Abstract
Foodborne diseases caused by bacterial pathogens pose a widespread and growing threat to public health in the world. Rapid detection of pathogenic bacteria is of great importance to prevent foodborne diseases and ensure food safety. However, traditional detection methods are time-consuming, labour intensive and expensive. In recent years, many attempts have been made to develop alternative methods for bacterial detection. Biosensors integrated with molecular imprinted polymers (MIPs) and various transducer platforms are among the most promising candidates for the detection of pathogenic bacteria in a highly sensitive, selective and ultra-rapid manner. In this review, we summarize the most recent advances in molecular imprinting for bacterial detection, introduce the underlying recognition mechanisms and highlight the applications of MIP-based biosensors. In addition, the challenges and future perspectives are discussed with the aim of accelerating the development of MIP-based biosensors and extending their applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Foodborne diseases due to consumption of food contaminated with harmful chemicals or microbes have been considered as the most vital food safety issues and pose threat to the public health. According to the World Health Organization (WHO), there are a projected 600 million cases of foodborne illnesses accompanied with 420,000 deaths annually [1]. In the USA, it is estimated that the cost due to foodborne illnesses is over $50 billion annually [2]. The most commonly reported foodborne pathogenic bacteria (e.g., Escherichia coli, Listeria monocytogenes, Salmonella) [3] are responsible for more than 91% of foodborne outbreaks in the USA [4]. Thus, development of rapid and sensitive methods to detect and identity foodborne pathogenic bacteria is of great importance.
Detection of foodborne pathogenic bacteria mainly relies on laboratorial tests such as conventional culturing method [5], polymerase chain reaction (PCR) [6] and enzyme-linked immunosorbent assay (ELISA) [7]. Although these methods have a high sensitivity and specificity, their application is still limited because they are time-consuming, labour intensive and expensive. Recently, molecular imprinting technology coupled with biosensor has been realized as a promising approach to detect pathogenic bacteria because it can detect target bacteria in a portable sensing system with a quick response and high specificity.
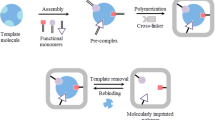
Molecular imprinting polymers (MIPs) are recognition materials prepared by polymerization of monomers, crosslinkers and other necessary constituents (i.e., initiators and porogens) in the presence of the target analyte called “template” [8]. After the removal of the template, free cavities that conserve the size, shape and chemical functionality [9] of the analyte are created, allowing specific recognition of the target analyte. In contrast to natural receptors such as antibodies or enzymes, MIPs are highly physically and chemically stable and can be reused for a long period. Therefore, MIP-based sensors have been employed for the detection of a large variety of chemical and biological hazards in food products, ranging from small molecules (mycotoxins [10, 11] and illegal food additives [12, 13]) to macromolecules (allergenic proteins and bacterial exotoxins) [14, 15] and even larger analytes (pathogenic bacteria and viruses) [16,17,18].
A series of reviews in the literature comprehensively discussed the fundamental aspects of molecular imprinting [19,20,21]. Reviews on the application of molecular imprinting technology in macromolecule and cell imprinting can also be found [22, 23]. However, to the best of our knowledge, MIP coupled with biosensor for the detection of pathogenic bacteria has not been reviewed yet. Numerous studies regarding bacterial imprinting have been conducted [24,25,26,27,28] and need to be summarized to get the up-to-date knowledge of this innovative detection method.
This review gives an overview of current trends in the development of MIP-based biosensors for the detection of foodborne pathogenic bacteria. Recognition mechanisms of MIP and its interaction with bacteria are introduced first. Then, we summarize various bacterial imprinting methods with an emphasis on surface imprinting. After that, key applications of MIP-based biosensors to detect pathogenic bacteria are highlighted. Finally, we propose the remaining challenges and limitations of current imprinting methods as well as future directions to improve molecular imprinting technology that can assist the detection of pathogenic bacteria.
Principle of MIPs in bacterial recognition
MIPs, known as “artificial receptors” or “plastic antibodies”, are functional porous materials with tailor-made binding sites that can selectively recognize template and discriminate it from the co-existing structural analogs. These binding sites are prepared by polymerization of crosslinkers and the pre-polymerized complex (i.e., functional monomers and template). During the process of pre-polymerization, functional monomers and template co-assemble through covalent/non-covalent interactions [19]. The polymerization process (usually heating or UV initiated) starts upon the addition of the crosslinker. Therefore, pre-polymerized complex will be immobilized in the highly cross-linked matrix, resulting in “freezing” of the orientation of functional groups of the template. After removal of the template, the binding sites that memorize the geometry and functional groups of target analyte will be generated. Then, the recognition of target analyte will be achieved through a combination of complementary size and morphology as well as chemical interactions (i.e., covalent, non-covalent and semi-covalent interactions [29]) (Fig. 1). The formation of chemical interactions requires specific intermolecular distance; therefore, recognition site that has similar geometrical properties to the template can facilitate this process. At the same time, the arrangement of functional groups can rule out non-specific binding that based on size and shape similarity.
In contrast to small molecules, bacteria are complexes that contain a large variety of molecules. Therefore, it is challenging to design specific covalent bindings. In addition, covalent interaction requires precise binding and association of the target, resulting in a longer response time. To achieve easier binding and dissociation between MIPs and bacteria, non-covalent interactions [30] such as hydrogen bonds, electrostatic affinity, ionic and hydrophobic interactions, van der Waals forces, and π–π interactions are preferred [23]. Since these chemical interactions occur between functional groups of monomer and molecules on the cell surface, the selection of appropriate monomers that have complementary functional groups is of great importance. Bacteria are negatively charged with different functional groups (e.g., hydroxyl, carboxyl, phosphoryl and amide) existing on the cell surface [31]. Thus, electrostatic interaction and hydrogen bonds are the most common chemical interactions that can be used to build recognition sites. Taking advantage of electrostatic interaction, various cationic monomers have been used by several researchers to develop polymers with high affinity and selectivity to bacteria. For instance, the charge heterogeneity of bacterial surface was encoded into polymer matrix using cationic monomer (methacrylatoethyl trimethyl ammonium chloride) and zwitterionic monomer [3-dimethyl (methacryloyloxyethyl) ammonium propane sulfonate] [28]. During self-assembly, cationic monomer interacted with negative groups on bacterial surface while zwitterionic monomer filled in the gap between the recognition sites. Possessing chemical information of the template, the polymer could differentiate target bacteria from three other different bacterial species, indicating its high affinity and selectivity. Recently, Gür and co-authors used Cu(II)-bounded Au nanoparticle as the monomer to imprint E. coli [32]. The specific recognition was achieved by electrostatic interaction between Cu(II) and negative groups of bacterial surface. In addition to electrostatic interaction, a large variety of monomers with functional groups (e.g., amino, hydroxyl, vinyl, carbonyl and carboxyl) have been selected to form hydrogen bond with molecules on bacterial surface. Methacrylic acid is a frequently used hydrogen bonding monomer because carboxyl group can act as hydrogen donor and acceptor at the same time [33]. Taking advantage of this, Razavilar and co-workers developed a MIP using methacrylic acid as the functional monomer for the detection of Salmonella enterica serotype Typhi [17]. Another functional monomer, dopamine, can form hydrogen bond with bacteria due to the existence of amino groups [34]. Recently, Bezdekova and others prepared magnetic particles coated with polydopamine (PDA) for the detection of Staphylococcus aureus [35]. The obtained polymer particles could specifically bind target bacteria, indicating their high bacterial affinity and selectivity.
Molecular imprinting methods in sensing pathogenic bacteria
According to the size of the template, different imprinting strategies such as bulk imprinting and surface imprinting can be selected to prepare MIPs. For bulk imprinting, cavities (recognition sites) are distributed all over the polymer matrix [36], which favours the absorption and release of small molecules. However, this approach is not suitable for bacterial imprinting because the analytes are large in size and could be hindered in the highly cross-linked matrix. Therefore, surface imprinting is more appropriate for bacterial imprinting.
Currently, surface imprinting can be divided into two categories according to the recognition unit of bacteria (Fig. 2). One is the cell membrane molecular impinging that selectively imprints cell membrane subunits, such as glycans and proteins [23]. In contrast, whole cell imprinting method imprints whole cell membrane instead of specific molecules, which conserves more surface information of the template [23]. Currently, the creation of bacterial recognition sites in most MIPs is achieved by using whole cell imprinting method. Both advantages and disadvantages of different imprinting methods are compared in Table 1 [37,38,39].
Whole cell imprinting
Microcontact/stamp imprinting
Microcontact imprinting has been extensively used to prepare MIPs that can detect different types of pathogenic bacteria [16, 40,41,42]. The processing procedure is straightforward: (1) to deposit bacterial suspension onto the top of a flat solid support substrate (stamp); (2) to deposit a thin layer of viscous prepolymer onto the surface of transducer; (3) to bring the stamp into contact with the polymer and cure the sandwich construction; and (4) to remove the stamp and template. Therefore, the obtained cavities on the surface of the polymer matrix can be used for bacterial recognition. A typical stamp imprinting procedure is shown in Fig. 3a.
Stamp imprinting method was first introduced to imprint yeast (Saccharomyces cerevisiae) cells on the surface of thin polyurethane layer by Dickert and Hayden in 2001 [44]. With the development of imprinting technology, this method was successfully applied for bacterial imprinting by Zare’s group [45]. Taking this a step further, Idil and co-workers [41] developed a capacitive biosensor with a stamp-imprinted gold electrode to detect E. coli. The stamp was prepared by depositing E. coli suspension onto the 3-aminopropyltriethoxysilane-modified glass slide. Subsequently, this stamp was brought into contact with the polytyramine gold electrode covered with reaction mixture (2-hydroxyethyl methacrylate, ethylene glycol dimethacrylate, N-methacryloyl-L-histidine methyl ester-Cu (II) complex, α-α'-azoisobutyronitrile) and polymerized under UV light. After treated with phosphate-buffered saline and lysozyme, the surface of prepared gold electrode was characterized by cyclic voltammetry, atomic force microscopy (AFM) and scanning electron microscopy (SEM). Using cyclic voltammetry, the successful insulation of gold electrode was confirmed. In addition, the results obtained from AFM and SEM indicated that the imprinting process was successful. The linear relationship between changes in capacitance and E. coli concentration was obtained from 1.0×102–1.0×107 CFU/mL, with a limit of detection (LOD) of 70 CFU/mL.
A further advancement in stamp imprinting is the use of artificial stamp. Latif and co-authors [46] prepared an artificial stamp by casting uncured polydimethylsiloxane (PDMS) onto the surface of bacteria-imprinted polyurethane. After polymerization, a replica stamp of E. coli was obtained, and it was used for further imprinting. Even though the recognition sites were prepared using replica stamp instead of bacterial stamp, they can distinguish between E. coli W strain (ATCC 9637) and E. coli B strain (EC 11303). There are a variety of bacteria that are highly pathogenic and not suitable for direct imprinting; therefore, not needing to apply real pathogenic bacteria is the main advantage of artificial stamp. In addition, artificial stamp is more robust compared to natural bacterial cells and can be used to produce numerous MIPs by using a single stamp.
Drop coating
Drop coating (sediment imprinting) is a fast surface imprinting approach using sedimentation by gravity and it has been applied in imprinting fragile bioanalytes, such as cells. Generally, the prepolymer is deposited and spin coated on the support substrate, after which the template solution is dropped onto the prepolymer, resulting in the generation of recognition sites. This method was first developed by Seifner and others to imprint red blood cells on the surface of a thin polyvinylpyrrolidone layer [47]. Recently, an example of quartz crystal microbalance (QCM) sensing system using drop coating for E. coli detection was presented by Samardzic and co-authors [48]. In their experiment, diphenylmethane-4,4′-diisocyanate, poly-(4-vinylphenol) and phloroglucinol were dissolved in tetrahydrofuran, and this mixture was then pre-polymerized at 70 °C for 15 min to form a gel-like oligomer. The oligomer was spin coated onto the electrode to form a layer of 300 nm height, onto which the bacterial suspension was dropped coated. After polymerization for 48 h, the template was removed by immersing the electrode in water for an hour. Using AFM imaging, cavities and trapped E. coli could be observed on the imprinted polyurethane, indicating the successful imprinting of the target. The MIP was used as the recognition element in the QCM sensor and real-time detection for E. coli was achieved by monitoring the changes in the frequency. The detection limit was 1.6 × 108 cells/mL and this sensor also showed a high selectivity towards the imprinted template (E. coli B strain) and a very close bacterial strain (E. coli W strain).
Spieker and co-authors implemented this technique into a QCM sensor for the detection of Bacillus cereus [49]. In this study, the affinity of five different polymers (polystyrene, polyacrylate, polyvinylpyrrolidone, polyacrylamide and polyurethane) towards B. cereus was screened at first. After imprinting, the number of recognition sites on the surface of polymer film was observed by light microscope. Taking these two factors into consideration, polyurethane with the highest affinity and the greatest number of recognition sites was used for imprinting. MIPs prepared by stamp imprinting and drop coating were observed using light microscope. Bacteria were more evenly distributed on the stamp-imprinted polyurethane, indicating that stamp imprinting was superior to drop coating in this condition.
Pickering emulsion interfacial imprinting
Pickering emulsion is a particle-stabilized emulsion, which is either oil-water or water-oil [50]. Bacteria tend to assemble at oil-water interface. Taking advantage of this, Shen and co-workers [43] developed an imprinting method by using template bacteria as the stabilizer in the emulsion polymerization process (Fig. 3b). After imprinting, the polymer beads could be used to capture and detect bacteria. In this study, negatively charged bacteria templates (E. coli and Micrococcus luteus) were assembled with the positively charged prepolymer that contains vinyl groups, and the obtained bacteria-prepolymer complexes were used as the stabilizer to form a stable emulsion (crosslinkers in water). Polymerization was then initiated by free radicals, resulting in the combination of prepolymer and polymer beads. After the removal of bacteria template, recognition sites were left on the surface of polymer beads. Zhao and others developed a similar system in which L. monocytogenes-imprinted water-soluble CdTe quantum dots (QDs) were formed using Pickering emulsion polymerization method [50]. Both N-acrylchitosan and QDs were used to form a complex via amide bond, which was obtained through the interaction between amino bond in N-acrylchitosan and carboxyl group in the functionalized QDs. Positively charged N-acrylchitosan-QD complex was assembled with negatively charged L. monocytogenes, and the resulting network was used to stabilize the Pickering emulsion in which the oil phase contains crosslinkers (trimethylolpropane trimethacrylate and divinylbenzene) and initiator (benzoyl peroxide). Two phases were mixed well through vigorous manual shaking, after which, a stable emulsion was formed and used to form solid polymer beads. The obtained beads were washed with 10% acetic acid, 1% sodium dodecyl sulphate (SDS), water and methanol to remove L. monocytogenes. Numerous recognition sites were formed on spherical-shaped MIPs (polymer beads). In addition, a MIP-based sensor was used to monitor the fluorescence intensity so as to detect and quantify L. monocytogenes. The limit of detection was 103 CFU/mL and further confirmed in real food samples (milk and pork), in which the visible colour changes were observed along with the increased concentration of L. monocytogenes.
Electropolymerization
Electropolymerization is a process that electroactive monomers are polymerized in the presence of the template [51], forming a uniform MIP layer on the surface of the electrode. The template bacteria are doped into the polymer matrix, and the recognition sites remain on the surface of the electrode after the removal of the template. Therefore, MIPs obtained using this method are coupled with electrochemical sensor platform for the detection of target analyte. This imprinting method is highly useful because the thickness of the film can be easily manipulated by controlling the electrochemical conditions, such as the number of cycles, scan rate and potential range [52]. Electroactive materials with different conducting abilities such as conductive polymers, insulators or non-conductive polymers [52] can be used for electropolymerization. Rebinding of the analyte will change electron transfer behaviour between the recognition sites and electrode [53], resulting in changes in capacitance and/or resistance that can be easily monitored. A schematic overview of this procedure is shown in Fig. 3c.
One of the first approaches to integrate electropolymerization into bacterial imprinting was proposed by Tokonami and co-workers in 2013 [54]. In this study, negatively charged Pseudomonas aeruginosa were introduced into a polypyrrole layer on the surface of the electrode, which was incorporated with a QCM sensor. To achieve the successful removal of the template, this film was pretreated with lysozyme and octyl phenyl ether and then overoxidized in sodium hydroxide solution (0.1 M). This system could detect P. aeruginosa in the range of 103–109 CFU/mL in sterilized water within 3 min. However, after application in real food sample (apple juice), the linear range decreased to 107–109 CFU/mL, which might be due to the interference of sample matrix components (e.g., sodium and carbohydrate).
In another study, E. coli O157:H7 were imprinted on PDA using electropolymerization method, and the resulted film was integrated into an electrochemiluminescence (ECL) biosensor for selective capture of the analyte [34]. To quantitatively detect the bacteria, E. coli O157:H7 polyclonal antibodies tagged N-doped graphene quantum dots were used and they could specifically bind to E. coli O157:H7 on the surface of the electrode. In the presence of K2S2O8, an ECL signal could be generated and detected. A good linear relationship between ECL intensity and E. coli O157:H7 concentration was obtained between 101 and 107 CFU/mL, with the LOD of 8 CFU/mL.
Colloidal imprinting
Colloidal imprinting is a method that bacterial cells can be imprinted by encapsulation in inorganic shells [55]. This process is achieved by depositing silica layer on bacterial surface using sol-gel process. Therefore, the obtained silica shell can maintain the size and shape of the template. After fragmentation, shell fragments that possess antibody-like ability can be used for bacterial recognition. Borovička and co-workers reported the first example of colloidal imprinting for creating yeast imprints [56]. In this study, detection of yeast cells could be visualized by tagging silica shells with 3-aminopropyltriethoxysilane and rhodamine B isothiocyanate as well as staining the yeast cells with perylene. The same group further developed “photothermal colloidal antibody” by depositing gold nanoparticles before the formation of the silica layer [57]. The obtained imprints could selectively destroy the yeast cell in a non-toxic way without using antimicrobials.
Recently, Zhang and co-authors fabricated artificial antibodies using surface imprinting method and then set up a sandwich ELISA for the detection and disinfection of S. aureus [58]. Capture antibodies (cAbs) were prepared by electrochemically polycondensation and detection antibodies (dAbs) were fabricated through colloidal imprinting. After encapsulation of S. aureus using silica, the surface of the shell was then deposited with CeO2 nanoparticles. The templates were removed by calcination, after which the hollow shells were cracked by ultrasonic treatment. The ability of these artificial antibodies to recognize S. aureus was evaluated using sandwich ELISA. The target pathogen was captured by cAbs and then recognized by dAbs. Successful detection could be visualized by the occurrence of blue colour and the amount of S. aureus could be indicated by the changes in colour intensity. Moreover, the captured bacteria could be disinfected in situ by electrochemical oxidation.
Cell membrane molecular imprinting
The inherent properties of bacteria such as large molecular size, complexity of the components and heterogeneous shapes limit the binding affinity of whole cell imprinting method. In contrast to whole cell imprinting, cell membrane molecular imprinting method uses the most characteristic components on the cell surface instead of the whole cells. This method mimics the natural antibody-antigen interaction [19, 59] that uses epitope to recognize its receptor; as a result, the obtained MIPs can detect the analyte. A representative structure of S. aureus, namely protein A (SpA), was selected as the imprinting template by several researchers for the detection of S. aureus. Xue and co-workers prepared SpA-imprinted polyacrylamide gel beads (SpA-IPGB) using inverse-phase suspension polymerization method [60]. SEM results showed that SpA-IPGB had spherical shape with numerous macropores on the side surface, indicating the successful removal of the template. Both binding capability and selectivity of the obtained gel beads towards SpA were evaluated by comparing the amount of SpA and its analogs in the solution before and after measuring the absorption by a UV spectrophotometer. In addition, plating assay showed that SpA-IPGB had a high recognition ability and specificity towards S. aureus compared with other types of bacteria (e.g., E. coli and Streptococcus thermophilus), with an absorption quantity of 103–104 CFU/g of gel beads.
Recently, Khan and others designed an electrochemical biosensor based on MIP for the detection of SpA [61]. In their experiment, SpA was electropolymerized with 3-aminophenol using cyclic voltammetry and the polyaminophenol film was formed on top of single-walled carbon nanotube film that was attached to the screen-printed electrodes. The electrochemical impedance spectroscopy was used to evaluate the electrochemical conditions of polymerization, after which the number of cyclic voltammetry cycles was adjusted to get the optimal film thickness. After imprinting, the template was successfully removed using proteinase K and the cavities on the surface could be used to selectively capture SpA. This sensor reached a LOD of 0.60 nM in 2-(N-morpholino)ethanesulphonic acid monohydrate buffer. A relatively higher detection limit of 16.83 nM was determined in tap water, indicating that inorganic ions could interfere with the performance of this sensor. The successful development of this sensor demonstrated its potential in the detection of S. aureus.
Analytical detection methods coupled with molecular imprinting systems
A biosensor is an analytical device that is capable of capturing bioanalytes and producing measurable signal for detection and quantification. Recognition and transduction elements are two major components of a biosensor [62]. Bioreceptors such as antibodies, aptamers and bacterial phages are used as recognition units. Due to its superior sensitivity, selectivity and stability, MIP has been widely used for the construction of biosensors. The analytical properties between these types of biosensor and MIP-based biosensor are compared and summarized in Table 2 [30, 63,64,65,66]. For MIP-based biosensor, the target analyte can be recognized and captured by imprinted cavities (recognition element), after which the transducer will transfer the biochemical information to quantifiable output signals. MIP-based biosensor can be classified into optical, piezoelectric, electrochemical biosensors [67] according to the signals generated by the transducer. A variety of MIP-based biosensors for the detection of pathogenic bacteria have been summarized in Table 3.
Optical biosensor
Optical biosensors have been extensively used for the detection of pathogenic bacteria due to its high sensitivity, accuracy, simplicity and cost-effectiveness. The specific biorecognition events on the surface of the optical transducer result in changes in absorption, transmission, reflection, refraction, phase, frequency or polarization of light [74]. Generally, optical biosensors can be divided to label-free and label-based modes [75]. For a label-free detection, generation of optical signal is based on direct interaction of analyte complex and the transducer. For a label-based mode, the analyte needs to be labelled first and optical signal could be measured using a colorimetric, fluorescent or luminescent method [75]. In recent years, optical biosensors coupled with different transducers have been used to detect bacterial pathogens. Among these, surface plasmon resonance and fluorescence are more frequently used due to their outstanding sensitivity.
Fluorescence biosensor
MIP-based fluorescence biosensor monitors the changes in fluorescence signal (brightness, characteristic wavelength, anisotropy and lifetime) during biomolecular interaction between the analyte and MIPs [76]. Usually, fluorescent intensity is measured and the intensity is proportional to the concentration of the analyte, thus realizing quantification of the target. Based on the non-fluorescent characteristic of bacteria, indirect fluorescence detection [19] is used for detection and quantification of the analyte.
One method is to label the analyte using a fluorescent dye and monitor the changes in fluorescence intensity at specific excitation and emission wavelength. Shan and co-authors manufactured polypyrrole-coated microspheres imprinted with E. coli O157:H7, in which specific binding of the analyte was confirmed by monitoring the changes in fluorescence intensity in bacterial supernatant [77]. E. coli O157:H7 was labelled with fluorescent dye, namely 5′-bi-1H-benzimidazole, which could preferentially bind to bacterial double-strained DNA. The imprinted microbeads absorbed E. coli O157:H7 from bacterial suspension, reducing the fluorescence intensity of the supernatant. In addition, the imprinted microspheres could distinguish E. coli O157:H7 from serotype E. coli O157:HNM because of its high selectivity. Taking this a step further, the same group optimized the imprinting method based on bacteria-imprinted 96-well microplates, which enabled rapid and high throughput detection of E. coli O157:H7 [78]. In this study, E. coli O157:H7 was labelled with SYTO 9 and fluorescence intensity for each well was monitored. Recently, Bezdekova and others coated PDA layer on the surface of magnetic particles for specific detection of S. aureus [35]. Bacteria bound to the particle were stained with propidium iodide, wherein the changes in fluorescence intensity could indicate bacterial adsorption as well as bacterial concentration in the sample (Fig. 4). The developed magnetic particles were applied to detect S. aureus in milk from cow with mastitis, leading to a low detection limit of 1 × 103 CFU/mL.
Molecular imprinted polymer (MIP)-based fluorescence biosensor for the detection of pathogenic bacteria. a Fluorescence intensity of MIPs or non-imprinted polymers versus Staphylococcus aureus concentration (I) or binding time (II). Schematic procedure for imprinting is shown at the right side of the figure. b Detection of S. aureus in buffer (I, II), milk (III, IV), milk from healthy cow (V, VI), milk from cow with mastitis (VII, VIII), and spiked cooked rice (IX, X) using polydopamine coated non-imprinted or S. aureus surface imprinted magnetic particles (Reproduced with permission from [35], copyright 2020, Elsevier)
Another method is to incorporate fluorophores/fluorescent monomer into the polymer matrix [19]. Therefore, the fluorescent characteristics of the polymer will be changed upon its interaction with the analytes. The unique optical properties of QDs and gold nanoparticles make them attractive and these two materials have been used by several researchers for the development of fluorescence biosensors. Zhao and colleagues fabricated a polymer with CdTe QDs as the recognition element for a fluorescence biosensor using Pickering emulsion polymerization method [50]. Visualized quantification of L. monocytogenes was achieved by measuring fluorescence intensity of bacterial solutions with different concentrations. Chen and others developed a ECL biosensor based on PDA layer and nitrogen-doped graphene quantumdots (N-GQDs) [34]. E. coli O157:H7 was first captured by PDA and then detected by polyclonal antibody–labelled N-GQDs. Quantification of E. coli O157:H7 was achieved by measuring ECL signal generated by N-GQDs with K2S2O8. Based on the ability of gold nanoparticles to separate excitation and emission wavelengths, Gültekin and colleagues fabricated a fluorescence biosensor for the detection of B. cereus spores [79]. As the main component of bacterial endospores, dipicolinic acid (DPA) could specifically bind to the recognition site. Therefore, the number of spores could be quantified by measuring the changes in fluorescence intensity.
Surface plasmon resonance biosensor
Surface plasmon resonance (SPR) biosensor is a label-free optical sensor that can offer real-time quantification of the analyte based on the detection of refractive index changes [75] at the surface of a conductive interface [19]. SPR biosensors have been employed to detect various pathogenic bacteria [80, 81,82,83] and the recognition element for these biosensors is mainly antibody. Recently, Gür and co-workers prepared a MIP-based SPR nanosensor for the detection of E. coli [32]. The sensing process was achieved by the interaction between E. coli and gold nanoparticles on the surface of gold SPR chip (Fig. 5a). The authors examined the response of the sensor by using different concentrations of E. coli (0.5 × 101–1 × 103 CFU/mL) and the changes in refractive index were proportional to cell concentration (Fig. 5b). Taking advantage of surface-imprinted chip and SPR technology, this method achieved a low detection limit at 1 CFU/mL. In addition, different bacteria solutions (S. aureus, Klebsiella pneumoniae and P. aeruginosa) were applied to the biosensor to assess its selectivity towards the analyte. As expected, the addition of E. coli produced a much greater response than any other bacteria species. In another study, Perçin and others developed a SPR nanobiosensor for selective detection of Salmonella enterica serotype Paratyphi A using microcontact imprinting method [42]. N-Methacryloyl L-histidine methyl ester-Cu(II) complex and SPR chip were separately used as the recognition and transduction elements in the sensing system, providing high selectivity and sensitivity to this device. The linear range between 2.5 × 106 and 15 × 106 CFU/mL was obtained with a LOD of 1.4 × 106 CFU/mL. The ratio of SPR response (△R) for S. Paratyphi A-MIP and -NIP SPR chips was 81, indicating the successful imprinting. This SPR sensor also had a high specificity for S. Paratyphi A against other bacteria, including E. coli, S. aureus and Bacillus subtilis.
MIP-based surface plasmon resonance (SPR) biosensor for the detection of pathogenic bacteria. a Schematic illustration of a MIP-based SPR biosensor for the detection of Escherichia coli. b Concentration dependent real-time sensorgram of MIP-based SPR biosensor (I) and comparison of non-imprinted and E. coli surface imprinted biosensors towards E. coli (II) (Reproduced with permission from [32], copyright 2019, Elsevier)
Electrochemical biosensor
Electrochemical biosensors are devices that contain electrochemical transducers and transform the biochemical interaction between analyte and recognition element into measurable electrochemical signals. Electrochemical biosensors can be separated into amperometric, potentiometric, impedimetric/capacitance and conductometric subgroups based on the measurement mode and response signals [62]. Among electrochemical biosensors, amperometric and impedimetric/capacitance sensors are the most commonly reported sensor types due to their outstanding properties in terms of rapid response, high sensitivity, cost-effectiveness and ease of production. Amperometric biosensors operate under conditions of constant potential and measure current variations due to oxidation or reduction of an electroactive species [84]. In impedimetric/capacitance-based detection, the changes in dielectric constant or resistance can be measured when biorecognition takes place on the surface of the electrode.
Mugo and colleagues prepared a capacitive sensor based on layer by layer assembly for rapid detection of E. coli K12 [68]. The electrochemical transducer was fabricated by depositing polyaniline-doped phenylboronic acid onto the surface of multi-walled carbon nanotube/cellulose nanocrystal nanoporous conductive film. The transducer was then coated with E. coli-imprinted poly(methacrylic acid) and the whole construction was used as working electrode in the sensing system (Fig. 6a). The impedance decreased along with the concentrations of E. coli ranging from 0 to 6.22 × 106 CFU/mL, with an LOD of 8.7 ± 0.5 CFU/mL (Fig. 6b). Depending on its high sensitivity for the detection of E. coli in spiked orange juice (3.84 × 106 and 9.6 × 106 CFU/mL), this sensor demonstrated a significant potential to detect E. coli in real beverage samples. In another study, Lahcen and others reported the development of overoxidized polypyrrole film for selective recognition of B. cereus spore. This film was then integrated onto the surface of carbon paste electrode so as to establish an electrochemical biosensor [73]. In this study, cyclic voltammetry was used to monitor the process of electropolymerization as well as specific binding of B. cereus spores. Under optimized polymerization conditions, current intensity of the redox probe decreased significantly along with the increased number of spores ranging from 102 to 105 CFU/mL. This is ascribed to the binding of spores in polypyrrole film that blocks electron transfer between working electrode and redox probe. Based on the hypothesis that phage-imprinted cavities could resemble phage receptors and recognize phage and host bacteria, Ertürk and co-workers made use of microcontact imprinting to construct a MIP-based capacitive biosensor for the detection of E. coli phage and E. coli [69]. The changes in total capacity offered a satisfactory linearity with E. coli phage and E. coli concentrations at 1.0 × 101–1.0 × 105 PFU/mL and 1.0 × 102–1.0 × 107 CFU/mL and with LOD values of 10 PFU/mL and 100 CFU/mL, respectively.
a Schematic illustration of MIP-based electrochemical biosensing system. b Cyclic voltammetry of E. coli imprinted electrode at different bacterial concentrations (I) and linear relationship between capacitance changes and E. coli concentration (II) (Reproduced with permission from [68], copyright 2020, John Wiley and Sons)
Piezoelectric biosensor
Quartz crystal microbalances (QCMs) have been used as the transduction element in biosensors based on measuring changes in resonance frequency of the quartz crystal resonator in response to mass changes caused by specific binding of the analyte to the recognition element [51]. MIP-based QCM sensor is prepared by depositing MIP layer onto the surface of QCM. This biosensing platform has been widely used for the detection and quantification of different types of bioanalytes [85, 86,87,88]. Detection of pathogenic bacteria based on QCM biosensor has been reported by several groups. For example, Tokonami and others combined QCM with P. aeruginosa-imprinted polypyrrole film to construct a QCM sensor [54]. To improve the ability of MIP to capture P. aeruginosa, the authors used dielectrophoresis to concentrate the analyte in the vicinity of the recognition sites. This design exhibited excellent sensitivity and it could detect P. aeruginosa at 103 CFU/mL within 3 min. Furthermore, this MIP-based biosensor demonstrated a high selectivity because it could discriminate P. aeruginosa from a bacterial mixture. Latif and co-authors used a MIP film developed by stamp imprinting of polyurethane on the surface of a QCM to sense E. coli and B. subtilis [46]. The frequency of the sensor decreased immediately upon exposure to different concentrations of E. coli, demonstrating the high sensitivity of this device. In the meanwhile, germination process of B. subtilis spore was monitored by the QCM sensor, in which the frequency continuously decreased because more B. subtilis were included in the cavities as the function of time. In another study, Yilmaz and co-authors prepared QCM and SPR biosensors by microcontact imprinting for the detection of E. coli (Fig. 7a) [72]. A linear response was obtained in the range of 0.5–4.0 and 0.5–3.0 McFarland for QCM and SPR sensors, respectively. The number of bacteria could be determined by comparing the turbidity of bacterial suspension to that of a series of McFarland standards. The matched McFarland value could then be used to represent bacterial concentration. The sensitivity of the developed sensor was investigated by comparing response time and detection limit towards the analyte. QCM sensor had a shorter response time within 56 s and a lower LOD of 3.72 × 105 CFU/mL, indicating that QCM is superior to SPR in terms of sensitivity (Fig. 7b).
a Schematic illustration of the preparation of SPR and quartz crystal microbalance (QCM) sensors using microcontact imprinting method. b Real-time responses and calibration curves of SPR (I, II) and QCM (III, IV) biosensors at different E. coli concentrations (Reproduced with permission from [72], copyright 2015, Elsevier)
Thermal sensing platforms
Heat-transfer method (HTM) has been used to develop biosensor platforms in combination with MIPs for detection of neurotransmitters [89, 90], drugs [91] and microorganisms [70, 92]. The changes in thermal resistance can be measured when target molecules bind to MIP layer at solid-liquid interface (Fig. 8) [92, 93]. Taking advantages of this label-free detection method, Grinsven and co-authors developed a biomimetic sensor integrated with a polyurethane surface-imprinted aluminium chip for the detection of E. coli [70]. The designed sensor platform showed a high selectivity and was able to distinguish live E. coli from dead E. coli as well as S. aureus. In addition, the authors monitored the response of this sensor by using different bacterial concentrations in the range of 1.0 × 104 to 1.0 × 107 CFU/mL and demonstrated the high sensitivity of this platform (Fig. 9a). The detection limit was reported to be 3.5 × 104 CFU/mL. HTM has various favourable traits such as simplicity, cost-effectiveness and straightforward data interpretation. However, the inherent noise of this platform may affect the thermal signal, resulting in a relatively high detection limit [92]. To have a better sensitivity for the detection of E. coli, the same group utilized thermal wave transport analysis (TWTA) method to construct a thermal sensing system with the same imprinting method [92]. TWTA examines the changes in phage and amplitude for thermal wave instead of thermal resistance, resulting in a less response time and a higher sensitivity compared to HTM [94]. Nine bacteria species were used as the templates to prepare the MIP layer. A measurable phase shift in the propagated wave was observed when analytes specifically bound to the surface of the polyurethane-coated aluminium chip (Fig. 9b). This biosensor exhibited a high sensitivity (LOD: 1 × 104 CFU/mL) and high selectivity towards the analyte. It could distinguish the target bacteria in the presence of 99-fold excess of mixed bacterial solution. In another study, Cornelis and co-workers proposed a new strategy to improve the performance of HTM sensor by replacing the heating unit with a planar meander-type metallic structure, which served as both a heater and a temperature sensor [71]. In this study, a surface-imprinted polyurethane stainless steel chip was used as the recognition element. Using the modified HTM method, E. coli was determined with a detection limit of 100 CFU/mL and a working range between 102 and 106 CFU/mL in both phosphate-buffered saline and apple juice. Moreover, this biosensor showed a high specificity to E. coli O157:H7 as other bacteria (i.e., Citrobacter freundii, Hafnia alvei, Serratia marcescens and E. blattae) did not produce any significant change in the sensorgram.
Schematic illustration of MIP-based thermal sensing platform used for bacterial detection (Reproduced with permission from [92], copyright 2017, American Chemical Society)
a Detection of E. coli based on heat-transfer method. Real-time thermal resistance upon exposure to solutions with different E. coli concentrations (I) and the calibration curve of thermal resistance changes versus the concentration of bacterial cells (II) (Reproduced with permission from [70], Copyright 2016, American Chemical Society). b Detection of E. coli based on thermal wave transport analysis. Real-time temperature response upon exposure to solutions with different E. coli concentrations (I) and the calibration curve of phase shift changes versus the concentration of bacterial cells (II) (Reproduced with permission from [92], copyright 2017, American Chemical Society)
Challenges and future perspectives
To date, great advances have been achieved of using MIPs as specific recognition element for the detection of pathogenic bacteria. However, this technique remains a scientific challenge and a practical problem of great importance. To address the existing problems and make this method a truly practical approach, a few recommendations are proposed.
First, the intrinsic properties (e.g., large size, fragile and complex structure, fluidity and poor stability in organic solvents) of bacteria make the molecular imprinting process more challenging compared to that of small molecules. Researchers have implemented new imprinting strategies, such as stamp imprinting and cell membrane molecular imprinting, together with novel MIP materials (e.g., nanoparticles, quantum dots) to improve the selectivity and sensitivity of bacteria-imprinted polymers. The uniqueness of each target requires tailor-made polymer and imprinting method so that rational design for MIPs at molecular level is of great importance, which is currently absent in the literature. The interaction between polymer and bacteria are assumed to be non-covalent, such as hydrogen bond, electrostatic affinity, as well as ionic and hydrophobic interaction. Therefore, the underlying mechanisms of bacterial recognition by MIPs need to be further elucidated.
Second, food matrices are complex systems containing various molecules that can affect bacterial detection especially when bacteria appear at relatively low concentrations. These molecules derived from food matrices can be absorbed to the recognition sites, changing the physical/chemical properties of the recognition element, thereby reducing the selectivity and sensitivity of the receptor. In many previous studies, MIP-based sensors were applied for bacterial detection in buffer solutions rather than real food samples. Thus, the capability (e.g., sensitivity, selectivity) of these platforms to detect pathogenic bacteria in food commodities remains largely unknown. Additional studies need to be performed to test the impact of food components on the performance of MIP-based sensing platforms.
Lastly, only a few studies used cell membrane molecular imprinting to prepare MIPs for bacterial detection, wherein protein was selected as the template. However, protein has poor stability and is easy to lose its spatial structure during the harsh imprinting process, resulting in rearrangement of functional groups. Therefore, other stable molecules such as glycans and epitope peptides on the cell membrane that can be chemically synthesised are promising candidates for molecular imprinting. To identify appropriate glycans and epitope peptides for imprinting process, more studies on cell biology are required to fully understand the structure and components of bacteria cell membrane.
Conclusion
Pathogenic bacteria in agri-food system generate a great concern to food industry and public health, calling for rapid detection methods and innovative controlling strategies. Mimicking natural molecular recognition events, molecular imprinted artificial receptors have been used as the recognition elements in biosensing systems for bacterial detection. Along with rapid development of molecular imprinting technology, various imprinting methods have been developed to generate bacteria-imprinted polymers. These methods can be divided into whole cell imprinting and cell membrane molecular imprinting according to the template used (whole cell or specific cell membrane molecule). The selectivity of MIPs prepared by whole cell imprinting is reliant on both chemical recognition and physical recognition, while the ones prepared by cell membrane molecular imprinting are mainly based on chemical interaction. Different optical, electrochemical and piezoelectric biosensors have been integrated with MIPs to construct biosensors for bacterial detection. High sensitivity and selectivity of the MIP-based platform demonstrated the great potential to detect bacteria in various systems, including food commodities. MIP-based biosensors can be used for industrial and commercial applications for bacterial detection.
References
World Health Organization. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiolgy reference group 2007–2015. Geneva: World Health Organization; 2015.
Scharff RL. Economic burden from health losses due to foodborne illness in the United States. J Food Prot. 2012;75:123–31. https://doi.org/10.4315/0362-028X.JFP-11-058.
Law JWF, Mutalib NSA, Chan KG, Lee LH. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2014;5:770. https://doi.org/10.3389/fmicb.2014.00770.
Yang L, Bashir R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol Adv. 2008;26:135–50. https://doi.org/10.1016/j.biotechadv.2007.10.003.
Arthur TM, Bosilevac JM, Nou X, Koohmaraie M. Evaluation of culture- and PCR-based detection methods for Escherichia coli O157:H7 in inoculated ground beef. J Food Prot. 2005;68:1566–74. https://doi.org/10.4315/0362-028X-68.8.1566.
Zhang Y, Zhu L, Zhang Y, He P, Wang Q. Simultaneous detection of three foodborne pathogenic bacteria in food samples by microchip capillary electrophoresis in combination with polymerase chain reaction. J Chromatogr A. 2018;1555:100–5. https://doi.org/10.1016/j.chroma.2018.04.058.
Pang B, Zhao C, Li L, Song X, Xu K, Wang J, et al. Development of a low-cost paper-based ELISA method for rapid Escherichia coli O157:H7 detection. Anal Biochem. 2018;542:58–62. https://doi.org/10.1016/j.ab.2017.11.010.
Eersels K, Lieberzeit P, Wagner P. A review on synthetic receptors for bioparticle detection created by surface-imprinting techniques - from principles to applications. ACS Sensors. 2016;1:1171–87. https://doi.org/10.1021/acssensors.6b00572.
Ertürk G, Mattiasson B. Molecular imprinting techniques used for the preparation of biosensors. Sensors. 2017;17:288. https://doi.org/10.3390/s17020288.
Sergeyeva T, Yarynka D, Piletska E, Linnik R, Zaporozhets O, Brovko O, et al. Development of a smartphone-based biomimetic sensor for aflatoxin B1 detection using molecularly imprinted polymer membranes. Talanta. 2019;201:204–10. https://doi.org/10.1016/j.talanta.2019.04.016.
Guo W, Pi F, Zhang H, Sun J, Zhang Y, Sun X. A novel molecularly imprinted electrochemical sensor modified with carbon dots, chitosan, gold nanoparticles for the determination of patulin. Biosens Bioelectron. 2017;98:299–304. https://doi.org/10.1016/j.bios.2017.06.036.
Hu X, Cai Q, Fan Y, Ye T, Cao Y, Guo C. Molecularly imprinted polymer coated solid-phase microextraction fibers for determination of Sudan I-IV dyes in hot chili powder and poultry feed samples. J Chromatogr A. 2012;1219:39–46. https://doi.org/10.1016/j.chroma.2011.10.089.
Feng F, Zheng J, Qin P, Han T, Zhao D. A novel quartz crystal microbalance sensor array based on molecular imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Talanta. 2017;167:94–102. https://doi.org/10.1016/j.talanta.2017.02.001.
Ashley J, Shukor Y, D’Aurelio R, Trinh L, Rodgers TL, Temblay J, et al. Synthesis of molecularly imprinted polymer nanoparticles for α-casein detection using surface plasmon resonance as a milk allergen sensor. ACS Sensors. 2018;3:418–24. https://doi.org/10.1021/acssensors.7b00850.
Ahari H, Hedayati M, Akbari-adergani B, Kakoolaki S, Hosseini H, Anvar A. Staphylococcus aureus exotoxin detection using potentiometric nanobiosensor for microbial electrode approach with the effects of pH and temperature. Int J Food Prop. 2017;20:S1578–87. https://doi.org/10.1080/10942912.2017.1347944.
Fu K, Zhang H, Guo Y, Li J, Nie H, Song X, et al. Rapid and selective recognition of Vibrio parahaemolyticus assisted by perfluorinated alkoxysilane modified molecularly imprinted polymer film. RSC Adv. 2020;10:14305–12. https://doi.org/10.1039/d0ra00306a.
Razavilar V, Ahari H, Akbari Adergani B, Anvar AA. A central composite face-centered design for optimizing the detection of Salmonella typhi with a fluorescence nanobiosensor using the microcontact method. Int J Environ Sci Technol. 2019;16:4637–46. https://doi.org/10.1007/s13762-018-1871-z.
Sykora S, Cumbo A, Belliot G, Pothier P, Arnal C, Dudal Y, et al. Virus-like particles as virus substitutes to design artificial virus-recognition nanomaterials. Chem Commun. 2015;51:2256–8. https://doi.org/10.1039/c4cc08843c.
Chen L, Wang X, Lu W, Wu X, Li J. Molecular imprinting: perspectives and applications. Chem Soc Rev. 2016;45:2137–211. https://doi.org/10.1039/c6cs00061d.
Chen L, Xu S, Li J. Recent advances in molecular imprinting technology: current status, challenges and highlighted applications. Chem Soc Rev. 2011;40:2922–42. https://doi.org/10.1039/c0cs00084a.
Vasapollo G, Del Sole R, Mergola L, Lazzoi MR, Scardino A, Scorrano S, et al. Molecularly imprinted polymers: present and future prospective. Int J Mol Sci. 2011;12:5908–45. https://doi.org/10.3390/ijms12095908.
Piletsky S, Canfarotta F, Poma A, Bossi AM, Piletsky S. Molecularly imprinted polymers for cell recognition. Trends Biotechnol. 2020;38:368–87. https://doi.org/10.1016/j.tibtech.2019.10.002.
Pan J, Chen W, Ma Y, Pan G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem Soc Rev. 2018;47:5574–87. https://doi.org/10.1039/c7cs00854f.
Tokonami S, Nakadoi Y, Nakata H, Takami S, Kadoma T, Shiigi H, et al. Recognition of gram-negative and gram-positive bacteria with a functionalized conducting polymer film. Res Chem Intermed. 2014;40:2327–35. https://doi.org/10.1007/s11164-014-1609-6.
Starosvetsky J, Cohen T, Cheruti U, Dragoljub D, Armon R. Effects of physical parameters on bacterial cell adsorption onto pre-imprinted sol-gel films. J Biomater Nanobiotechnol. 2012;3:499–507. https://doi.org/10.4236/jbnb.2012.324051.
Namvar A, Warriner K. Microbial imprinted polypyrrole/poly(3-methylthiophene) composite films for the detection of Bacillus endospores. Biosens Bioelectron. 2007;22:2018–24. https://doi.org/10.1016/j.bios.2006.08.039.
Mankar JS, Sharma MD, Rayalu SS, Krupadam RJ. Molecularly imprinted microparticles (microMIPs) embedded with reduced graphene oxide for capture and destruction of E. coli in drinking water. Mater Sci Eng C. 2020;110:110672. https://doi.org/10.1016/j.msec.2020.110672.
Bao H, Yang B, Zhang X, Lei L, Li Z. Bacteria-templated fabrication of a charge heterogeneous polymeric interface for highly specific bacterial recognition. Chem Commun. 2017;53:2319–22. https://doi.org/10.1039/c6cc09242j.
Idil N, Mattiasson B. Imprinting of microorganisms for biosensor applications. Sensors. 2017;17:708. https://doi.org/10.3390/s17040708.
Kryscio DR, Peppas NA. Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater. 2012;8:461–73. https://doi.org/10.1016/j.actbio.2011.11.005.
Jiang W, Saxena A, Song B, Ward BB, Beveridge TJ, Myneni SCB. Elucidation of functional groups on gram-positive and gram-negative bacterial surfaces using infrared spectroscopy. Langmuir. 2004;20:11433–42. https://doi.org/10.1021/la049043+.
Gür SD, Bakhshpour M, Denizli A. Selective detection of Escherichia coli caused UTIs with surface imprinted plasmonic nanoscale sensor. Mater Sci Eng C. 2019;104:109869. https://doi.org/10.1016/j.msec.2019.109869.
Yilmaz E, Schmidt RH, Mosbach K. The noncovalent approach. In: Yan M, Ramström O, editors. Molecularly imprinted materials: science and technology. New York: Marcel Dekker; 2005. pp. 25–57.
Chen S, Chen X, Zhang L, Gao J, Ma Q. Electrochemiluminescence detection of Escherichia coli O157:H7 based on a novel polydopamine surface imprinted polymer biosensor. ACS Appl Mater Interfaces. 2017;9:5430–6. https://doi.org/10.1021/acsami.6b12455.
Bezdekova J, Zemankova K, Hutarova J, Kociova S, Smerkova K, Adam V, et al. Magnetic molecularly imprinted polymers used for selective isolation and detection of Staphylococcus aureus. Food Chem. 2020;321:126673. https://doi.org/10.1016/j.foodchem.2020.126673.
Schirhagl R. Bioapplications for molecularly imprinted polymers. Anal Chem. 2014;86:250–61. https://doi.org/10.1021/ac401251j.
Jia M, Zhang Z, Li J, Ma X, Chen L, Yang X. Molecular imprinting technology for microorganism analysis. TrAC - Trends Anal Chem. 2018;106:190–201. https://doi.org/10.1016/j.trac.2018.07.011.
Zhou T, Zhang K, Kamra T, Bülow L, Ye L. Preparation of protein imprinted polymer beads by Pickering emulsion polymerization. J Mater Chem B. 2015;3:1254–60. https://doi.org/10.1039/c4tb01605j.
Crapnell RD, Hudson A, Foster CW, Eersels K, van Grinsven B, Cleij TJ, et al. Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection. Sensors. 2019;19:1204. https://doi.org/10.3390/s19051204.
Poller AM, Spieker E, Lieberzeit PA, Preininger C. Surface imprints: advantageous application of ready2use materials for bacterial quartz-crystal microbalance sensors. ACS Appl Mater Interfaces. 2017;9:1129–35. https://doi.org/10.1021/acsami.6b13888.
Idil N, Hedström M, Denizli A, Mattiasson B. Whole cell based microcontact imprinted capacitive biosensor for the detection of Escherichia coli. Biosens Bioelectron. 2017;87:807–15. https://doi.org/10.1016/j.bios.2016.08.096.
Perçin I, Idil N, Bakhshpour M, Yılmaz E, Mattiasson B, Denizli A. Microcontact imprinted plasmonic nanosensors: powerful tools in the detection of Salmonella paratyphi. Sensors. 2017;17:1375. https://doi.org/10.3390/s17061375.
Shen X, Svensson Bonde J, Kamra T, Bülow L, Leo JC, Linke D, et al. Bacterial imprinting at Pickering emulsion interfaces. Angew Chem - Int Ed. 2014;53:10687–90. https://doi.org/10.1002/anie.201406049.
Hayden O, Dickert FL. Selective microorganism detection with cell surface imprinted polymers. Adv Mater. 2001;13:1480–3. https://doi.org/10.1002/1521-4095(200110)13:19<1480::AID-ADMA1480>3.0.CO;2-V.
Ren K, Zare RN. Chemical recognition in cell-imprinted polymers. ACS Nano. 2012;6:4314–8. https://doi.org/10.1021/nn300901z.
Latif U, Qian J, Can S, Dickert FL. Biomimetic receptors for bioanalyte detection by quartz crystal microbalances-from molecules to cells. Sensors. 2014;14:23419–38. https://doi.org/10.3390/s141223419.
Seifner A, Lieberzeit P, Jungbauer C, Dickert FL. Synthetic receptors for selectively detecting erythrocyte ABO subgroups. Anal Chim Acta. 2009;651:215–9. https://doi.org/10.1016/j.aca.2009.08.021.
Samardzic R, Sussitz HF, Jongkon N, Lieberzeit PA. Quartz crystal microbalance in-line sensing of Escherichia coli in a bioreactor using molecularly imprinted polymers. Sens Lett. 2014;12:1152–5. https://doi.org/10.1166/sl.2014.3201.
Spieker E, Lieberzeit PA. Molecular imprinting studies for developing QCM-sensors for Bacillus cereus. Proc Eng. 2016;168:561–4. https://doi.org/10.1016/j.proeng.2016.11.525.
Zhao X, Cui Y, Wang J, Wang J. Preparation of fluorescent molecularly imprinted polymers via Pickering emulsion interfaces and the application for visual sensing analysis of Listeria monocytogenes. Polymers (Basel). 2019;11:984. https://doi.org/10.3390/polym11060984.
Boysen RI, Schwarz LJ, Nicolau DV, Hearn MTW. Molecularly imprinted polymer membranes and thin films for the separation and sensing of biomacromolecules. J Sep Sci. 2017;40:314–35. https://doi.org/10.1002/jssc.201600849.
Frasco MF, Truta LAANA, Sales MGF, Moreira FTC. Imprinting technology in electrochemical biomimetic sensors. Sensors. 2017;17:523. https://doi.org/10.3390/s17030523.
Li R, Feng Y, Pan G, Liu L. Advances in molecularly imprinting technology for bioanalytical applications. Sensors. 2019;19:177. https://doi.org/10.3390/s19010177.
Tokonami S, Nakadoi Y, Takahashi M, Ikemizu M, Kadoma T, Saimatsu K, et al. Label-free and selective bacteria detection using a film with transferred bacterial configuration. Anal Chem. 2013;85:4925–9. https://doi.org/10.1021/ac3034618.
Zhang N, Zhang N, Xu Y, Li Z, Yan C, Mei K, et al. Molecularly imprinted materials for selective biological recognition. Macromol Rapid Commun. 2019;40:1900096. https://doi.org/10.1002/marc.201900096.
Borovička J, Stoyanov SD, Paunov VN. Shape recognition of microbial cells by colloidal cell imprints. Nanoscale. 2013;5:8560–8. https://doi.org/10.1039/c3nr01893h.
Borovička J, Metheringham WJ, Madden LA, Walton CD, Stoyanov SD, Paunov VN. Photothermal colloid antibodies for shape-selective recognition and killing of microorganisms. J Am Chem Soc. 2013;135:5282–5. https://doi.org/10.1021/ja400781f.
Zhang Z, Guan Y, Li M, Zhao A, Ren J, Qu X. Highly stable and reusable imprinted artificial antibody used for in situ detection and disinfection of pathogens. Chem Sci. 2015;6:2822–6. https://doi.org/10.1039/c5sc00489f.
Haupt K. Imprinted polymers - tailor-made mimics of antibodies and receptors. Chem Commun. 2003;2:171–8. https://doi.org/10.1039/b207596b
Xue X, Pan J, Xie H, Wang J, Zhang S. Specific recognition of Staphylococcus aureus by Staphylococcus aureus protein A-imprinted polymers. React Funct Polym. 2009;69:159–64. https://doi.org/10.1016/j.reactfunctpolym.2008.12.013.
Khan MAR, Moreira FTC, Riu J, MG FS. Plastic antibody for the electrochemical detection of bacterial surface proteins. Sensors Actuators B Chem. 2016;233:697–704. https://doi.org/10.1016/j.snb.2016.04.075.
Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C. An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv. 2010;28:232–54. https://doi.org/10.1016/j.biotechadv.2009.12.004.
Kintzios S, Banerjee P. Mammalian cell-based sensors for high throughput screening for detecting chemical residues, pathogens, and toxins in food. In: Bhunia AK, kim MS, Taitt CR, editors. High throughput screening for food safety assessment: biosensor technologies, hyperspectral imaging and practical applications. Cambridge: Woodhead Publishing; 2015. pp. 123-146.
Wu W, Yu C, Wang Q, Zhao F, He H, Liu C, et al. Research advances of DNA aptasensors for foodborne pathogen detection. Crit Rev Food Sci Nutr. 2020;60:2353–68. https://doi.org/10.1080/10408398.2019.1636763.
Janczuk-Richter M, Marinović I, Niedziółka-Jönsson J, Szot-Karpińska K. Recent applications of bacteriophage-based electrodes: a mini-review. Electrochem Commun. 2019;99:11–5. https://doi.org/10.1016/j.elecom.2018.12.011.
Farooq U, Yang Q, Ullah MW, Wang S. Bacterial biosensing: recent advances in phage-based bioassays and biosensors. Biosens Bioelectron. 2018;118:204–16. https://doi.org/10.1016/j.bios.2018.07.058.
Saylan Y, Yilmaz F, Özgür E, Derazshamshir A, Yavuz H, Denizli A. Molecular imprinting of macromolecules for sensor applications. Sensors. 2017;17:898. https://doi.org/10.3390/s17040898.
Mugo SM, Lu W, Dhanjai D. A pathogen imprinted hybrid polymer capacitive sensor for selective Escherichia coli detection. Med Devices Sensors. 2020;3:e10071. https://doi.org/10.1002/mds3.10071.
Ertürk G, Lood R. Bacteriophages as biorecognition elements in capacitive biosensors: phage and host bacteria detection. Sensors Actuators B Chem. 2018;258:535–43. https://doi.org/10.1016/j.snb.2017.11.117.
van Grinsven B, Eersels K, Akkermans O, Ellermann S, Kordek A, Peeters M, et al. Label-free detection of Escherichia coli based on thermal transport through surface imprinted polymers. ACS Sensors. 2016;1:1140–7. https://doi.org/10.1021/acssensors.6b00435.
Cornelis P, Givanoudi S, Yongabi D, Iken H, Duwé S, Deschaume O, et al. Sensitive and specific detection of E. coli using biomimetic receptors in combination with a modified heat-transfer method. Biosens Bioelectron. 2019;136:97–105. https://doi.org/10.1016/j.bios.2019.04.026.
Yilmaz E, Majidi D, Ozgur E, Denizli A. Whole cell imprinting based Escherichia coli sensors: a study for SPR and QCM. Sensors Actuators B Chem. 2015;209:714–21. https://doi.org/10.1016/j.snb.2014.12.032.
Ait Lahcen A, Arduini F, Lista F, Amine A. Label-free electrochemical sensor based on spore-imprinted polymer for Bacillus cereus spore detection. Sensors Actuators B Chem. 2018;276:114–20. https://doi.org/10.1016/j.snb.2018.08.031.
Malhotra BD, Ali MA. Nanomaterials in biosensors: fundamentals and applications. In: Malhotra BD, Ali MA, editors. Nanomaterials for biosensors. Amsterdam: William Andrew Publishing; 2018. pp. 1–74.
Damborský P, Švitel J, Katrlík J. Optical biosensors. Essays Biochem. 2016;60:91–100. https://doi.org/10.1042/EBC20150010.
Yang Q, Li J, Wang X, Peng H, Xiong H, Chen L. Strategies of molecular imprinting-based fluorescence sensors for chemical and biological analysis. Biosens Bioelectron. 2018;112:54–71. https://doi.org/10.1016/j.bios.2018.04.028.
Shan X, Yamauchi T, Yamamoto Y, Niyomdecha S, Ishiki K, Le DQ, et al. Spontaneous and specific binding of enterohemorrhagic Escherichia coli to overoxidized polypyrrole-coated microspheres. Chem Commun. 2017;53:3890–3. https://doi.org/10.1039/c7cc00244k.
Shan X, Yamauchi T, Yamamoto Y, Shiigi H, Nagaoka T. A rapid and specific bacterial detection method based on cell-imprinted microplates. Analyst. 2018;143:1568–74. https://doi.org/10.1039/c7an02057k.
Gültekin A, Ersöz A, Hür D, Sariözlü NY, Denizli A, Say R. Gold nanoparticles having dipicolinic acid imprinted nanoshell for Bacillus cereus spores recognition. Appl Surf Sci. 2009;256:142–8. https://doi.org/10.1016/j.apsusc.2009.07.097.
Subramanian A, Irudayaraj J, Ryan T. A mixed self-assembled monolayer-based surface plasmon immunosensor for detection of E. coli O157:H7. Biosens Bioelectron. 2006;21:998–1006. https://doi.org/10.1016/j.bios.2005.03.007.
Subramanian A, Irudayaraj J, Ryan T. Mono and dithiol surfaces on surface plasmon resonance biosensors for detection of Staphylococcus aureus. Sensors Actuators B Chem. 2006;114:192–8. https://doi.org/10.1016/j.snb.2005.04.030.
Leonard P, Hearty S, Quinn J, O’Kennedy R. A generic approach for the detection of whole Listeria monocytogenes cells in contaminated samples using surface plasmon resonance. Biosens Bioelectron. 2004;19:1331–5. https://doi.org/10.1016/j.bios.2003.11.009.
Zhang X, Tsuji S, Kitaoka H, Kobayashi H, Tamai M, Honjoh KI, et al. Simultaneous detection of Escherichia coli O157:H7, Salmonella enteritidis, and Listeria monocytogenes at a very low level using simultaneous enrichment broth and multichannel SPR biosensor. J Food Sci. 2017;82:2357–63. https://doi.org/10.1111/1750-3841.13843.
Thévenot DR, Toth K, Durst RA, Wilson GS. Electrochemical biosensors: recommended definitions and classification. Biosens Bioelectron. 2001;16:121–31. https://doi.org/10.1016/S0956-5663(01)00115-4.
Ayankojo AG, Reut J, Boroznjak R, Öpik A, Syritski V. Molecularly imprinted poly(meta-phenylenediamine) based QCM sensor for detecting amoxicillin. Sensors Actuators B Chem. 2018;258:766–74. https://doi.org/10.1016/j.snb.2017.11.194.
Lin TY, Hu CH, Chou TC. Determination of albumin concentration by MIP-QCM sensor. Biosens Bioelectron. 2004;20:75–81. https://doi.org/10.1016/j.bios.2004.01.028.
Gültekin A, Karanfil G, Kuş M, Sönmezoǧlu S, Ri S. Preparation of MIP-based QCM nanosensor for detection of caffeic acid. Talanta. 2014;119:533–7. https://doi.org/10.1016/j.talanta.2013.11.053.
Hussain M, Kotova K, Lieberzeit PA. Molecularly imprinted polymer nanoparticles for formaldehyde sensing with QCM. Sensors. 2016;16:1011. https://doi.org/10.3390/s16071011.
Casadio S, Lowdon JW, Betlem K, Ueta JT, Foster CW, Cleij TJ, et al. Development of a novel flexible polymer-based biosensor platform for the thermal detection of noradrenaline in aqueous solutions. Chem Eng J. 2017;315:459–68. https://doi.org/10.1016/j.cej.2017.01.050.
Peeters MM, Van Grinsven B, Foster CW, Cleij TJ, Banks CE. Introducing thermal wave transport analysis (TWTA): a thermal technique for dopamine detection by screen-printed electrodes functionalized with molecularly imprinted polymer (MIP) particles. Molecules. 2016;21:552. https://doi.org/10.3390/molecules21050552.
Betlem K, Mahmood I, Seixas RD, Sadiki I, Raimbault RLD, Foster CW, et al. Evaluating the temperature dependence of heat-transfer based detection: a case study with caffeine and molecularly imprinted polymers as synthetic receptors. Chem Eng J. 2019;359:505–17. https://doi.org/10.1016/j.cej.2018.11.114.
Steen Redeker E, Eersels K, Akkermans O, Royakkers J, Dyson S, Nurekeyeva K, et al. Biomimetic bacterial identification platform based on thermal wave transport analysis (TWTA) through surface-imprinted polymers. ACS Infect Dis. 2017;3:388–97. https://doi.org/10.1021/acsinfecdis.7b00037.
van Grinsven B, Eersels K, Peeters M, Losada-Pérez P, Vandenryt T, Cleij TJ, et al. The heat-transfer method: a versatile low-cost, label-free, fast, and user-friendly readout platform for biosensor applications. ACS Appl Mater Interfaces. 2014;6:13309–18. https://doi.org/10.1021/am503667s.
van Grinsven B, Betlem K, Cleij TJ, Banks CE, Peeters M. Evaluating the potential of thermal read-out techniques combined with molecularly imprinted polymers for the sensing of low-weight organic molecules. J Mol Recognit. 2017;30:e2563. https://doi.org/10.1002/jmr.2563.
Funding
This work was supported by the fund awarded to X.L. by McGill University new faculty start-up grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interests.
Additional information
Published in the topical collection Analytical Chemistry for Infectious Disease Detection and Prevention with guest editors Chaoyong Yang and XiuJun (James) Li.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, J., Wang, Y. & Lu, X. Molecular imprinting technology for sensing foodborne pathogenic bacteria. Anal Bioanal Chem 413, 4581–4598 (2021). https://doi.org/10.1007/s00216-020-03138-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-03138-x