Abstract
The ascidian Ecteinascidia turbinata (Herdman) is a colonial sea squirt found in the Caribbean and Mediterranean Seas. In the present study, the bacterial complement of E. turbinata has been assessed by 16S rRNA gene analysis and the most commonly occurring strains identified by restriction fragment length polymorphism. Three strains were found to predominate using this approach, with one representing >50% of clones from both larval and adult material. The 16S rRNA gene sequence of the most commonly occurring strain did not match with any known bacterial sequences and could only be assigned to the γ-proteobacteria subdivision. The two other frequently occurring strains were assigned to the Mollicutes. In situ hybridisation analysis with eubacterial probes to 16S rRNA revealed the presence of apparently endosymbiotic bacteria in adult and larval tissue, and electron microscopy confirmed the presence of putative bacteriocytes in the larval tissue. The presence of the same bacteria in the brooded larvae suggested that they were vertically transmitted from parent to offspring. Further hybridisation using a novel probe designed to be specific to the 16S rRNA sequence of the dominant strain, highlighted the same cell types as that revealed by the eubacterial probe. The results suggest that the bacteria represent a novel strain, denoted "Candidatus Endoecteinascidia frumentensis", and that they may have an important role in the biology of E. turbinata.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ascidian Ecteinascidia turbinata (Herdman) is a colonial tunicate from the family Perophoridae found in the Caribbean and Mediterranean Seas (Berrill 1932). As an important component of the benthic ecology of the Caribbean mangroves, E. turbinata has been the subject of various studies examining settlement, species succession and larval behaviour (Young and Bingham 1987; Bingham and Young 1991, 1995). The colony-forming individuals, or zooids, are approximately 30 mm in length and are linked by a basal vessel termed the stolon. This creeping structure allows blood flow throughout the colony, buds to form new zooids during asexual reproduction and acts as a storage organ during colony regression. Embryos are brooded in the zooid and released as larvae when fully developed. In the 1960s interest in this species was heightened when an extract of the animal was found to have potent cytotoxic properties. It was not until 1990 that the compounds conferring these properties, the ecteinascidins, were identified and characterised (Rinehart et al. 1990). Little is known about the production of these secondary metabolites or what function, if any, they play in the animal.
Bioactive secondary metabolites are found in many marine invertebrates, especially sponges, molluscs, bryozoans and ascidians (Rinehart 1992; Faulkner 2000). As yet, there is little direct evidence for the function these compounds may have in their hosts, although some investigations support a role in chemical defence, as anti-fouling, anti-infective or anti-predation agents (Pennings et al. 1994; Waddell and Pawlik 2000). Similarly, marine larvae of chemically defended adults also possess anti-predatory compounds (Lindquist et al. 1992; Lindquist and Hay 1996; Bullard et al. 1999). As soft-bodied, sessile marine invertebrates, adult ascidians are especially susceptible to predation, and to fouling of their external surfaces. The production of potentially defensive secondary metabolites is widespread among certain groups of ascidians, and feeding studies have indicated these metabolites may confer anti-predatory protection (Paul et al. 1990; Davis 1991; Vervoort et al. 1998; Pisut and Pawlik 2002). Many ascidians, including E. turbinata, produce large, conspicuous larvae, which are released in daylight hours, enabling them to search for optimal settlement sites, but leaving them exposed to predators (Svane and Young 1989). Many such larvae may be chemically defended, being unpalatable to predators when presented in feeding experiments (Young and Bingham 1987; Lindquist et al. 1992).
Although there is little known of the mechanisms of production of secondary metabolites in marine invertebrates, there is evidence to support the theory that symbionts (especially bacterial ones) associated with the invertebrate host produce at least some of these compounds, either on their own, or in conjunction with their host (Bewley et al. 1996; Haygood et al. 1999; Davidson et al. 2001). For example, the contribution of prokaryotic, in this case, epibionts to surface anti-fouling by the production of bioactive substances has been particularly clearly displayed in studies by Holmström et al. (2002). This theory is also based on the fact that some secondary metabolites show close structural similarities to compounds produced by bacteria (Proksch et al. 2002). This is true of the ecteinascidins found in E. turbinata, which are structurally similar to the bacterially produced compound safracin B.
Although there are many secondary metabolite-producing ascidians, there are few studies on the presence of microorganisms associated with ascidians, and even fewer on their potential role in metabolite production (Wahl 1995; Lambert et al. 1996; Hirose et al. 1996, 1998; Rottmayr et al. 2001). However, there are now many descriptions of associations between bacteria and other marine invertebrate phyla, addressing both bacterial complement and activity. For example bacterial associations or specific symbioses have been described in sponges (Vacelet and Donadey 1977; Santavy et al. 1990; Schmidt et al. 2000; Webster et al. 2001), molluscs (Distel et al. 1991; McFall-Ngai 1999; Nyholm et al. 2000), bryozoans (Woollacott 1981; Haygood and Davidson 1997) and echinoderms (Holland and Nealson 1978; Kelly and McKenzie 1995). If E. turbinata contains a population of bacterial symbionts then it represents a potentially tractable model organism for investigating the role of bacteria in compound production and chemical defence, as secondary metabolites have been discovered (Rinehart et al. 1990) and its larvae are chemically protected (Young and Bingham 1987).
The aim of the present study was to analyse the bacterial flora associated with E. turbinata, and identify the most commonly occurring types both in larval and adult tissues. Molecular methods were employed in an effort to ensure that all associated bacteria were represented, not just those which could be grown by conventional bacterial isolation and culture techniques. In situ hybridisation analysis using probes to the 16S rRNA was carried out on larval and adult tissues of E. turbinata to provide localisation information, in order to confirm the specificity of the associated bacteria and to identify potentially symbiotic strains. This study was intended to provide an essential basis for any further investigation of the putative role of bacteria in secondary metabolite production.
Materials and methods
Tissue preparation and dissection
Ecteinascidia turbinata colonies were obtained from the Mediterranean Sea (Formentera, Spain) during the summers of 1999 and 2000, and from the Atlantic Ocean (Cádiz, Spain) in July 2000. They were transferred to a marine aquarium at a water temperature of 24°C. The tissue types used represented three different developmental states, as defined in Table 1. Embryos and larvae were obtained from ripe colonies, which were undergoing larval release, or by dissection from these colonies. They were rinsed several times in sterile, filtered seawater before DNA extraction or fixation. Zooids were pinched off the colony at the base and incubated in sterile seawater at 24°C for 24 h, with one change of water. They were then given a final rinse in sterile seawater before use. The stolon was rinsed in sterile seawater and prepared for fixation, or gently homogenised to liberate cells. The cells were washed by centrifugation, the supernatant discarded and the cells finally pelleted, ready for DNA extraction. All material for DNA extraction was snap frozen in liquid nitrogen and either processed immediately or stored at −80°C. Scallop muscle (Chlamys opercularis) was removed from an animal held in the aquarium, rinsed in sterile seawater and used as a control for 16S analysis.
DNA isolation, 16S rDNA PCR amplification and cloning
All DNA manipulations were performed essentially as described by Sambrook et al. (1989). DNA extraction from whole E. turbinata and larval tissue was carried out by homogenisation of tissue frozen in liquid N2 with a sterile mortar and pestle (except the stolon cells which were used directly). Lysis buffer (100 mM Tris-HCl, 10 mM EDTA, 150 mM NaCl, 100 µg RNase A ml−1) was added to the pulverised tissue and allowed to thaw. The homogenised tissue was immediately transferred to a sterile, 1.5-ml centrifuge tube, to which lysozyme (200 µg ml−1 final concentration) was then added, and the sample was incubated for 10 min at 37°C. Proteinase K and SDS (400 µg ml−1 and 1% final concentrations, respectively) were added, and the suspension was incubated for a further 30 min at 37°C. DNA was purified by phenol:chloroform extraction and ethanol precipitation. The DNA pellet was air-dried and stored in TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA) at −20°C.
Universal 16S rDNA bacterial PCR primers [forward, 8-AGRGTTTGATCMTGGCTCAG-27; reverse 1509-GKTACCTTGTTACGACTT-1494; primer position is according to Escherichia coli; Weisburg et al. (1991)] were used to amplify total bacterial 16S rDNA from the extracted DNA using a DNA engine (MJ Instruments, USA) thermocycler [cycled as follows: 94°C for 2 min, followed by 30 cycles of 55°C (30 s), 72°C (1 min 30 s) and 94°C (15 s), and a final elongation step of 72°C for 10 min]. 16S rDNA was amplified from several DNA dilutions to obtain optimal results, and a "no-DNA" control was run for each PCR mix. The PCR product (approximately 1400 base pairs) was confirmed by 1% agarose gel electrophoresis and ethidium bromide staining.
The PCR products from three separate reactions were purified from the agarose gel by the use of a centrifugal purification kit (Millipore Ultrafree-DA columns, USA) and ligated into pGEM-T Easy (Promega, USA) overnight at 4°C and then transformed into competent E. coli DH5α (Life Technologies, UK). Transformants were spread on LB agar, containing 50 µg ampicillin ml−1, 0.2 mM X-Gal and 0.16 mM IPTG for blue-white screening, and were incubated overnight at 37°C.
RFLP analysis of selected clones and sequencing
A total of 96 clones from each of the larval, zooid and stolon 16S rDNA PCR amplifications, and 40 from the scallop-control material, were picked at random and emulsified into a 50 µl PCR reaction containing M13/pUC universal primers (forward 5′-GTTTTCCCAGTCACGAC-3′; reverse 5′-CAGGAAACAGCTATG AC-3′). PCR reactions were cycled as follows: 94°C for 5 min, followed by 30 cycles of 50°C (30 s), 72°C (1 min 30 s), 94°C (15 s), and a final elongation step of 72°C for 10 min. Positive PCR products were ethanol precipitated and resuspended in sterile dH2O. Restriction fragment length polymorphism (RFLP) analysis was carried out using the restriction enzymes HaeIII and HhaI (Promega, USA). The restricted PCR products were electrophoresed through a 1× Tris-acetate (40 mM), EDTA (1 mM), 3% wide-range agarose gel (Sigma, USA) and stained with ethidium bromide. Kodak 1-D system and software (Kodak, USA) was used to capture the resultant RFLP patterns. Clones representative of each different pattern observed were isolated and grown up in LB broth with ampicillin (75 µg ml−1) overnight and stored frozen in 10% glycerol at −80°C. Plasmid DNA was isolated from selected clones for DNA sequence analysis according to the manufacturer's instructions (Qiagen Spin Mini-Preps, UK). DNA sequencing was performed using the M13 universal primers and ABI BigDye chemistry (PE Applied Biosystems, USA) and analysed on an ABI 377 DNA sequencer (PE Applied Biosystems, USA).
Phylogenetic analysis
Compiled DNA sequences from two or more independent clones of each RFLP type sequenced were aligned with the RDP II database (Maidak et al. 2001) using SEQUENCE MATCH and checked with CHIMERA CHECK to ensure that no sequence was chimeric. Phylogenetic inference of the novel 16S rDNA sequences was achieved by aligning the compiled 16S rDNA sequences with representative sequences obtained from the RDP II (Madiak et al. 2001) and GenBank using CLUSTALX (Thompson et al. 1997). Alignments were manually corrected and ambiguous positions removed. PAUP*4.0b8 (Swofford 2001) was used to infer phylogenetic trees using parsimony and neighbour-joining analysis according to the Kimura two-parameter model of nucleotide substitution (Kimura 1980). Bootstrap support for the inferred tree was established following re-sampling of 100 data sets based on neighbour-joining analysis.
In situ hybridisation with a universal probe
In situ hybridisation was carried out using standard methods (Amann et al. 1990; Manz et al. 1992; Wilkinson 1994). Tissue was fixed in 4% paraformaldehyde (PFA) in TBS (0.1 M Tris-HCl, 0.9% NaCl) with 0.1 M MOPS (pH 7.6) at room temperature for 30 min to 1 h. It was then embedded in paraffin wax, sectioned, and sections were mounted on gelatin-coated slides. Two panels of sections were mounted on each slide, placing alternate sections on each panel, to provide a test and control panel of the same area of the tissue. Prior to hybridisation, the slides were treated as follows. In brief, following rehydration, slides were rinsed in TBS and incubated in TBS containing 1 µg proteinase K ml−1 for 30 min. Slides were then post-fixed in 4% PFA in TBS for 10 min, rinsed and treated with 0.25% acetic anhydride in 0.1 M triethanolamine-HCl (pH 8.0), dehydrated, air dried and stored at −80°C.
Initial in situ hybridisation studies were carried out using a biotinylated universal bacterial 16S rRNA probe, EUB338, to identify sites of potential interest (Table 2), and a control probe, non-EUB338 (Table 2) (Amann et al. 1990). Other controls consisted of incubations with no probe and panels of attached bacterial cells (E. coli), which had been fixed and processed as per the sections. Hybridisation was carried out at 45°C for 3 h in buffer [0.9 M NaCl, 20 mM Tris-HCl (pH 7.2), 1× Denhardts, 0.1% SDS, 5 mM EDTA, 0.1 mg poly(A) RNA ml−1] with 2.5 ng µl−1 of probe. Hybridisation chambers with dual ports were used to prevent evaporation (Grace BioLabs, USA). Following hybridisation, the slides were washed two times for 15 min in wash buffer (0.9 M NaCl; 20 mM Tris-HCl, pH 7.2; 0.1% SDS) at 48°C. Binding of probe was visualised using FITC-Avidin-DN (Vector Labs, USA). Sections were mounted in Vectorshield (Vector Labs) and viewed on a Zeiss Axioskop with fluorescent attachments. For visualisation with alkaline phosphatase (AP), the slides were washed after hybridisation and incubated overnight in AP-conjugated anti-biotin antibody (Vector Labs) at 4°C. They were then washed and visualised using a BCIP/NBT–alkaline phosphatase substrate kit (Vector Labs).
Specific probes: dot blots and in situ hybridisation
Probes were designed to unique regions of the newly generated 16S rDNA sequences (Table 2; Table 3 for percentage dominance of these sequences). Probe specificity was examined by submitting the probe sequence to SEQUENCE MATCH and PROBE MATCH on the RDP II database (Maidak et al. 2001) and compared with the E. coli 16S rRNA secondary structure to check for optimal probe regions (Amann et al. 1995; Zheng et al. 1996).
Conditions for probe binding were confirmed by dot blot analysis. 16S rDNA PCR products from plasmid clones of the four most common strains (Table 3) (50 ng) and E. coli DNA were spotted onto a positively charged nylon membrane as per manufacturer's instructions (Schleicher and Schuell, Germany). The membranes were prehybridised at 46°C for 1 h in hybridisation buffer (as previously plus 2% non-fat milk powder), and then hybridised at 46°C for 3 h with 0.5 ng μl−1 of probe. Membranes were washed two times for 15 min at 50°C (wash buffer as before), blocked in NaPBS (20 mM sodium phosphate, 0.9% NaCl) with 1% BSA for 1 h and visualised using anti-biotin alkaline phosphatase and the BCIP/NBT-AP substrate kit (Vector Labs).
Following dot blots, hybridisation with the biotinylated EUB338, non-EUB338 and a novel probe to the sequence representing the most common strain (denoted type I-4; Table 2) was carried out on sections. Hybridisation conditions were 46°C for 3 h, in hybridisation buffer, containing 0%, 5% and 10% formamide, plus a no probe control of each, to optimise the stringency for each probe. Washes were carried out two times for 15 min (as previously) at 50°C. Slides with bacteria (E. coli) were also used as controls. Stolon tissue was screened on sections as this provided a more uniform basis for comparisons between the tests and controls. Based on the results of this initial hybridisation, further hybridisation studies were then carried out on whole embryos using a 5′-digoxigenin (DIG)-labelled type I-4 probe to improve clarity and consolidate the observations made with the specific probe. Hybridisation and wash buffers, and temperatures, were as above, with visualisation using anti-DIG conjugated to AP (Roche) and BCIP/NBT substrate reaction (Holland 1996).
Electron microscopy
Electron microscopy was carried out on larvae. Larvae were fixed in 1.5% glutaraldehyde in 0.2 M sodium cacodylate with 1.2% NaCl, post-fixed in 1% osmium tetroxide in the same buffer and embedded in Spurr's resin. The blocks were orientated so that sections were cut from the pharynx, an area potentially containing the cells of interest as identified by in situ hybridisation, and sectioned using an LKB ultramicrotome. Sections were mounted on grids and stained using uranyl acetate and lead citrate. Observations were made using a JEOL 100S transmission electron microscope.
Results
16S rDNA analysis and RFLPs
To minimise the contribution of non-specifically associated bacteria contaminating the total DNA pool, tissue was cleaned as far as possible before DNA extraction. The incubation of live animals in sterile seawater helped to depurate Ecteinascidia turbinata, which as a filter feeder would have contained large numbers of non-specifically associated bacteria. Near full-length 16S rRNA gene fragments were amplified from the total DNA extracted from zooid, larval and stolon tissues and from the control, scallop adductor muscle. After cloning and RFLP analysis, percentage dominance was assigned to the patterns observed in the three different tissues in order to determine any commonly occurring types. Four types were identified as occurring in all three E. turbinata tissues. These four patterns, denoted "types I, III, IV and VI", also represented the most abundant RFLP types (Table 3; Fig. 1). The type I pattern was the most common, representing between 42% and 67% of RFLP patterns observed in the three ascidian tissue types. Types VI, III and IV were the remaining three most abundant patterns, respectively (Table 3; Fig. 1). The scallop control tissue showed no type I, III or VI patterns, but a type IV pattern was observed. This, together with the sequence analysis, identified type IV as more likely to be a commonly occurring marine bacterium, rather than having a specific relationship with E. turbinata.
HaeIII RFLP (restriction fragment length polymorphism) analysis of cloned 16S rDNA. In both panels, lane 1 (lanes numbered from left to right) is a 1 kb DNA ladder. A RFLPs from larval tissue showing the common type I (lanes 2–5, 7–9, 12), III (lane 10) and IV (lane 11) RFLP patterns. B RFLPs from stolon tissue showing the type I (lanes 4, 6–8, 10), III (lane 12), IV (lane 5) and VI (lanes 3, 9, 11) patterns
Phylogenetic Inference
Representative clones of the commonly occurring type I, III, IV and VI patterns were sequenced. Accession numbers for each of the sequences denoted by these patterns are shown in Figs. 2 and 3. Alignment of these sequences with the RDP II database indicated that the 16S rRNA gene sequences of types I, III and VI were unrelated to any particular bacterial species in the database. The type IV pattern was shown to have 98% sequence similarity with Pseudomonas fluorescens (D86001). Type I could only be reliably assigned as a member of the Gram-negative, γ-proteobacteria. Within this group type I showed a closer relationship to members of the Legionellales (Fig. 2). However, the unsupported branching order of the tree and long branch length of this sequence make this a tentative assignment. The closest sequence affiliation to type I was a Coxiella-like symbiont (AF521888) harboured by Haemaphysalis concinnae ticks, which shared 80.6% similarity across 1044 bp (excluded from phylogenetic analysis due to the short sequence length), and an unidentified γ-proteobacteria (AB015255) isolated from the sediment of a deep-sea trench, which shared 80.3% similarity across 1451 bp. Types III and VI shared the greatest nucleotide sequence similarity with each other (92.2%), while the next highest sequence similarity was with members of the Spiroplasmataceae (81.5% and 77.6%, respectively). Phylogenetically, they could be reliably assigned as members of the Mollicutes, but only with weak support as members of the Spiroplasmataceae (Fig. 3). We have tentatively assigned type III and VI as Spiroplasma-like.
Neighbour-joining analysis of 1288 bp of unambiguously aligned 16S rDNA gene sequences of type I and IV, with representative members of the γ-proteobacteria and an α-proteobacterial outgroup. The values (>50%) at the nodes show the bootstrap support based on neighbour-joining analysis of 100 re-sampled data sets. The scale bar represents 0.01 substitutions per nucleotide position. Nucleotide accession numbers are shown alongside each representative organism
Neighbour-joining analysis of 1289 bp of unambiguously aligned 16S rRNA gene sequences of type III and VI, with representative members of the Mollicutes and Bacilli as an outgroup. The values (>50%) at the nodes show the bootstrap support based on neighbour-joining analysis of 100 re-sampled data sets. The scale bar represents 0.1 substitutions per nucleotide position. Nucleotide accession numbers are shown alongside each representative organism
In situ hybridisation
Hybridisation of the universal bacterial EUB338 16S rRNA probe was observed in larval sections (Fig. 4A–E), stolon material and at low levels in zooid sections. Positive in situ hybridisation was found in cells of approximately 10–12 µm diameter, which demonstrated a granular appearance (Fig. 4B, D, E). These cells usually had an unstained area at one pole, which was thought to be the host cell nucleus. In larvae, the cells were predominantly found in the pharynx (Fig. 4A, C) and appeared to be freely circulating, rather than associated with large tissue masses. In zooid tissue, only a few scattered positively hybridising cells were observed. These cells were located in the region of the pharynx and possibly associated with the blood vessels. As in the larvae, they did not appear to be anchored to basement membranes or other cells. The background fluorescence was relatively high, although hybridisation-positive cells were easily distinguishable on the test sections, but were clearly not present in the controls.
Ecteinascidia turbinata. In situ hybridisation on sections of larvae. A A longitudinal section through a whole larva stained with haematoxylin and eosin (HE) (major anatomical features visible: s siphons; t tail; p pharynx; g gut). Cells which hybridised with probe EUB338 were often located within the pharyngeal area (boxed) or the body sinuses. B, D, E In situ hybridisation of larvae (pharyngeal area) with the probe EUB338. Fluorescent cells indicate areas of probe binding (arrows). B A cluster of cells in the pharynx with fluorescent inclusions. C HE-stained section to indicate the location of the hybridised cells in the pharyngeal primordia. The section is through a similar area to the hybridised section shown in panel B, and to that boxed in panel A. D Individual fluorescent inclusions are visible in these host cells. E Hybridised cell with individual inclusions. Scale bars: 250 μm (A), 10 μm (B, D, E), 75 μm (C)
Dissected portions of stolon and budding tissue (asexual reproduction) were also examined by fluorescent in situ hybridisation. With these preparations, the background fluorescence under the FITC filter was too high to facilitate accurate interpretation of hybridisation. Consequently, subsequent hybridisation experiments used the histochemical AP visualisation system. Hybridising cells were found scattered throughout the stolon and bud tissue. The hybridisation-positive cells in the stolon and bud tissue had the same general appearance as observed in the larvae, with the hybridised part of the cell exhibiting granular staining and an unstained, approximately polar region, presumably representing the host nucleus (see Fig. 6A). The non-EUB338 probe gave very low levels of background binding using the AP system, such that it was almost indiscernible when compared to the intensity of EUB338 binding. In all cases where the control was buffer without probe, the cells hybridised in the test sections could not be observed in the control sections.
Dot blots and specific probes
All probe sequences are shown in Table 2. Probes were named according to the RFLP pattern type that they represented. The dot blots indicated the type I-4 probe was specific for the type I sequence, binding only to type I 16S rDNA and not to that of the other common types isolated from E. turbinata, or to E. coli DNA, under standard hybridisation conditions (Fig. 5). Therefore, this probe was used for subsequent experiments. Probes to the type III sequence (probe type III-4 and type III-1) were also positive in dot blots, although one showed additional cross reactivity (Fig. 5). These probes have not yet been characterised on tissue. The type I-4 probe tested on stolon sections bound to very similar cells as those highlighted by the universal eubacterial probe EUB338 (Fig. 6A), suggesting that these were the same bacteria and that they possessed the type I sequence (Fig. 6B). Subsequent hybridisation with DIG-labelled type I-4 probe gave a positive reaction in whole mounts of embryos and larvae (Fig. 6C, D, E), providing further evidence that these bacteria represented the type I strain. The cells containing bacteria in the embryos are larger than those observed in larval and adult tissues (approximately 30 µm, although size is variable), but they still have the same granular appearance and the bacterial inclusions are of a similar size (Fig. 6D, E). The cells appeared to be generally distributed in the embryo, rather than in a particular region.
Dot blot hybridisation of oligonucleotide probes. Each panel was spotted with 50 ng 16S rDNA from one of the commonly occurring bacterial types from Ecteinascidia turbinata (I, III, IV, VI), from Escherichia coli, or TE buffer (neg), as denoted in panel A. A Hybridisation with EUB338. Probe has hybridised to all the DNA. B Hybridisation with probe type I-4 showing a reaction only with the type I DNA. C Hybridisation with probe type III-1. This probe hybridises with type III and VI DNA, but also with type I DNA. D Hybridisation with probe type III-4. This probe hybridises with type III and VI DNA only
Ecteinascidia turbinata. In situ hybridisation with EUB338 and type I-4 specific probe using alkaline phosphatase visualisation. A In situ hybridisation of stolon sections with the EUB338 probe. B Hybridisation of stolon sections with type I-4 specific probe. A black deposit shows the area of probe binding in similar cells to the EUB338 probe (arrows). C Whole mount in situ hybridisation of an E. turbinata embryo using DIG-labelled type I-4 probe. Large cells in the embryo are positively stained (arrow). D High magnification of cells in an embryo hybridised with the type I-4 probe. The cells are large, with a granular appearance and an unstained area probably representing the host nucleus. E High magnification of a disrupted host cell, showing putative bacteria hybridised with the type I-4 probe. Scale bars: 10 μm (A, B), 200 μm (C), 30 μm (D, E)
Electron microscopy
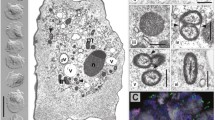
Putative cells in the larval pharynx, which correlated with those observed by fluorescent in situ hybridisation, were identified and termed "bacteriocytes" (Fig. 7A). They had a very similar morphology to the other major type of blood cells, except for the presence of putative bacteria. The bacteriocytes frequently had cytoplasmic protrusions extending from the cell surface and all appeared to be free in the blood vessels or body sinuses. In most cells the nuclei were heterochromatic and unremarkable. Mitochondria were frequently observed. There were some obvious strands of rough endoplasmic reticulum, but the cells were most notable for the high densities of what are apparently polyribosomes distributed throughout the cytoplasm (Fig. 7A, B). There was no obvious Golgi apparatus. The putative bacteria were rounded and approximately 1–2 µm in diameter (Fig. 7A). Their membrane structure suggested that they were Gram-negative bacteria. The chromosomal DNA was usually well distributed and only partially condensed. The bacteria appeared to be intracellular symbionts rather than pathogens or ingested bacteria. This is suggested from a number of observations: (1) there is no obvious endocytotic behaviour by the host cells, (2) no bacteria were observed being lysed, (3) there is no obvious pathology of the host cell in the smaller bacteriocytes, (4) the presence of so many polyribosomes suggests a large amount of synthetic activity by the cell that is intended for internal consumption rather than export, and (5) the bacteria appear to be of a uniform type and the same type has been observed in different individual larvae.
Ecteinascidia turbinata. Transmission electron microscopy of the pharyngeal region of a larva. A Larval cell ("bacteriocyte") showing numerous putative bacterial inclusions (arrows) (n nucleus of host cell). B Higher magnification of inclusions showing membrane detail. Scale bars: 2 μm (A), 1 μm (B)
Discussion
This study provides one of the first reports of putative endosymbiotic bacteria in an ascidian. The combination of 16S rRNA gene analysis, in situ hybridisation and electron microscopy, provides strong evidence for a symbiotic association between these bacteria and the host cells. The observation of identical bacterial strains, with similar levels of dominance, and of similar cell types, in both adult and larval tissues suggests that these cells are a consistent and important feature of Ecteinascidia turbinata. In situ hybridisation, based on the obtained sequence data of the type I RFLP, strongly supports the contention that the type I strain is the observed bacterium. Under the naming convention for so far uncultivable, novel bacterial species, the type I bacteria has been assigned "Candidatus Endoecteinascidia frumentensis".
The 16S rDNA sequence analysis of dominant bacterial strains in E. turbinata has identified three novel strains. "Candidatus Endoecteinascidia frumentensis" falls into the γ-proteobacteria subdivision. Although it is only distantly affiliated to any other known strains within this group, it bears similarities to species in the Legionella and Oceanospirillum subgroups. Many previously described marine endosymbionts belong to the γ-proteobacteria subdivision, with representatives from sponges (Webster et al. 2001), bivalves (Distel et al. 1988; Durand et al. 1996; Sipe et al. 2000), oligochaetes (Dubilier et al. 2001) and bryozoans (Haygood and Davidson 1997), covering a range of ecological niches and putative activities.
The type III and VI RFLP patterns appear to represent novel "Spiroplasma-like" bacteria. Mollicutes are often found as parasitic organisms in vertebrates and invertebrates, although there is increasing evidence to support both commensal and saprophytic roles. There are few records of them in marine invertebrates, although they have been identified in a bryozoan (Boyle et al. 1987). The significance of finding a Gram-negative bacterium (the γ-proteobacterium), together with two Spiroplasma-like bacteria, in E. turbinata is not known, but a similar co-occurrence has been reported in aphids (Fukatsu et al. 2000, 2001). Localisation of the Mollicutes in E. turbinata would be a valuable step toward understanding if they are simply saprophytic, opportunistic pathogens or have a more profound role as secondary symbionts in conjunction with the identified putative endosymbiont. The current study focussed on what at this time appears to be the primary symbiont, as this clearly represented a specific and consistent association, with high numbers of bacteria present in the tissue.
The use of 16S rRNA clone libraries to study bacterial communities is now a standard procedure for analysis of bacteria in environmental samples and has enabled much of the current progress in the area of bacterial symbiosis. Whilst being suitable for surveying of bacterial community composition, this technique can lead to misinterpretation if attempting to quantify those types present, and any analysis of results must take note of a number of possible failings. For example, Polz and Cavanaugh (1998) found that templates containing GC-rich combinations in the priming sites might preferentially amplify. However, the results presented here indicate such a high predominance of one novel strain, in three different tissues, that it is unlikely this could be explained by PCR bias. Similarly, bacterial types possess different copy numbers of the 16S rRNA gene, leading to potential weighting of amplification results. For example, the spiroplasma-like strains identified probably possess only one copy of the gene, whereas "Candidatus Endoecteinascidia frumentensis" may contain two or more. This could also lead to amplification bias, and results must be considered with this in mind. In the case of E. turbinata, the high predominance of strains in all tissues examined, and the subsequent use of in situ hybridisation, helps to negate the possibility of interpretation errors.
In situ hybridisation using universal and specific probes suggested that bacteria were located within host cells in adult and larval tissues. It appears that these cells are free, possibly in the blood in zooids, and present at higher densities in the embryos and larvae. In embryos they are present in larger cells throughout the developing tissue. In larvae, they are clustered in the pharynx and body sinuses. This high density of bacteria supports the dominance observed in the 16S analysis of larval tissue, and this result, together with in situ hybridisation using the specific probe to the type I sequence, provides strong evidence that "Candidatus Endoecteinascidia frumentensis" is the strain identified in situ.
The significance of the high numbers of bacteriocytes in the pharynx is not clear, as the larvae are not feeding, but it suggests that the bacteria may be present at sites of optimal nutrition (gaseous and nutrient exchange) as soon as the larva has settled and the siphons have opened. However, it is also possible that a high density in this area is a side effect of the developing blood system and that the bacteriocytes are clustered here due to blind-ending vessels that are still growing to form the pharynx. Higher densities of putative bacteriocytes are also found in the stolon, which becomes engorged with cells as colonies regress. Few cell types are present in this tissue, suggesting that cells have de-differentiated, or are primarily for storage, ready to support the next phase of budding. Hybridised cells are much harder to detect in zooids, where tissue is diffuse and cell density low. However, several putative bacteriocytes have been observed by in situ hybridisation, with similar characteristics as before, that is, not anchored, but freely circulating, possibly in the blood. The variability in distribution and abundance between different tissues of the host species indicates the importance of targeting a range of tissues for analysis, providing a clearer overall assessment of the potential host–symbiont relationship.
The larval pharynx was the target tissue for TEM studies based on the in situ hybridisation results. The presence of bacteriocytes, of similar size and apparent morphology as the cells illuminated by hybridisation in sections of the pharynx, suggests that the TEM results involve the same cells as those seen by hybridisation. A double membrane appears to surround the bacteria, suggesting that they are Gram-negative, in concurrence with the phylogenetic analysis. However, the method of fixation was not optimal, and further investigations of the fine structure of the bacteriocytes would help to confirm this observation.
Intracellular symbionts can be transmitted to the next generation directly through reproductive tissue (vertical transmission), or from a free-living form (environmental transmission), and both mechanisms have been described in marine invertebrates. One of the most extensive studies of environmental transmission of symbionts has been carried out on the association between the squid, Euprymna and Vibrio bacteria (McFall-Ngai 1999; Nyholm et al. 2000). Vertical transmission has been observed in the ascidian Diplosoma similes (Hirose 2000) and in some bivalves (Cary and Giovannoni 1993; Krueger et al. 1996; Sipe et al. 2000). The presence of a consistent bacterial association through many of the developmental stages of E. turbinata, strongly suggests that these bacteria are vertically transmitted, although the exact mechanism of transfer of bacteria from adult to oocyte or embryo is not yet known. Intraovarial transmission of symbionts is also observed in insects, and the changes in morphology between the bacteria-containing cells in E. turbinata, from embryos through to larvae and zooids, is also similar to that observed in a number of insect groups (Douglas 1989; Sauer et al. 2002).
That "Candidatus Endoecteinascidia frumentensis" is endocytotic and likely to be vertically transmitted also suggests that it is unlikely to be cultivable in simple media. Culturing experiments, followed by 16S RFLP analysis of 22 bacterial strains derived from larvae and 9 from zooids, indicated that bacteria similar to the type I, III and VI strains did not grow under standard conditions (data not shown). The difficulty in culturing symbiotic bacteria is a barrier to the further elucidation of their roles, and methods to overcome this problem are required. Schmidt et al. (2000) have managed to grow a symbiont of Theonella using specific culture conditions. Efforts to culture intracellular symbionts have been limited, and attempts to culture host cells have generally failed. Ways of re-addressing this problem would greatly improve the understanding of bacterial symbioses in general.
As yet, we cannot assign a role for "Candidatus Endoecteinascidia frumentensis", although the presented evidence supports a symbiotic relationship with E. turbinata. Many marine symbionts are thioautotrophs from anoxic environments, oxidising sulphur and fixing CO2. Indeed a complete sulphur cycle involving two symbionts has recently been elegantly demonstrated in a marine oligochaete (Dubilier et al. 2001). In the case of E. turbinata it is possible that utilisation of certain nutrients in a potentially anoxic mangrove environment may be advantageous, although this species is known to grow and thrive in a range of habitats. E. turbinata larvae and adults are vulnerable to predation and/or fouling, and the production of noxious compounds may act as an effective defensive strategy. The fact that this species is chemically defended, known to contain secondary metabolites and has now also been shown by the current study to house a large population of potentially endosymbiotic bacteria may be circumstantial. However, it is possible that a symbiosis between the ascidian and a secondary metabolite-producing bacterium may be an important factor in the biology of E. turbinata. There is little evidence to support the possibility that the bacteria identified are responsible for the production of the secondary metabolites found in E. turbinata, and this is currently under further investigation.
References
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA–targeted oligonucleotide probes with flow cytometry for analysing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Amann RI, Ludwig W, Schleifer K-H (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Berrill NJ (1932) Ascidians of the Bermudas. Biol Bull (Woods Hole) 62:77–88
Bewley CA, Holland ND, Faulkner DJ (1996) Two classes of metabolites from Theonella swinhoei are localised in distinct populations of bacterial symbionts. Experentia 52:716–722
Bingham BL, Young CM (1991) Larval behaviour of the ascidian Ecteinascidia turbinata Herdman: an in situ experimental study of the effects of swimming on dispersal. J Exp Mar Biol Ecol 145:189–204
Bingham BL, Young CM (1995) Stochastic events and dynamics of a mangrove root epifaunal community. Mar Ecol 16:145–163
Boyle PJ, Maki JS, Mitchell R (1987) Mollicute identified in novel association with aquatic invertebrate. Curr Microbiol 15:85–89
Bullard SG, Lindquist NL, Hay ME (1999) Susceptibility of invertebrate larvae to predators: how common are post-capture larval defenses. Mar Ecol Prog Ser 191:153–161
Cary SC, Giovannoni SJ (1993) Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc Natl Acad Sci USA 90:5695–5699
Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG (2001) Evidence for the biosynthesis of bryostatins by the bacterial symbiont "Candidatus Endobugula sertula" of the bryozoan Bugula neritina. Appl Environ Microbiol 67:4531–4537
Davis AR (1991) Alkaloids and ascidian chemical defense: evidence for the ecological role of natural products from Eudistoma olivaceum. Mar Biol 111:375–379
Distel DL, Lane DJ, Olsen GJ, Giovannoni SJ, Pace B, Pace NR, Stahl DA, Felbeck H (1988) Sulphur-oxidising bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol 170:2506–2510
Distel DL, DeLong EF, Waterbury JB (1991) Phylogenetic characterisation and in situ localisation of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridisation. Appl Environ Microbiol 57:2376–2383
Douglas AE (1989) Mycetocyte symbiosis in insects. Biol Rev 64:409–434
Dubilier N, Mulders C, Ferdelman T, de Beer D, Pernthaler A, Klein M, Wagner M, Erseus C, Thierman F, Krieger J, Giere O, Amann R (2001) Endosymbiotic sulphate-reducing and sulphide-oxidising bacteria in an oligochaete worm. Nature 411:298–302
Durand P, Gros O, Frenkiel L, Prieur D (1996) Phylogenetic characterization of sulfur-oxidizing bacterial endosymbionts in three tropical Lucinidae by 16S rDNA sequence analysis. Mol Mar Biol Biotechnol 5:37–42
Faulkner DJ (2000) Marine pharmacology, vol 77. Antonie van Leeuwenhoek, pp 135–145
Fukatsu T, Nikoh N, Kawai R, Koga R (2000) The secondary endosymbiotic bacterium of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol 66:2748–2758
Fukatsu T, Tsuchida T, Nikoh N, Koga R (2001) Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol 67:1284–1291
Haygood MG, Davidson SK (1997) Small subunit rRNA genes and in situ hybridisation with oligonucleotides specific for the bacterial symbionts of the larvae of the bryozoan Bugula neritina and proposal of 'Candidatus Endobugula sertula'. Appl Environ Microbiol 6:4612–4616
Haygood MG, Schmidt EW, Davidson SK, Faulkner DJ (1999) Microbial symbionts of marine invertebrates: opportunities for microbial biotechnology. J Mol Microbiol Biotechnol 1:33–43
Hirose E (2000) Plant rake and algal pouch of the larvae in the tropical ascidian Diplosoma similis: an adaptation for vertical transmission of photosynthetic symbionts Prochloron sp. Zool Sci (Tokyo) 17:233–240
Hirose E, Aoki M, Chiba K (1996) Fine structures of tunic cells and distribution of bacteria in the tunic of luminescent ascidian Clavelina miniata (Ascidiacea, Urochordata). Zool Sci (Tokyo) 13:519–523
Hirose E, Maruyama T, Cheng L, Lewin RA (1998) Intra- and extracellular distribution of photosynthetic prokaryotes, Prochloron sp., in a colonial ascidian: ultrastructural and quantitative studies. Symbiosis 25:301–310
Holland ND, Nealson KH (1978) The fine structure of the echinoderm cuticle and subcuticular bacteria of echinoderms. Acta Zool 59:169–185
Holland PWH (1996) Whole mount in situ hybridisation applicable to Amphioxus and other small larvae. In: Ferraris JD, Palumbi SR (eds) Molecular zoology. Advances, strategies and protocols. Wiley-Liss, New York, pp 476–483
Holmström C, Egan S, Franks A, McCloy S, Kjelleberg S (2002) Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol Ecol 41:47–58
Kelly MS, McKenzie JD (1995) Survey of the occurrence and morphology of sub-cuticular bacteria in shelf echinoderms from the north east Atlantic Ocean. Mar Biol 123:741–756
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Krueger DM, Gustafson RG, Cavanaugh CM (1996) Vertical transmission of chemoautotrophic symbionts in the bivalve Solemya velum (Bivalvia: Protobranchia). Biol Bull (Woods Hole) 190:195–202
Lambert G, Lambert CC, Waaland JR (1996) Algal symbionts in the tunics of six New Zealand ascidians (Chordata, Ascidiacea). Invertebr Biol 115:67–78
Lindquist N, Hay ME (1996) Palatability and chemical defense of marine invertebrate larvae. Ecol Monogr 66:431–450
Lindquist N, Hay ME, Fenical W (1992) Defense of ascidians and their conspicuous larvae: adult vs. larval chemical defenses. Ecol Monogr 62:547–568
Maidak BL, Cole JR, Lilburn TG, Parker CT Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res 29:173–174
Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H (1992) Phylogenetic oligodeoxynucleotide probes for the major subclasses of Protobacteria: problems and solutions. Syst Appl Microbiol 15:593–600
McFall-Ngai MJ (1999) Consequences of evolving with bacterial symbionts: insights from the squid–Vibrio associations. Annu Rev Ecol Syst 30:235–256
Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ (2000) Establishment of an animal–bacterium association: recruiting symbiotic Vibrios from the environment. Proc Natl Acad Sci USA 97:10231–10235
Paul VJ, Lindquist N, Fenical W (1990) Chemical defense of the tropical ascidian Atapazoa sp. and its nudibranch predators Nembrotha spp. Mar Ecol Prog Ser 59:109–118
Pennings SC, Pablo SR, Paul VJ, Duffy JE (1994) Effects of sponge secondary metabolites in different diets in feeding by three groups of consumers. J Exp Mar Biol Ecol 180:137–149
Pisut DP, Pawlik JR (2002) Anti-predatory chemical defenses of ascidians: secondary metabolites or inorganic acids? J Exp Mar Biol Ecol 270:203–214
Polz MF, Cavanaugh CM (1998) Bias in template-to-product rations in multitemplate PCR. Appl Environ Microbiol 64:3724–3730
Proksch P, Edrada RA, Ebel R (2002) Drugs from the sea—current status and microbiological implications. Appl Microbiol Biotechnol 59:125–134
Rinehart KL (1992) Secondary metabolites from marine organisms. In: Chadwick DJ, Whelan J (eds) Secondary metabolites: their function and evolution. Ciba Found Symp 171:236–254
Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG (1990) Ecteinascidin 729, 743, 745, 759A, 759B and 770, potent antitumour agents from the Caribbean tunicate Ecteinascidia turbinata. J Org Chem 55:4512–4515
Rottmayr EM, Steffan B, Wanner G (2001) Pigmentation and tunic cells in Cystodytes dellechiajei (Urochordata, Ascidiacea). Zoomorphology 120:159–170
Sambrook E, Fritsch F, Maniatis T (1989) Molecular cloning. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
Santavy DL, Willenz P, Colwell RR (1990) Phenotypic study of bacteria associated with the Carribean sclerosponge, Ceratoporella nicholsoni. Appl Environ Microbiol 56:1750–1762
Sauer C, Dudaczek D, Holldobler B, Gross R (2002) Tissue localisation of the endosymbiont bacterium "Candidatus Blochmannia floridanus" in adults and larvae of the carpenter ant Camponotus floridanus. Appl Environ Microbiol 68:4187–4193
Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG (2000) Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel delta-proteobacterium, "Candidatus Entotheonella palauensis". Mar Biol 136:969–977
Sipe AR, Wilbur AE, Cary SC (2000) Bacterial symbiont transmission in the wood-boring shipworm Bankia setacea (Bivalvia: Teredinidae). Appl Environ Microbiol 66:1685–1691
Svane IB, Young CM (1989) The ecology and behaviour of ascidian larvae. Oceanogr Mar Biol Annu Rev 27:45–90
Swofford DL (2001) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer, Sunderland, Mass.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Vacelet J, Donadey C (1977) Electron microscope study of the association between some sponges and bacteria. J Exp Mar Biol Ecol 30:301–314
Vervoort HC, Pawlik JR, Fenical W (1998) Chemical defense of the Caribbean ascidian Didemnum conchyliatum. Mar Ecol Prog Ser 164:213–220
Waddell B, Pawlik JR (2000) Defense of Caribbean sponges against invertebrate predators. I. Assays with hermit crabs. Mar Ecol Prog Ser 195:125–132
Wahl M (1995) Bacterial epibiosis on Bahamian and Pacific ascidians. J Exp Mar Biol Ecol 191:239–255
Webster NS, Wilson KJ, Blackall LL, Hill RT (2001) Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl Environ Microbiol 67:434–444
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:607–703
Wilkinson DG (1994) In situ hybridisation: a practical approach. Rickwood D, Hames BD (eds) Practical approaches series. IRL Press, Oxford
Woollacott RM (1981) Associations of bacteria with bryozoan larvae. Mar Biol 65:155–158
Young CM, Bingham BL (1987) Chemical defense and aposematic coloration in larvae of the ascidian Ecteinascidia turbinata. Mar Biol 96:539–544
Zheng D, Alm EW, Stahl DA, Raskin L (1996) Characterisation of universal small-subunit rRNA hybridisation probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol 62:4504–4513
Acknowledgements
This work was funded under the EU MAST-CRAFT scheme, project code MAS3-CT098-0179. Thanks to A. Keay, C. Barbero, S. Martin, H. Margot and B. Kukurtku for help with maintenance and supply of animals.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.P. Thorpe, Port Erin
Rights and permissions
About this article
Cite this article
Moss, C., Green, D.H., Pérez, B. et al. Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: phylogenetic and in situ hybridisation analysis. Marine Biology 143, 99–110 (2003). https://doi.org/10.1007/s00227-003-1060-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-003-1060-5