Abstract
Environmental salinity is important in defining Brachionus plicatilis sibling species distributions. However, while salinity influences distributions, sibling species often co-exist. Three different mechanisms potentially account for the partial co-occurrence of sibling species: (1) siblings have differing salinity tolerances that partially overlap; (2) siblings physiological tolerances may be commonly broad, but relatively small differences in tolerances differentiate distributions via interactions e.g. competition; or (3) siblings distributions may be influenced by physical factors other than salinity. Here, we assess the extent of salinity tolerance in three B. plicatilis sibling species (B. plicatilis 6TUR, B. plicatilis IOM and B. rotundiformis 6TOS) by measuring population growth rate (μ, day−1) and egg development time in response to salinity (5–60‰) and salinity fluctuations (≤ Δ40‰). Sibling species were identified by analysis of the mitochondrial COI gene, and salinity responses were compared by regression analysis. Responses differed significantly between siblings, although the broad trends were similar. Positive growth occurred at all salinities, and highest growth rates ranged between 0.93 and 1.08 day−1 at 16–18‰. Rapid changes in salinity reduced growth rates, but net mortality occurred only in one treatment (100% mortality on transfer from 10 to 40‰). Egg development time was largely invariant with salinity except for B. plicatilis IOM and where rotifers were transferred from 30 to 60‰. We indicate that several siblings are similarly euryhaline and tolerate salinity fluctuations. Undoubtedly, wide tolerances in B. plicatilis are adaptations to ephemeral and seasonally variable habitats. Given common broad salinity tolerances, it is unlikely that the differential distributions of sibling species are a direct result of physiological constraints. Instead, we illustrate using a simple model that subtle differences in physiological tolerances may have important impacts on interactions between sibling species, which may in turn influence distributions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biodiversity is an integral component of ecosystem function. However, a large proportion of biodiversity is potentially unaccounted for as it is now recognised that cryptic species complexes are common for many taxa (Knowlton 1993). For instance, the rotifer Brachionus plicatilis, an important component of microbial food-webs in continental aquatic systems (Walker 1981), which was considered a cosmopolitan euryoecious species, is now recognised as a species complex (Ciros-Pérez et al. 2001b; Gomez et al. 2002). The existence of such complexes raises important questions regarding the extent of niche separation required to maintain highly similar species and the mechanisms that result in cryptic species formation (Knowlton 1993).
The B. plicatilis complex consists of at least seven sibling species (Ortells et al. 2003), three of which are currently described as separate siblings (B. plicatilis, B. rotundiformis, B. ibericus; Ciros-Perez et al. 2001b). B. plicatilis sibling species are common in ephemeral and highly variable brackish coastal marshes and inland salt lakes (Gomez and Snell 1996; Walker 1981). Field based studies have indicated that the distributions of siblings are differentially influenced by a small number of environmental variables, primarily salinity (Gomez et al. 1995), such that different physiological tolerances confine siblings to different spatial and temporal distributions, allowing sibling species co-existence (Gomez et al. 1997). However, while it is clear that distributions are strongly dependent on salinity subsequent studies have demonstrated that sibling species distributions commonly overlap (Ortells et al. 2003).
These more recent data raise important ecological questions concerning the role of environmental variables in determining the distributions and co-existence of sibling species. Broadly three potential scenarios may explain the observed patterns of sibling species distributions in natural habitats: (1) sibling species may have widely differing salinity tolerances that only partially overlap, (2) the physiological tolerances of sibling species may be similarly broad, and relatively small differences in physiological responses account for partially overlapping distributions via biological interactions, e.g. local competitive exclusion, or (3) sibling species distributions may be influenced by factors other than salinity.
Current studies indicate that numerous environmental factors influence sibling species distributions (e.g. Gomez et al. 1995; Ortells et al. 2003) and provide evidence of competitive exclusion between siblings (e.g. Ciros-Pérez 2001a), thus it is becoming clear that a potentially complex series of interactions may ultimately determine distributions in natural environments. However, details of how underlying ecophysiological responses (e.g. growth rate) and environmental tolerances (e.g. salinity) differ between siblings are scarce, this is partially because such details cannot easily be inferred from field studies and partially because a large number of the experimental studies assessing physiological tolerances in B. plicatilis predate the discovery of cryptic species in this taxon (e.g. Miracle and Serra 1989, but see Gomez et al. 1997; Lowe et al. 2005a, b). As a result, our knowledge of the extent of differentiation in physiological tolerances between sibling species is limited. Such information is critical to understand the relative roles of environmental factors and sibling species interactions in determining distributions. If, for example, physiological tolerances overlap extensively, interactions between sibling species may be common, such a scenario raises important questions regarding sibling species co-occurrence and sympatric speciation.
Here we focus on rigorously characterising salinity tolerance in three defined sibling species, to assess the extent of overlap in physiological tolerances. We also extend studies of salinity tolerance in these organisms, by assessing the potential effect of fluctuations in environmental salinity on the ecology of B. plicatilis sibling species. Such fluctuations, and episodic changes, in environmental variables are potentially important parameters shaping the distributions of, and interactions between, organisms (Chesson and Huntley 1997); this may be especially important in ephemeral and high disturbance environments, such as the shallow brackish water habitats where B. plicatilis sibling species are common and salinity may fluctuate widely (Miracle and Serra 1989).
In this study, we use an experimental approach to compare ecophysiological responses of three B. plicatilis sibling species to both a broad range of salinities (5–60‰) and to rapid salinity changes (Δ10–40‰). First, sibling species identities were confirmed by analysing fragments of the mitochondrial COI gene. Then, differences in salinity responses were identified using estimates of population growth rate and egg development time. Finally, we illustrate, using our data and a simple population growth model, the potential influence of small differences in physiological tolerances on the occurrence of sibling species in natural habitats.
Materials and methods
Maintenance of rotifer cultures
Three monoclonal isolates from the Brachionus plicatilis complex were cultured. B. plicatilis IOM is used in commercial aquaculture and was provided by N. Fullerton (Larval rearing centre, Port Erin marine laboratory, Isle of man). B. plicatilis and B. rotundiformis were environmental isolates provided by M. Serra, University of Valencia, Spain. All isolates were maintained on a diet of the flagellate Dunaliella salina, as previously described (Lowe et al. 2005a).
Mitochondrial COI sequencing and analysis
The identity and phylogenetic positions of the three isolates were examined using sequence data for a fragment of the mitochondrial cytochrome oxidase one gene (following Gomez et al. 2002). For DNA extraction, rotifers were washed on Nitex mesh with sterile saline and left for 12 h to process ingested food. Rotifers were then washed again, concentrated into 2 ml screw top cryotubes (Nalgene), and pelleted by centrifugation. Pellets were stored at −80 °C until processed. Genomic DNA was extracted using a high salt protocol (Sambrook and Russell 2001) and a fragment of the mitochondrial COI gene amplified by PCR using the following primers: forward GGT CAA CAA TCA TAA AGA TAT TGG, and reverse TAA ACT TCA GGG TGA CCA AAA AAT CA. PCR products were sequenced using the BigDye Terminator v3.1 sequencing kit on the ABI 3,100 genetic analysis system.
Sequences were aligned using DNA Star and subjected to maximum likelihood analyses (PAUP, V4.0b10). An optimal model (P = 0.05) was determined using Modeltest V3.06 (Posada and Crandall 1998). The best-fit model was the transversional model of evolution with estimated base frequencies and a substitution rate model; the proportion of invariant sites was 0.5542, and a gamma distribution (shape parameter = 2.0307) was invoked on the remaining sites. Parameters were added prior to maximum likelihood runs, and heuristic searches were performed. To examine robustness, bootstrap values were calculated by stepwise addition (1,000 replicates).
The effect of salinity on growth rate
For acclimated growth experiments, rotifers were maintained at one of 12 salinities between 5 and 60‰ for > 2 weeks. Experiments were conducted as described by Lowe et al. (2005a). Specific growth rate (μ, day−1) was calculated, from the change in total number of females as μ = ln(Nt/N0)/t, where N0 is the number of females at the start of the growth incubation, Nt is the number of females after 3 days, and t is time in days. During all acclimation and experimental incubations food remained replete.
To examine the response of rotifers when transferred between different salinities, three un-acclimated growth responses were examined. Rotifers acclimated at 10‰ were transferred directly to increasing salinities (20, 30, 40‰); rotifers acclimated at 50‰ were transferred to decreasing salinities (40, 30, 10‰); finally, rotifers acclimated at 30‰ were transferred to increasing and decreasing salinities (20, 10, 40, 50, 60‰). Experiments on un-acclimated rotifers were otherwise identical to the acclimated experiments. For each of the three transfer experiments an aliquot of rotifers was transferred to the salinity, at which it was acclimated, to test for the effects of the transfer procedure.
Statistical analysis
Predictive models were fit to the growth rate data to examine responses to salinity and to compare responses between siblings. Responses for all three siblings were characterised by fitting polynomial models, using the non-linear curve fitting function in SAS (v8.02). The complexity (e.g. linear, 2nd, 3rd or 4th order polynomial) of each model for each sibling was determined by sequentially adding parameters; whether the addition of a parameter significantly improved the fit was then tested using a sum of squares reduction test (Zar 1999).
Comparisons between growth rate responses for each sibling were made in a similar fashion. Growth responses for the three siblings were combined and two models were fitted to the data: (1). a full regression model, in which each component of the model was allowed a separate parameter for each sibling (e.g. y = b1x2 + b2x2 + b3x2, where b1–3 are separate parameters for each sibling, for the component bx2); and (2). a reduced regression model, in which each component of the model was constrained to a single parameter (e.g. y = bnX2 where bn is a single parameter for all siblings, for the component bx2). Thus, if all responses are the same, no difference between the full and reduced regression models occurs. Differences between models were again analysed using the sum of squares reduction test.
Egg development time was calculated, for both acclimated and transfer treatments, using model II linear regression (Sokal and Rohlf 1995), as the slope of growth rate vs. mean number of eggs female−1 as μ = ln(E + 1)/EDT, where EDT is egg development time, and E is the average number of amictic eggs female−1.
Results
Phylogeny
The three Brachionus plicatilis isolates used in this study were placed in a phylogeny of the B. plicatilis species complex (Fig. 1). Phylogenetic analysis and nucleotide blast searches (Altschul et al. 1997) supported B. plicatilis to be an isolate from Estany den Turies, Spain (isolate shared 96% identity with Genbank entries AF266853, AF266854, and AF266855 from Estany den Turies) and B. rotundiformis as an isolate from Poza Sur (isolate shared 99% identity with Genbank entries AY785221 and AY785222, from Poza Sur). From here on we refer to these isolates as B. plicatilis 6TUR and B. rotundiformis 6TOS (following Gomez et al. 2000). The identity of the B. plicatilis isolate provided by the larval rearing centre, Isle of Man (referred to as B. plicatilis IOM) was unknown; the mitochondrial COI data place this isolate within the ‘Austria’ clade of B. plicatilis, which is likely a discrete sibling species (Gomez et al. 2002).
The phylogenetic positions of the Brachionus plicatilis sibling species used in this study based on maximum likelihood analysis of a fragment the mitochondrial COI gene. The phylogeny presented is a subset of the B. plicatilis species complex and does not include all strains or sibling species for which sequences are available (e.g. B. ibericus). Numbers above selected nodes are bootstrap values (1,000 replicates) and are given where support was > 50%. Designations are as defined by Gomez et al. (2002). The GenBank accession number for the previously unstudied B. plicatilis IOM is DQ140386
The effect of salinity on growth rate and egg development time
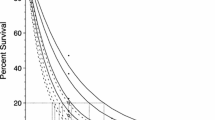
Over the range of salinities tested (5–60‰), all three acclimated isolates achieved positive population growth. The modelled responses are given in Fig. 2 and the parameters summarised in Table 1. The growth rate responses were significantly different between the three siblings (F7,95 = 12.05, P < 0.001). At all salinities B. rotundiformis 6TOS achieved the highest growth rate. B. plicatilis 6TUR exhibited the lowest growth rate (0.26 day−1) at a salinity of 60‰. The growth rate responses for B. plicatilis 6TUR and B. plicatilis IOM intersected; at low salinities B. plicatilis 6TUR achieved higher growth rates, above 10‰ the relationship was reversed.
The response of growth rate (μ, day−1) to salinity for B. plicatilis 6TUR (a), B. plicatilis IOM (b), and B. rotundiformis 6TOS (c). Polynomial models (solid lines) were fitted to each response. The parameters for each model are given in Table 1 and a summary of statistical comparisons in Table 3. The relationship between μ and ln(E + 1) for B. plicatilis 6TUR (d), B. plicatilis IOM (e), and B. rotundiformis 6TOS (f). Egg development time is given as the slope of μ versus ln(E + 1)
The shapes of the growth rate responses differed for each sibling as indicated by the complexity of the polynomial regressions fitted to each response (Fig. 2; Table 1). Growth rate for B. plicatilis 6TUR was relatively constant at low salinities; maximum growth rate (0.93 day−1) occurred at 16‰ and decreased rapidly at high salinity. For B. plicatilis IOM and B. rotundiformis 6TOS distinct decreases in growth rate occurred below 15‰; growth maximum occurred at 16‰ for B. plicatilis IOM (1.05 day−1) and 18‰ for B. rotundiformis 6TOS (1.08 day−1), followed by a less pronounced, relative to B. plicatilis 6TUR, reduction in growth rate with increasing salinity
A strong positive relationship between ln(E + 1) and μ for B. plicatilis 6TUR and B. rotundiformis 6TOS (R2 = 0.61 and 0.76 respectively) indicates that salinity had only a small affect on egg development time (calculated as the slope of ln(E + 1) vs. μ, Fig. 2). For B. plicatilis IOM the relationship between ln(E + 1) and μ was weak (R2 = 0.23) suggesting that egg development time varied with salinity. Shortest egg development time (0.6 day) occurred at 20‰ and longest development at 60‰ (average 1.0 day, data not shown).
The effect of rapid changes in salinity on growth rate
In all cases changes in salinity significantly altered growth rate relative to acclimated responses (Fig. 3; Tables 2, 3). Both the direction of the transfer and the starting salinity influenced growth rate. When transferred upward, from low salinities (10‰) growth rate decreased (Fig. 3), and at 40‰ all individuals died immediately on transfer. Transfers upward from 30‰ also resulted in a decreased growth rate in comparison to acclimated responses, although the effect was smaller than that for transfers upward from 10‰. In contrast, decreases in salinity had a lesser impact on growth rates. When rotifers were transferred to decreasing salinities (from 30 to 50‰), growth rates increased (from 30‰) or did not significantly decrease (from 50‰).
The effects of transfer between salinities on population growth rates for B. plicatilis 6TUR (row a), B. plicatilis IOM (row b), and B. rotundiformis 6TOS (row c). Data for each isolate, for transfer experiment, are also plotted together for comparison (row d). Polynomial or linear (B. plicatilis 6TUR shift from 10‰) models (solid lines) were fitted to each response. The parameters for each model are given in Table 2 and a summary of statistical comparisons in Table 3. Cases where 100% mortality occurred on transfer to a new salinity are indicated (x)
Responses to salinity transfers were similar, for all three siblings. No significant difference in growth rate responses between B. plicatilis IOM and B. rotundiformis 6TOS occurred for any of the transfer treatments (Table 3). In all transfer experiments B. plicatilis 6TUR achieved lowest growth rates (Fig. 3; Table 3).
In contrast to growth rate, there was no pronounced relationship between changes in salinity and egg development time. For all siblings and treatments egg development time was constant with salinity (Fig. 4a–c), and the relationship between μ and ln(E + 1) was linear (data not shown). One exception occurred, for all three siblings transfers from 30 to 60‰ resulted in a 2.4×, 3.4× and 2.5× increase in egg development time for B. plicatilis 6TUR, B. plicatilis IOM, and B. rotundiformis 6TOS, respectively.
The effect of salinity transfer on egg development time for B. plicatilis 6TUR (a), B. plicatilis IOM (b), and B. rotundiformis 6TOS (c). In each case the relationship between μ and ln(E + 1) was linear (see Materials and methods), thus egg development time was assumed to be constant with salinity. Numbers 10, 30 and 50 refer to the starting salinities (‰) for the three transfer experiments
Discussion
Sibling species of the Brachionus plicatilis complex are common brackish water organisms (King and Serra 1998), and field studies indicate that different sibling species display differing temporal and spatial distributions, which correlate with salinity variation (Ortells et al. 2003). Initial field observations suggested that unique adaptations partition siblings and facilitate their co-occurrence (Gomez et al. 1995, 1997). However, more recent field and ecophysiological data indicate substantial overlap in the ecological tolerances of siblings, forcing a re-examination of the evolutionary mechanisms allowing sibling species coexistence (Ciros-Pérez et al. 2001a; Lapesa et al. 2004; Ortells et al. 2003).
Here we compared salinity tolerance in three sibling species by measuring growth rate and egg development time over salinities between 5 and 60‰ and compared the effects of sudden transfer between salinities on these life history parameters. To our knowledge this is the broadest range of salinities over which the tolerances of defined B. plicatilis sibling species have been assessed (for previous studies, see Miracle and Serra 1989; Lubzens et al. 2001; Serra et al. 1998). In addition to providing a rigorous characterisation of ecophyioslogical responses, the use of a broad range of salinities and the statistical comparison of salinity tolerances, using predictive models, provides useful data for future studies modelling the effects of environmental variation on sibling species distributions.
In nearly all cases, the shape and magnitude of salinity responses significantly differed between sibling species. However, the overall trends in responses to salinity were similar: all were tolerant to a broad salinities range and were robust to sudden salinity changes. In acclimated experiments, all three sibling species achieved maximum growth between 16 and 18‰ and maintained positive growth between 5 and 60‰. When transferred between salinities, sibling species maintained net positive growth over large decreases (Δ 40‰) and increases (Δ 30‰) in salinity. Net mortality occurred only in one case (transfer from 10 to 40‰) where all individuals died on transfer. Further, egg development time was invariant with changes in salinity except for B. plicatilis IOM in acclimated experiments and for all three sibling species where rotifers were transferred from 30 to 60‰. Below, we assess the likely relevance of the observed similarities and differences in salinity tolerance between siblings in determining their distributions.
Broad, but varying, salinity tolerances may be common in the B. plicatilis complex-common, broad salinity responses, and tolerance to salinity fluctuations, suggest that narrow non-overlapping tolerances as a mechanism for isolating different sibling species is unlikely. Admittedly, we have examined only a single isolate from each of three sibling species, and ideally more isolates should be assessed to support our results. Nevertheless, the isolates examined were from phylogenetically diverse sibling species suggesting that broad salinity tolerances, and tolerance to fluctuating salinities, might be common traits in the B. plicatilis species complex.
Undoubtedly, there are strong selection pressures to maintain wide physiological tolerances. B. plicatilis rotifers are common in continental brackish water environments, which experience broad seasonal changes in physical factors (King and Serra 1998). In such ephemeral habitats, long-term survival of rotifers depends on the production of resting eggs via sexual reproduction (Gomez et al. 1995; King and Serra 1998). However, short term survival and colonisation of new environments is likely to depend on physiological tolerance. For example, in small aquatic invertebrates, such as rotifers, the rapid establishment of large populations following dispersal to novel environments potentially “buffers” habitats from further colonisation by competitors (De Meester et al. 2002). As such, wide environmental tolerances, allowing rapid growth in new environments, are likely to be important components of dispersal and colonisation and in determining sibling species distributions.
Information on how wide salinity tolerances, and tolerances to salinity fluctuations, are maintained by B. plicatilis rotifers is limited, but understanding how physiological adaptations differ between siblings may elucidate the causes of their distributions. It was previously held that B. plicatilis was an osmoconformer with limited ability to adapt to salinity changes (Epp and Winston 1977). However, we have previously indicated that B. plicatilis sibling species are likely to be osmoregulators, based on the activity of the osmoregulatory enzyme Na+/K+ ATPase (Lowe et al. 2005a). We have further indicated that Na+/K+ ATPase activity correlates with differences in growth rate responses to salinity in siblings, and that the differences in ion regulatory activity were associated with variation in the expression of Na+/K+ ATPase gene transcripts that were highly similar between siblings (> 99% similarity, Lowe et al. 2005b). These data illustrate that differences in ecologically important adaptations between sibling species may be accounted for by relatively minor genetic variation.
How regulatory mechanisms associated with salinity tolerance differ in response to fluctuations in salinity has not been assessed, though here we indicate that growth rate differences between sibling species may be reduced by salinity fluctuations. For example, B. rotundiformis 6TUR achieved comparatively high-acclimated growth rates, but in all salinity transfer experiments growth rates for B. rotundiformis 6TOS and B. plicatilis IOM were the same; thus, B. rotundiformis was more susceptible to salinity fluctuation. Whether differences in responses to fluctuating environmental variables are ecologically important is, at this point, difficult to assess. In a general context, the potential consequences of environmental fluctuations for ecological communities have proved contentious (May and MacArthur 1972; Hebert and Crease 1980; Chesson and Huntley 1997). Nevertheless, many of the habitats in which B. plicatilis rotifers occur are ephemeral, and there is growing evidence suggesting strong interactions and competition between B. plcatilis sibling species (Ciros-Pérez et al. 2001a; Lapesa et al. 2004). Given these conditions, the reductions in growth rate associated with salinity changes, and the subsequent rate of recovery, are likely to influence the distributions of sibling species in natural environments.
Small physiological differences potentially affect sibling species distributions
Given the common broad salinity tolerances displayed by the sibling species examined in this study, the differences in the shapes of the growth rate responses, though significant, appear comparatively minor (Fig. 1). However, specific growth rate (μ) is an exponent of population growth, and as a result small differences in growth rates can result in large differences in subsequent population sizes over time. Thus, the apparently minor differences in growth rate responses to salinity assessed here have potentially significant ecological consequences. We illustrate this using a simple population model based on logistic growth, assuming that two sibling species have the same density-dependent effect on growth (i.e. a shared carrying capacity). Using STELLA 8 (High performance systems), population growth of B. plicatilis 6TUR and B. plicatilis IOM was simulated using the coupled equations dNTUR/dt = NTURμTUR [1−(NTUR+NIOM/K)]−NTURm and dNIOM/dt = NIOMμIOM [1−(NIOM+NTUR/K)]−NIOMm where N is population density (initially 1 for both sibling species; subscripts refer to rotifer siblings), μ is the specific growth rate (determined as a function of salinity, again subscripts refer to siblings), K is the carrying capacity (100 for both siblings) and m is a density dependant loss rate (akin to losses due to e.g. predation and set at 0.1, for both sibling species, Brandl 2005). The model was applied at three salinities (Fig. 5), using the growth rate response parameters given in Table 2.
The modeled population dynamics of Brachionus plicatilis IOM (solid lines) and B. plicatilis 6TUR (dotted lines) at salinities of 5, 15 and 55‰. Specific growth rate responses to salinity were determined using parameters from Table 1
The model outputs (Fig. 5) illustrate both the potentially large differences in population size resulting from relatively small differences in salinity tolerance and how different sibling species might dominate under differing conditions. Although these are simple simulations, they highlight an important point: the distribution of a sibling species may be constrained well within its potential distribution based on physiological tolerances by, for example, interactions with other sibling species. Notably, we have not tested for competition between siblings in this study, and the inclusion of an interaction between sibling species (i.e. shared density dependence) in the model is speculative. However, as sibling species are by definition highly similar and are likely to share many ecological niche parameters (Ortells et al. 2003; Serra et al. 1998), the assumption of competition between sibling species seems reasonable. Indeed, there is now strong evidence that B. plicatilis sibling species compete for resources and are capable of out-competing each other (Ciros-Pérez et al. 2001a). In this context, subtle differences in physiological tolerances (e.g. to salinity) might significantly influence sibling species distributions and abundance by mediating biological interactions. Future direct competition experiments to fully parameterise the impact of salinity and other key factors on sibling species interactions will undoubtedly provide information regarding the mechanisms for sibling species isolation.
The role of multiple environmental variables in determining distributions
Although salinity is undoubtedly a major factor influencing B. plicatilis sibling species distributions, ecological niches are potentially defined by a number of environmental variables. Multivariate analyses of sibling species abundances in natural habitats highlight temperature, alkalinity and oxygen concentrations (in addition to salinity) as principle parameters differentiating distributions (Gomez et al. 1995; Ortells et al. 2003), though it is now recognised that overlaps in sibling species distributions are common and substantial (Ortells et al. 2003). These field investigations require parallel multifactor experimental studies to provide precise ecophysiological responses to asses the impacts of both direct physiological limitation and potential biological interactions on distributions. Some multifactor experimental approaches have been attempted. For instance, Gomez et al. (1997) demonstrated that salinity and temperature have an interactive effect on the growth rates of B. plicatilis with interactive effects differing between sibling species. As Gomez et al. (1997) assessed interactions using only three salinities and three temperatures the data are insufficient to parameterise limits to physiological tolerance or build multifactor interactions into potential ecosystem models. Clearly the next step will be to combine field data with multifactor experimental approaches to assess the interactive effects of key environmental variables on sibling species distributions (e.g. Kimmance et al. 2006).
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Brandl Z (2005) Freshwater copepods and rotifers: predators and their prey. Hydrobiologia 546:475–489
Chesson P, Huntley N (1997) The roles of harsh and fluctuating conditions on the dynamics of ecological communities. Am Nat 150(5):519–553
Ciros-Pérez J, Carmona MJ, Serra M (2001a) Resource competition between sympatric sibling rotifer species. Limnol Oceanogr 46:1511–1523
Ciros-Pérez J, Gomez A, Serra M (2001b) On the taxonomy of three sympatric sibling species of the Brachionus plicatilis (Rotifera) complex from Spain, with the description of B. ibericus n. sp. J Plank Res 23(12):1311–1328
De Meester L, Gomez A, Okamura B, Schwenk K (2002) The monopolization hypothesis and the dispersal–gene flow paradox in aquatic organisms. Acta Oecol 23:121–135
Epp RW, Winston PW (1977) Osmotic regulation in the brackish water rotifer Brachionus plicatilis. J Exp Biol 68:151–156
Gomez A, Snell TW (1996) Sibling species and speciation in the Brachionus species complex (rotifera). J Evol Biol 9:953–964
Gomez A, Temprano M, Serra M (1995) Ecological genetics of a cyclical parthenogen in temporary habitats. J Evol Biol 8:601–622
Gomez A, Carmona MJ, Serra M (1997) Ecological factors affecting gene flow in the Brachionus plicatilis complex (rotifera). Oecologia 111:350–356
Gomez A, Carvalho GR, Lunt DH (2000) Phylogeography and regional endemism of a passively dispersing zooplankter: mitochondrial DNA variation in rotifer resting egg banks. Philos Trans R Soc Lond B 267:2189–2197
Gomez A, Serra M, Carvalho GR, Lunt DH (2002) Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis (rotifera). Evolution 56:1431–1444
Hebert PDN, Crease TJ (1980) Clonal coexistence in daphnia pulex (Leydig) another planktonic paradox. Science 207:1363–1365
Kimmance SA, Atkinson D, Montagnes DJS (2006) Do Temperature-food interactions matter? Responses of production and its components in the model heterotrophic flagellate Oxyrrhis marina. Aquat Microb Ecol 42:63–73
King C, Serra M (1998) Seasonal variation as a determinant of population structure in rotifers reproducing by cyclical partenogenesis. Hydrobiologia 387/388:361–372
Knowlton N (1993) Sibling species in the sea. Annu Rev Ecol Syst 24:189–216
Lapesa S, Snell TJ, Fields DM, Serra M (2004) Predatory interactions between a cyclopoid copepod and three sibling rotifer species. F Biol 47(9):1685–1689
Lowe CD, Kemp SJ, Bates AD, Montagnes DJS (2005a) Evidence that the rotifer Brachionus plicatilis is not an osmoconformer. Mar Biol 146:923–929
Lowe CD, Kemp SJ, Montagnes DJS (2005b) An interdisciplinary approach to assess the functional diversity of free-living microscopic eukaryotes. Aquat Microb Ecol 41:67–77
Lubzens E, Zmora O, Barr Y (2001) Biotechnology and aquaculture of rotifers. Hydrobiologia 446/447:337–353
May RM, MacArthur RH (1972) Niche overlap as a function of environmental variability. Proc Nat Acad Sci 69(5):1109–1113
Miracle MR, Serra M (1989) Salinity and temperature influence in rotifer life history characteristics. Hydrobiologia 186/187:81–102
Ortells R, Gomez A, Serra M (2003) Coexistence of cryptic rotifer species: ecological and genetic characterisation of Brachionus plicatilis. Freshwater Biol 48:2194–2002
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Serra M, Gomez A, Carmona MJ (1998) Ecological genetics of Brachionus sympatric sibling species. Hydrobiologia 387/388:373–384
Sokal RR, Rohlf FJ (1995) Biometry, 3rd edn. W. H. Freeman, New York
Walker KF (1981) A synopsis of ecological information on the saline lake rotifer Brachionus plicatilis Muller 1786. Hydrobiologia 81:159–167
Zar JH (1999) Biostatistical analysis, 4th edn. Prentice Hall, London
Acknowledgments
This study forms part of a PhD thesis by C. D. Lowe at the University of Liverpool, funded by the Natural Environment Research Council (Grant No. NER/S/A/2001/0630). Pierre Ferrer and Stephanie Swift provided valuable assistance with experimental work. Drs. I. Saccheri and P. Watts provided valuable discussion.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.P. Thorpe, Port Erin
Rights and permissions
About this article
Cite this article
Lowe, C.D., Kemp, S.J., Díaz-Avalos, C. et al. How does salinity tolerance influence the distributions of Brachionus plicatilis sibling species?. Mar Biol 150, 377–386 (2007). https://doi.org/10.1007/s00227-006-0366-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-006-0366-5