Abstract
Chitinases are involved in defense against chitinaceous pathogens in both invertebrates and vertebrates. This study investigated whether sea fan corals, Gorgonia ventalina (Linnaeus) collected from the Florida Keys between the summer of 2002 and the summer of 2005 contain chitinases, and whether these enzymes could serve an analogous protective role against the fungal pathogen, Aspergillus sydowii (Thom et Church). Crude extracts of healthy sea fans contained detectible levels of exochitinase activity in an in vitro microplate assay using fluorogenic substrates. The exochitinase levels decreased upon injury, agitation, or manipulation of the tissue. A concurrent, transient increase of exochitinase in the surrounding water suggests that sea fans release chitinases as a response to these stresses. By contrast, endochitinase was detected in only 2 of 15 sea fans (13%), suggesting a high degree of variation for this enzyme. Sea fan chitinase-containing seawater and anion exchange chromatography fractions were both active against A. sydowii in an absorbance-based antifungal assay. The presence of chitinases in sea fan extracts, their release into the surrounding water upon stress, and their activity against A. sydowii suggests that further study of these enzymes in coral stress responses is warranted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Like other invertebrates, corals respond to potential pathogens and physical stress through innate defensive mechanisms such as physical barriers, shedding tissue (Nagelkerken et al. 1997; Petes et al. 2003; Alker et al. 2004), phagocytosis (Olano and Bigger 2000), and a battery of secondary metabolites (Jensen et al. 1996; Kim et al. 2000a, b). Coral colonies are made up of thousands of individual, physiologically integrated coral polyps that live in obligate symbiosis with carbon-fixing algae (zooxanthellae). In addition, they are encased in a dynamic, microbe-filled mucus layer that also contributes to the protection of the coral and influences nutrient intake (Knowlton and Rohwer 2003; Rohwer 2002; Ritchie and Smith 2004; Brown and Bythell 2005; Gil-Agudelo et al. 2006).
An outbreak of lesions and mortality in the Caribbean sea fan, Gorgonia ventalina, beginning in the mid 1990s, was found to be caused by the common terrestrial fungus Aspergillus sydowii (Smith et al. 1996; Nagelkerken et al. 1997; Geiser et al. 1998). Since several species of Aspergillus had been previously identified as opportunistic pathogens of many animals, including immune-compromised humans (Dixon and Walsh 1992; Shibuya et al. 1999), its involvement in a marine invertebrate disease raised questions about the immune status of sea fans, their mechanisms of defense against fungal pathogens, and how they compare to other invertebrates and vertebrates in their responses. In well-studied terrestrial plants and insects, response to fungal pathogens involves a myriad of complex constitutive and inducible components (Levin 1976; Gillespie et al. 1997; Agrawal et al. 1999; Berenbaum and Zangerl 1999). Studies of several plant and insect immune components have formed the basis for the targeted search for immunity related factors in non-model invertebrates such as sea fan corals.
Because chitin is a major component of fungal cell walls, chitin-degrading and modifying enzymes are among the better-characterized defense mechanisms against fungal pathogens (reviewed by Flach et al. 1992; Kramer and Muthukrishnan 1997; Tuzun and Bent 1999; Ng 2004). In chitin-containing organisms such as insects, chitin-modifying enzymes, normally involved in growth and morphological control, can also respond to fungal challenge (Kramer and Muthukrishnan 1997; Merzendorfer and Zimoch 2003). In organisms that do not contain chitin, such as plants and mammals, chitinases have been demonstrated to be involved in the digestion of chitinaceous food or pathogens and can be elevated in response to fungal infection (reviewed in Flach et al. 1992; Overdijk et al. 1999; Suzuki et al. 2001; Nagy et al. 2004). Although chitinases were shown to be widely distributed in marine invertebrates over three decades ago (Elyakova 1972), hydroids are the species most closely related to sea fans in which chitinases have undergone detailed analysis (Klug et al. 1984). Based on gene expression patterns, the Hydractinia echinata chitinase appears to be involved in both pattern formation and immunity in this chitinaceous organism (Mali et al. 2004).
In this study, we assayed extracts from the common sea fan, Gorgonia ventalina for chitinolytic activities and evaluated whether these chitinases are involved in antifungal defense in this coral. In testing the hypothesis that chitinase is an induced response to fungal infection, we also explored whether chitinase is primarily retained in tissues or released from sea fans and how other stressors, such as injury and agitation, influence this release.
Materials and methods
Collection of infected sea fans
Samples (∼5 cm × 8 cm) were cut from six sea fans (Gorgonia ventalina, Linnaeus) in the Florida Keys in the summer of 2002, both near, within 2 cm (diseased tissue), and 10 cm away (healthy tissue) from the margin of what was presumed to be an aspergillotic lesion (dark, purple regions surrounding necrotic tissue, Nagelkerken et al. 1997). The tissue was immediately frozen at −20°C, shipped to Cornell University on dry ice, and stored at −20°C until extracts were prepared for analysis.
Crude coral extraction
Frozen samples from field-infected or experimentally manipulated sea fans (details below) were blotted dry and a ∼1 cm × 1 cm piece was ground in liquid nitrogen with a mortar and pestle to a fine powder and transferred to a 15-ml tube on ice. Extraction buffer (100 mM sodium acetate, 0.1% SDS, 0.1% Triton X-100, 10 mM EDTA, 10 mM β-mercaptoethanol) was added to bring the sample to a concentration of 200 mg ml−1 and the samples were left on ice for 1 h. The extract was then removed from the settled coral skeleton and other particulates and frozen at −80°C. Protein concentration of each extract was determined by Biorad DC Protein assay with bovine gamma globulin as a standard.
Chitinase assay
Crude extracts were tested for their ability to hydrolyze two chitin-containing substrates that distinguish between exochitinase and endochitinase activity. Crude sea fan extracts were diluted 2,000-fold while the captured seawater and Tris buffer samples (described below) were left undiluted for fluorogenic chitinase activity assays (modified from Tronsmo and Harman 1993). Thirty microliter of each sample was combined with 30 μl of the fluorogenic substrates for exochitinase (0.2 mM 4-methylumbelliferyl N-acetyl-β-d-glucosaminide, Sigma-Aldrich) or endochitinase (4-methylumbelliferyl N,N′-diacetyl-β-d-chitobioside, Sigma-Aldrich) in Extraction buffer in a black bottom, 96-well microtiter plate (NUNC). We defined exochitinase activity as the cleavage of terminal non-reducing ends of chitobiose, and endochitinase activity as random cleavage within chitin polymers. Plates were incubated for 30 min at 37°C and the reaction was terminated by addition of 30 μl of 0.5 M Na2CO3. Fluorescence of the liberated methylumbelliferone (MU) was detected on a PerkinElmer Fusion plate reader (excitation 365 nm, emission 460 nm) and compared with a dilution series of free MU (Sigma-Aldrich) in 0.5 M Na2CO3 to determine nanomolar concentrations. Chitinase activity of extracts was compared to 1 μg jack bean β-N-acetylhexsaminidase (β-Hex, Sigma-Aldrich) and 2.5 μg Streptomyces griseus chitinase (Sigma-Aldrich). Each extract was assayed at least three times for mean values. We confirmed that we were detecting enzymatic activity by boiling the crude extracts or commercially available enzymes for 10 min and showing that the activity disappeared.
Aquarium inoculation experiments
Six (∼5 cm × 8 cm) pieces of 15 healthy sea fans (Gorgonia ventalina) were collected on a patch reef near Looe Key, Florida USA (N 24° 34.171′, W 81° 22.914′) in October 2004, and transported in seawater filled bags in a cooler to Mote Marine Laboratory, Tropical Research Lab in Summerland Key. One piece of each fan was processed immediately to determine initial baseline levels of activity and the remainder were suspended in randomly chosen positions in 38-liter seawater tanks under shade cloth to remove 80% of the natural light. The tanks were cooled from outside by running seawater at ambient temperature (∼27°C). After 2 days of acclimation, two ∼3-cm slits were cut into each fan piece to hold a strip of Yeast Peptone Dextrose (YPD) agar infused gauze (1) alone (Control); (2) with Aspergillus sydowii, Florida Keys isolate 1 (FK1) from Garriet Smith (Fungus); or (3) with a small piece of Millepora sp. (Fire Coral). The gauze was prepared by immersing the strips in partially solidified YPD agar in a petri dish, inoculating the solidified plate with 100 μl of a 2.5 × 107 spore ml−1 of the fungus, and allowing the fungal hyphae to grow a lawn across the gauze strips (3 days at 30°C). One control and one fungus-exposed piece from each sea fan were processed after 2 days of treatment. A separate control, fungus, and fire coral-exposed piece from each fan was processed after 8 days of treatment. All sub-samples for chitinase analysis were frozen immediately in liquid nitrogen, shipped to Cornell on dry ice, and stored at −80°C.
Cut sea fan experiment
Sea fan pieces (∼5 cm × 8 cm) collected on a patch reef near Looe Key, Florida, USA in July 2004 were shipped to Cornell and acclimated in an artificial seawater (ASW) aquaculture system for at least one month (salinity 35, 27°C, 14 h light: 10 h dark cycle). A 1-cm circle was cut from three of these sea fan pieces and immediately frozen in liquid nitrogen for extraction. The sea fan pieces were then cut in half, placed in 5-l tanks of ASW under 55 Watt compact fluorescence lighting with gentle agitation of the water, and another 1-cm diameter sample taken from each piece after 4 days. The pre and post-cutting samples were extracted and assayed for chitinolytic activity as described above.
Capturing released sea fan chitinases
Sea fan pieces (∼3 cm × 6 cm) collected near Looe Key, Florida, USA in July 2004 were shipped to Cornell and acclimated to an ASW aquaculture system for at least 1 month. A 1-cm diameter circle was cut from four of these aquarium-acclimated sea fan pieces for crude protein extracts as described above. These sub-sampled sea fan pieces as well as three acclimated sea fan pieces that were not sub-sampled were then placed in presoaked pieces of 50 mm (flat width) dialysis tubing (MWCO 6000-8000), with ∼2 ml of ASW, and a 500-μl water sample taken for analysis. The sea fan pieces were maintained in the dialysis tubing in 5-l tanks of ASW under 55 Watt compact fluorescence lighting with gentle agitation of the water. Water samples (500 μl) were removed at 1, 24, and 48 h. The volume inside the dialysis tubing naturally equilibrated to ∼2 ml during the experiment.
Anion exchange chromatography
Two sea fan pieces (∼3 cm × 6 cm) that had acclimated to the ASW aquaculture system for 2 months were placed in presoaked pieces of 50 mm (flat width) dialysis tubing (MWCO 6000-8000) and submerged in 200 ml 25 mM Tris–HCL, pH 8.0. After 2 h at 25°C, the buffer inside the tubing (∼2 ml) was collected and refrigerated overnight. Four hundred microliters were loaded onto a Vivapure Q microcentrifuge column (Vivascience) and washed twice with 400 μl 25 mM Tris–HCL, pH 8. Bound molecules were eluted with 200 μl 0.5 M NaCl in 25 mM Tris–HCL, pH 8.0 then with 200 μl 1.0 M NaCl in 25 mM Tris–HCL, pH 8.0. Because they were essentially identical, chitinase and antifungal activity values (see below) of each anion exchange column fraction from the two sea fans were averaged for subsequent analysis. Cation exchange columns (Vivapure S columns from Vivascience) were also tested, but failed to fractionate chitinase or antifungal activity and the data therefore are not presented.
Antifungal assay
Antifungal activity was measured against the Florida Keys isolate 1 (FK1) of Aspergillus sydowii following methods modified from Thevissen et al. (2000). Ninety microliter of spore solution (105 spores ml−1 in half concentration yeast peptone glucose broth) or half concentration yeast peptone glucose broth alone was added to each well of a 96-well plate and the spores allowed to germinate for 18 h at 28°C. Ten microliter of each sample to be tested was added to each well and fungal growth monitored by absorbance at 595 nm on a Thermo Electron Multiskan Spectrum plate reader at 0, 24, and 48 h after addition. The initial absorbance (time 0) for each well was subtracted from the 48-h value to determine the increase in absorbance. The percent inhibition of growth was calculated as (1 − increase in absorbance/mean increase in absorbance of fungus alone) × 100. The mean percent inhibition by Extraction buffer (crude extract samples), seawater (capture experiment), Tris buffer (anion exchange load, flow-through and washes), Tris buffer with 0.5 or 1.0 M NaCl (Anion exchange elutions) alone was subtracted to give the values presented. Jack Bean β-N-acetylhexsaminidase (β-Hex, Sigma-Aldrich) and the known antimycotic, Hygromycin B (Sigma-Aldrich) were included as controls.
Data analysis
The data were tested for normality (Shapiro–Wilks test) and homogeneity of variances (Levene test) and Box–Cox transformed when necessary to allow the use of parametric statistics. In one case, the antifungal assay with the anion exchange chromatography fractions, transformation of the data did not produce a normal distribution. Because a one-way ANOVA gave a very low P value (<0.0001), the Shapiro–Wilks test is a conservative measure, and the data were homogeneous, we accepted the parametric statistics for this data set. Our data were analyzed with Statistical Analysis System (SAS version 8e, Copyright © 2001 SAS Institute Inc., Cary, NC, USA) and JMP (version 5.1.2, Copyright © 2004 SAS Institute Inc.).
Results
Demonstration of chitinolytic enzymes in sea fan extracts
Tissue samples from naturally infected Gorgonia ventalina contained measurable levels of exochitinase (Fig. 1). There was, however, no significant difference in this type of activity between samples taken from diseased tissue (2 cm from lesion margin) and healthy tissue (10 cm from lesion margin) (two-way ANOVA F = 0.0982, P = 0.7581). We found little or no endochitinase activity in either tissue from these six fans (Fig. 1). The crude extracts were compared to commercially available exochitinase (jack bean β-N-acetylhexsaminidase, β-Hex) and endochitinase (Streptomyces griseus chitinase) to verify the specificity of the substrates. Boiling of crude extracts or commercially available enzymes completely eliminated chitinolytic activity, confirming that we were detecting enzymatic activity (data not shown).
Gorgonia ventalina. Chitinase activity of field-collected sea fan extracts. Tissues from field collected visible lesions (Diseased, n = 6) and adjacent unaffected regions (Healthy, n = 6) were tested for ability to hydrolyze the fluorogenic substrates for exochitinase (4-methylumbelliferyl N-acetyl-β-d-glucosaminide) or endochitinase (4-methylumbelliferyl N,N′-diacetyl-β-d-chitobioside). Jack Bean β-N-acetylhexsaminidase (β-Hex, exochitinase, 1 μg) and Streptomyces griseus Chitinase (endochitinase, 2.5 μg) were included for comparison. Mean activity expressed as methylumbelliferone (MU) released mg protein−1 ± SE
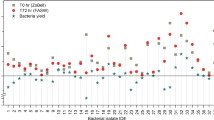
Analysis of a larger set of healthy sea fans collected for experimental infections with Aspergillus sydowii (Florida Keys isolate 1-FK1) revealed striking differences between the two types of chitinolytic activities. Initial levels of exochitinase were comparatively consistent for the 15 sea fans sampled while endochitinase was highly variable, with only two of the fans showing initial endochitinase activity well above background levels (Fig. 2). Changes in both types of activity were monitored through an 8-days fungal inoculation experiment, however, the low prevalence of sea fans with endochitinase made it impossible to draw statistically relevant comparisons between treatments for this activity (data not shown).
The low variability of exochitinase, by contrast, allowed comparison of the effect of 2 or 8 days of fungal exposure or 8 days of fire coral exposure to the minimal manipulation of controls (Fig. 3). Although there was no increase in exochitinase activity in the fungus-inoculated samples compared to the controls at either 2 or 8 days (treatment effect in three-way ANOVA F = 0.36, P = 0.5512), there was a significant decrease in exochitinase activity at 2 days in both the controls and fungus-inoculated samples (time effect in three-way ANOVA F = 39.86, P ≤ 0.0001). The control and fungus-exposed sea fan pieces left for 8 days before processing had exochitinase activity similar to initial levels but the exochitinase activity of pieces exposed to fire coral for 8 days (Millepora sp.) remained lower (Fig. 2, Tukey–Kramer post-hoc test P ≤ 0.0001). A similar decrease in exochitinase activity was induced by cutting aquarium-acclimated (for several months) sea fan pieces (n = 6, time effect in three-way ANOVA F = 63.28, P ≤ 0.0001). The mean exochitinase activity, mg protein−1, prior to cutting, 9.2705 ± 1.8588, decreased to 0.9316 ± 0.1664, 4 days after cutting.
Gorgonia ventalina. Time course of exochitinase activity of sea fan tissue exposed to Aspergillus sydowii or fire coral. Six clonal replicate pieces were cut from each of 15 healthy sea fans. One piece of each fan was processed immediately (Initials, presented individually in Fig. 2), two pieces were exposed to control gauze only (for 2 or 8 days), two pieces were exposed to gauze with fungus (for 2 or 8 days), and one piece was exposed to gauze with fire coral (for 8 days). Mean exochitinase activity mg protein−1 of three duplicate assays of all 15 sea fan extracts from each treatment ± SE
Capture of chitinolytic enzymes released by sea fans
Because our monitoring of exochitinase suggested a transient decrease in activity in the tissue when sea fan pieces were cut and manipulated (Fig. 3), we investigated whether there was a corresponding increase of activity in the surrounding seawater. Exochitinase activity increased transiently in the water surrounding both freshly cut and uncut sea fan pieces as soon as 1 h after placing them in dialysis tubing (Fig. 4, time effect in two-way ANOVA of cut F = 15.67, P = 0.0002 and uncut samples F = 6.91, P = 0.013). Endochitinase activity also increased in the water between 1 and 24 h and the increase was more pronounced for the uncut (time effect in two-way ANOVA F = 10.29, P = 0.0040) than for the cut sea fan pieces (time effect in two-way ANOVA F = 16.08, P = 0.0002). We further fractionated the chitinase activity released from the sea fans into Tris buffer by anion exchange chromatography. The exochitinase active compound(s) bound completely to the column and eluted with 0.5 and 1.0 M NaCl (Fig. 5).
Gorgonia ventalina. Chitinase activity in water surrounding sea fan samples. Aquarium-acclimated sea fan pieces were injured by cutting (n = 4) or left uncut (n = 3) and inserted into dialysis tubing to collect released proteins. Water samples were taken from inside tubing at times indicated and tested for exochitinase and endochitinase activity. Mean activity for each treatment ± SE
Antifungal activity of chitinase-containing fractions. Samples were tested for inhibitory effects on the growth of Aspergillus sydowii (strain FK1) in a liquid microplate absorbance assay. Two concentrations of the antimycotic, Hygromycin B (Hygro) and the exochitinase, Jack Bean β-N-acetylhexsaminidase (β-Hex) were compared to whole coral extracts (from 15 sea fan pieces exposed to fungus for 2 days in Fig. 3), water collected from around the uncut sea fans at 1 and 24 h (presented in Fig. 4), and anion exchange column fractions. Bars represent mean percent inhibition of six cultures after 24 h incubation minus mean inhibition due to the corresponding buffer alone ± SE. Positive values indicate inhibition of growth; negative values indicate increased growth. Black triangles represent exochitinase activity (nmols of MU released from 4-methylumbelliferyl N-acetyl-β-d-glucosaminide). Hygromycin B was not tested for chitinase activity and the values for β-Hex are single measurements. All other values are mean ± SE (whole coral extracts = 15, collected seawater = 3, anion exchange load = 2, flow-through = 4, washes = 7, elutions = 5)
Possible role of chitinases in sea fans
To determine if sea fan chitinases could be involved in defense against fungus we tested the antifungal properties of whole sea fan extracts, collected seawater, and anion exchange column fractions against Aspergillus sydowii (strain FK1). Jack bean β-N-acetylhexsaminidase (β-Hex) and the known antimycotic, Hygromycin B, showed increased antifungal activity with increasing concentration, while whole coral extracts and anion exchange column flow-through and washes actually encouraged fungal growth (Fig. 5, treatment effect in one-way ANOVA F = 102.05, P < 0.0001 and fraction effect in one-way AVOVA of anion exchange column data F = 81.11, P < 0.0001). However, the 1-h seawater collection and anion exchange column load and elution fractions inhibited fungal growth (Fig. 5, time effect in one-way ANOVA of collected seawater data F = 45.72, P = 0.0005 and fraction effect in one-way AVOVA of anion exchange column data F = 81.11, P < 0.0001).
Discussion
We established that sea fans contain at least two types of chitinolytic enzymes, one of which, exochitinase, was constitutively active in all fans assayed. We tested the hypothesis that challenging sea fans with fungus would cause an increase in chitinase activity and found, instead, that exochitinase activity decreased in sea fan tissue but increased in the surrounding water in response to several types of manipulation, including exposure to fungus. Although some components of immunity are actively synthesized in response to specific elicitors, others are pre-synthesized and are constitutively available under a variety of stresses (VanEtten et al. 1994). Our preliminary findings with sea fan exochitinase suggest that if it is a component of defense, it likely acts in the latter capacity, as a constitutive protection against opportunistic infections. The release of antifungal activity that we observed is consistent with studies of gorgonian and scleractinian corals demonstrating that antimicrobial activity is released into the surrounding seawater constitutively and in response to stress (Slattery et al. 1997; Geffen et al. 2005).
In contrast to the low variability in exochitinase activity among sea fans, there was striking variation in endochitinase activity. In the 2 of 15 sea fans with endochitinase activity, all of the pieces from these fans had measurable activity, regardless of their treatment or time in aquaria. This argues against the possibility that the variation is due to differences in handling of samples from different sea fans. It appears, instead, that the variation observed represents real physiological differences among individuals that could be genetically based, the result of different environmental histories, or differences between the microbial communities of individual sea fans. In future studies, we hope to elucidate the nature and significance of this individual variation and determine whether it is related to other factors such as fungal exposure, susceptibility to infection, or developmental stages.
One consideration in interpreting our results with sea fan chitinases is the complex nature of coral colonies. The coral tissue, the symbiotic zooxanthellae, or the microbial community of the mucus layer could be responsible for or contribute to the chitinase activity that we detected. Although it is possible to separate the coral host tissue and zooxanthellae by differential centrifugation, it is difficult to establish that isolated cells behave in a physiologically relevant manner outside of the tissue. Similarly, complete separation of the mucus microbial layer from the coral is not only technically problematic, but would likely also disrupt the normal physiology and composition of the microbial community and the coral cells. In addition, once chitinases are released into the water it is difficult to trace the activity back to a single source, especially if release is instigated by the stress of the separation process. Our current definition of “sea fan”, therefore, encompasses the entire, interdependent and intact coral/algal/microbial community. We are currently developing molecular, biochemical, and cell culture techniques to address the complex issue of where the chitinolytic activity originates within the coral colony.
In testing the antifungal properties of sea fan extracts, we observed that aqueous whole coral extracts slightly encouraged fungal growth. We suspect that this is an artifact of liberating large quantities of nutrients in the extraction procedure which masked the effect of specific antifungal compounds. The antifungal activities of the chitinase-containing seawater and Tris buffer samples with lower nutrients levels support this possibility. The lack of antifungal activity in those fractions lacking chitinase activity (as in the 24 h seawater collection and anion exchange flow-through) also suggests that sea fan chitinases could be responsible for the antifungal activity. Of course, the antifungal, chitinase-containing fractions in our assay still contain many other proteins and molecules that could be responsible for inhibiting the fungus in vitro. As we further fractionate the sea fan chitinases we will be able to test whether they are responsible for the antifungal activity that we observed. The data presented here also do not rule out the possibility that sea fan chitinases are involved in morphological or digestive processes in the sea fan. The rare endochitinase activity in the tissue, for example, could indicate individuals caught in an uncommon developmental stage or responding to a scarce chitinaceous food source. Our future studies will address this question by establishing whether sea fans themselves contain chitin and determining in which tissue and sub-cellular location chitinases reside.
In conclusion, the observation that exochitinase in sea fan tissue did not increase in response to fungal exposure argues against the hypothesis that this enzyme is a positively regulated fungus-specific response in this species. However, exochitinase is released into the surrounding seawater in response to injury or agitation of the coral and the released activity corresponds with antifungal activity. These findings suggest that this enzyme may play a role in the general response of sea fans to stress. Given the lytic nature and specificity of chitinases, and their demonstrated role in defense in many other organisms, our preliminary interpretation of these data is that sea fan exochitinase may serve as protection against opportunistic infection by chitinaceous pathogens during stress. The widely varying endochitinase activity of sea fans may prove a useful indicator of individual susceptibility to stressors, prior exposure to chitinaceous intruders/food sources, or may reflect different developmental stages of the host. Future studies will attempt to correlate endochitinase activity with disease and resistance measures and genomic markers to explore this possibility.
References
Agrawal AA, Tuzun S, Bent E (eds) (1999) Induced plant defenses against pathogens and herbivores: biochemistry, ecology, and agriculture. The American Phytopathological Society Press, St. Paul, Minnesota
Alker A, Kim K, Dube D, Harvell CD (2004) Localized induction of a generalized response against multiple biotic agents in Caribbean sea fans. Coral Reefs 23:397–405
Berenbaum MR, Zangerl AR (1999) Coping with life as a menu option: inducible defenses of the wild parsnip. In: Tollrian R, Harvell CD (eds) The ecology and evolution of inducible defenses. Princeton University Press, Princeton, New Jersey, pp 10–32
Brown BE, Bythell JC (2005) Perspectives on mucus secretion in reef corals. Mar Ecol Prog Ser 296:291–309
Dixon DM, Walsh TJ (1992) Human pathogenesis. Biotechnology 23:249–267
Elyakova LA (1972) Distribution of cellulases and chitinases in marine invertebrates. Comp Biochem Physiol 43:67–70
Flach J, Pilet PE, Jolles P (1992) What’s new in chitinase research? Experientia 48:701–716
Geffen Y, Rosenberg E (2005) Stress-induced rapid release of antibacterials by scleractinian corals. Mar Biol 146:931–935
Geiser DM, Taylor JW, Ritchie KB, Smith GW (1998) Cause of sea fan death in the West Indies. Nature 394:137–138
Gil-Agudelo DL, Myers C, Smith GW, Kim K (2006) Changes in the microbial communities associated with Gorgonia ventalina during Aspergillosis infection. Dis Aquat Org 69:89–94
Gillespie JP, Kanost MR, Trenczek T (1997) Biological mediators of insect immunity. Ann Rev Entomol 42:611–43
Jensen PR, Harvell CD, Wirtz K, Fenical W (1996) The incidence of anti-microbial activity among Caribbean gorgonians. Mar Biol 125:411–420
Kim K, Harvell CD, Kim PD, Smith GW, Merkel SM (2000a) Fungal disease resistance of Caribbean sea fan corals (Gorgonia spp.). Mar Biol 136:259–267
Kim K, Kim PD, Alker AP, Harvell CD (2000b) Antifungal properties of gorgonian corals. Mar Biol 137:393–401
Klug M, Tardent P, Smid I, Holstein T (1984) Presence and localization of chitinase in Hydra and Podocoryne (Cnidaria, Hydrozoa). J Exp Zool 229:69–72
Knowlton N, Rohwer F (2003) Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am Nat 162:S51–S62
Kramer KJ, Muthukrishnan S (1997) Insect chitinases: molecular biology and potential use as biopesticides. Insect Biochem Molec 27:887–900
Levin DA (1976) The chemical defenses of plants to pathogens and herbivores. Annu Rec Ecol Syst 7: 121-159
Mali B, Mohrlen F, Frohme M, Frank U (2004) A putative double role of a chitinase in a cnidarian: pattern formation and immunity. Dev Comp Immunol 28:973–981
Merzendorfer H, Zimoch L (2003) Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol 206:4393–4412
Nagelkerken I, Buchan K, Smith GW, Bonair K, Bush P, Garzón-Ferreira J, Botero L, Gayle P, Harvell CD, Heberer C, Kim K, Petrovic C, Pors L, Yoshioka P (1997) Widespread disease in Caribbean sea fans: II. Patterns of infection and tissue loss. Mar Ecol Prog Ser 160:255–263
Nagy NE, Fossdal CG, Dalen LS, Lonneborg A, Heldal I, Johnsen O (2004) Effects of Rhizoctonia infection and drought on peroxidase and chitinase activity in Norway spruce (Picea abies). Physiol Plant 120:465–473
Ng TB (2004) Antifungal proteins and peptides of leguminous and non-leguminous origins. Peptides 25:1215–1222
Olano CT, Bigger CH (2000) Phagocytic activities of the gorgonian coral Swiftia exserta. J Invertebr Pathol 76:176–184
Overdijk B, Van Steijn GJ, Odds FC (1999) Distribution of chitinase in guinea pig tissues and increases in levels of this enzyme after systemic infection with Aspergillus fumigatus. Microbiology 145:259–269
Petes L, Harvell CD, Peters EC, Webb MAH, Mullen KM (2003) Pathogens compromise reproduction and induce melanization in Caribbean sea fans. Mar Ecol Prog Ser 264:167–171
Ritchie KB, Smith GW (2004) Microbial communities of coral surface mucopolysaccharide layers. In: Rosenburg E, Loya Y (eds) Coral health and disease. Springer, Berlin Heidelberg New york, pp 259–263
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Shibuya K, Naoe S, Yamaguchi H (1999) Animal models of A. fumigatus infections. Contrib Microbiol 2:130–138
Slattery M, Hamann MT, McClintock JB, Perry TL, Puglisi MP, Yoshida WY (1997) Ecological roles for water-borne metabolites from Antarctic soft corals. Mar Ecol Prog Ser 161:133–144
Smith GW, Ives LD, Nagelkerken IA, Ritchie KB (1996) Caribbean sea fan mortalities. Nature 383:487
Suzuki M, Morimatsu M, Yamashita T, Iwanaga T, Syuto B (2001) A novel serum chitinase that is expressed in bovine liver. FEBS Lett 506:127–130
Thevissen K, Osborn RW, Acland DP, Broekaert WF (2000) Specific binding sites for an antifungal plant defensin from Dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol Plant Microbe Interact 13:54–61
Trench RK (1996) Specificity and dynamics of algal–invertebrate symbiosis. Symbiosis 96:16
Tronsmo A, Harman GE (1993) Detection and quantification of N-acetyl-beta-d-glucosaminidase, chitobiosidase, and endochitinase in solutions and on gels. Anal Biochem 208:74–79
Tuzun S, Bent E (1999) The role of hydrolytic enzymes in multigenic and microbially-induced resistance in plants. In: Agrawal AA, Tuzun S, Bent E (eds) Induced plant defenses against pathogens and herbivores. APS Press, St Paul, pp 95–115
VanEtten HD, Mansfield JW, Bailey JA, Farmer EE (1994) Two classes of plant antibiotics: phytoalexins versus phytoanticipins. Plant Cell 6:1191–1192
Acknowledgments
We thank K. Boor and J. Thaler for use of their microplate readers and we thank K. Rypien, J. Ward, and L. Mydlarz for critical reading. We also thank the staff of the Mote Marine Laboratories, Tropical Research Lab for their help in our experiments at their facility. This work was conducted under National Science Foundation grant number OCE-0326705 and collections were made under Florida Keys National Marine Sanctuary Research permit number FKNMS-2004-092.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.P. Grassle, New Brunswick.
Rights and permissions
About this article
Cite this article
Douglas, N.L., Mullen, K.M., Talmage, S.C. et al. Exploring the role of chitinolytic enzymes in the sea fan coral, Gorgonia ventalina . Mar Biol 150, 1137–1144 (2007). https://doi.org/10.1007/s00227-006-0444-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-006-0444-8