, Danchin E, Dhondt AA, Nichols JD (eds) Dispersal. Oxford University Press, New York
Wyatt ASJ, Hewitt CL, Walker DI, Ward TJ (2005) Marine introductions in the Shark Bay World Heritage Property, Western Australia: a preliminary assessment. Divers Distrib 11:33–44
Yamaguchi M (1975) Growth and reproductive cycles of marine fouling ascidians Ciona intestinalis, Styela plicata, Botrylloides violaceus, and Leptoclinum mitsukurii at Aburatsubo-Moroiso Inlet (Central Japan). Mar Biol 29:253–259
Yund PO, O’Neil PG (2000) Microgeographic genetic differentiation in a colonial ascidian (Botryllus schlosseri) population. Mar Biol 137:583–588
Acknowledgments
We are grateful to J. Wong and J. Torkkola for help with collecting samples in the field. Also, thanks to S. FitzGibbon, S. O. Polsky and three anonymous reviewers for valuable comments. The research undertaken in this study was funded by Australian Greenhouse Office (AGO) and Australian Research Council (ARC). However, the views herein may not reflect those of the AGO or ARC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Reusch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
David, G.K., Marshall, D.J. & Riginos, C. Latitudinal variability in spatial genetic structure in the invasive ascidian, Styela plicata . Mar Biol 157, 1955–1965 (2010). https://doi.org/10.1007/s00227-010-1464-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-010-1464-y