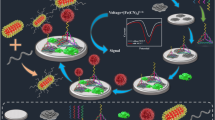
Abstract
In this study, we fabricated a high-sensitivity “signal-off” electrochemical aptasensing platform for quantifying circulating tumor cells (CTCs) based on target-triggered signal readout of methylene blue (MB). Au nanoparticles (AuNPs) were introduced to enlarge the specific surface area of the gold electrode (GE), which would immobilize homogeneous and more MB-aptamers. MB-modified and stem-loop-like aptamers were assigned as a recognition element with K562 cells. Thiolated complementary strands hybridized with MB-aptamers to form double-stranded DNA (dsDNA) conformation which were further self-assembled on the surface of AuNP-modified GE, leading to a marked current peak of MB signal. In the presence of K562 cells, the MB-aptamers preferred to recognize and bind with the cells, causing the disassembly of MB-aptamers from the GE surface. Therefore, the reduced value of MB signal was related to the number of K562 cells. With the proposed aptasensor, a dynamic linear range from 1 × 102 to 1 × 106 cells mL−1 was obtained with a detection limit of 23 cells mL−1. Moreover, the aptasensor showed good selectivity, stability, and reproducibility as well as potential use in the clinical setting. Meanwhile, characterization techniques such as field-emission scanning electron microscopy, energy-dispersive X-ray spectroscopy, atomic force microscopy, cyclic voltammetry, and electrochemical impedance spectroscopy were performed to analyze the evolution of the morphology and each fabricated step of the constructed aptasensor. Our proposed aptasensor could be designed as a universal platform for CTC determination by replacing tumor cell–specific aptamers, which is a promising strategy for basic research and clinical applications.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Liquid biopsy is a noninvasive analysis of tumor-related biomarkers in body fluids including circulating tumor DNA, circulating tumor cells (CTCs), and exosomes, and it holds promising potential for cancer management, such as early diagnosis, real-time monitoring of tumor progression, and treatment selection [1, 2]. CTCs are an important area of liquid biopsy, and those derived from orthotopic tumor circulate through the bloodstream [3, 4]. Therefore, CTCs could colonize in distant tissues and organs, and finally cause tumor recurrence, resulting in the death of a patient [5, 6]. Compared with tissue biopsy, determination of CTCs is a noninvasive, safe, and rapid diagnostic technique [7]. However, CTCs in blood are extremely rare, namely 1–100 cells mL−1 among billions of normal blood cells [8]. Therefore, highly sensitive and rapid determination of CTCs is still challenging in clinical applications.
To date, various technologies have been developed for CTC assay based on density gradient, microfiltration, microfluidic array, dielectrophoresis, and immunoaffinity [9, 10]. Despite its advantages of high efficiency, sensitivity, and specificity, there are still some problems with CTC assay including low purity, risk of losing and damaging small or large CTCs, relatively high cost and long detection time, and reliance on biomarkers [11, 12]. Therefore, simple and time-saving methods are urgently needed. In recent years, electrochemical sensors have garnered considerable interest in the determination of CTCs, owing to their inherent advantages of miniaturization, low cost, high sensitivity, and short detection time [13, 14]. Currently, there are two strategies to amplify signal for highly sensitive determination of CTCs with an electrochemical biosensor. The first strategy involves modification of a sensing platform using nanomaterials. The second strategy is cell-labeled signal amplification (e.g., rolling circle amplification and hybridization chain reaction). Using the first strategy, Yang et al. developed an electrochemical biosensor through Au nanocages/amino-functionalized multiwalled carbon nanotubes and 3D graphene for assay of breast cancer cells [15]. Ge et al. established a paper-based electrochemical sensor integrated with 3D Au nanoparticles/graphene and ionic liquid for assay of K562 cancer cells [16]. Using the second strategy, Sun et al. fabricated an electrochemical sensor with signal amplification and dual recognition to quantify HepG2 cells [17]. Chen et al. fabricated an electrochemical sensor with multibranched hybridization chain reaction amplification to quantify HepG2 cells [18]. In our previous study, a strategy comprising dual-aptamer recognition and rolling circle amplification was utilized to quantify CTCs [19]. These reported electrochemical biosensors have improved the development of CTC determination. However, complicated fabrication on the reaction area and cell-labeled strategy still exist.
“Signal-off” biosensors have the advantages of simple fabrication, sensitive detection, and a wide range of linear detection [20]. In “signal-off” biosensors, the signals (e.g., current, impedance, and electrochemiluminescence intensity) change from an “on” status into an “off” status in the presence of a target. Owing to their advantages, “signal-off” biosensors have been used to quantify various substances, such as proteins, amino acids, and insecticides [21,22,23]. Aptamers are promising alternatives to antibodies as a target recognition element for their advantages of easy synthesis and modification, long-term storage, and high stability [24, 25]. Furthermore, aptamers have minimal to no toxicity and immunity [26, 27]. Because of the high affinity between aptamers and targets, aptamers have been introduced into electrochemical sensors to quantify targets (e.g., proteins and adenosine) according to hybridization-induced conformational change, and the targets need not be labeled [28,29,30,31]. As far as we know, there has been no study of a target-triggered “signal-off” biosensor for CTC determination based on high affinity with aptamers, resulting in hybridization-induced conformational change.
Accordingly, in the present study, we developed a “signal-off” electrochemical aptasensing platform to quantify K562 cells with high sensitivity. Firstly, stem-loop-like aptamers that had high affinity to K562 cells were modified with methylene blue (MB) in the 5′ termini and assigned as the recognition element with K562 cells. Thiolated complementary strands (in the 3′ termini) hybridized with MB-aptamers to form double-stranded DNAs (dsDNAs). Next, the dsDNAs were fixed onto the gold electrode (GE) by Au–S bond, and the current peak of MB was introduced owing to the close proximity of the MB tags to the GE surface. For the purpose of immobilizing more MB-aptamers on the GE surface, Au nanoparticles (AuNPs) were electrodeposited on the GE to enlarge the specific surface area. In the presence of K562 cells, aptamers preferred to recognize and bind with K562 cells, causing the unwinding of dsDNAs, detachment of MB-labeled aptamers from the electrode, and decline of the MB signal. Therefore, the number of K562 cells was related to the degree of MB signal reduction. With the target-triggered conformational change and “signal-off” biosensor, the detection limit of K562 cells was 23 cells mL−1, and the linear range was 1 × 102–1 × 106 mL−1. Good selectivity, stability, reproducibility, and clinical applications were also presented. Moreover, there was no need to obtain CTCs by breaking the link between CTCs and reaction interface through enzymatic digestion or electrochemical methods, thus maintaining cell viability, which is substantially important to the study of the mechanisms underlying tumorigenesis and tumor development.
Experimental section
Reagents and materials
The aptamers [32, 33] and other oligonucleotides (see Electronic Supplementary Material (ESM) Table S1) were synthesized, purified, and modified by Takara Biomedical Technology Co., Ltd. (China). HAuCl4·3H2O (H821422), 30% H2O2 (H811240), glucose monohydrate (D810457), K4[Fe(CN)6]·3H2O (P816416), and K3[Fe(CN)6] (P816307) were obtained from Macklin Biochemical Co., Ltd. (China). Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, 646547), bovine serum albumin (BSA, ≥ 98%, V900933), and 6-mercapto-1-hexanol (MCH, 451088) were obtained from Sigma-Aldrich (USA). Penicillin-streptomycin (BL505A) was obtained from Biosharp (China). RPMI 1640 medium (01-100-1ACS) and fetal bovine serum (FBS, 04-001-1ACS) were obtained from Biological Industries (Israel). Calcium chloride dihydrate (20011160) was bought from Sinopharm Chemical Reagent Co., Ltd. (China). Magnesium chloride hexahydrate (05.001.1848) was bought from ChengDu Chron Chemicals Co., Ltd. (China). Saturated calomel reference electrode (R0232), platinum counter electrode (Pt120), and gold working electrode (Au120) were bought from TianJin AIDAhengsheng Science-Technology Development Co., Ltd. (China). Piranha solution (98% H2SO4 and 30% H2O2 with v/v = 3:1) and phosphate-buffered saline (PBS) (0.01 M, pH 7.4) were made in our laboratory. Healthy human sera were collected from Southwest Hospital (China). Ultrapure water was produced with a resistance of 18.2 MΩ cm from the water purification instrument (RephiLe Bioscience, Ltd., China).
Apparatus
Alternating current voltammetry (ACV) was conducted with a 1040C electrochemical system (Shanghai CH Instruments, China). Cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) were conducted with a 760E electrochemical system (Shanghai CH Instruments, China). The electrode system consisted of a saturated calomel reference electrode, a platinum counter electrode, and a gold working electrode. ACV was scanned from − 0.5 to − 0.1 V (amplitude 25 mV, increment 4 mV, and frequency 25 Hz) in PBS containing 0.1 M NaClO4. CV and EIS were scanned in 10 mM [Fe(CN)6]3−/4− with 0.1 M KCl (prepared with PBS) as the reversible redox probe. CV was scanned from − 0.2 to + 0.6 V under a scan rate of 0.1 V s−1. EIS was scanned in the range of 0.1 to 105 Hz (amplitude 5 mV) under open circuit potential. Flow cytometry was performed using a Navios system (Beckman Coulter, Inc., USA). Energy-dispersive X-ray spectroscopy (EDS) was performed with XFlash6|10 (Bruker, Germany). Field-emission scanning electron microscopy (FE-SEM) was operated using Inspect F50 (FEI, USA). Atomic force microscopy (AFM) was conducted with the Dimension Icon system (Bruker, Germany).
Cell culture and preparation
K562, Jurkat, and U937 cells were bought from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (China). The cell lines were grown in RPMI 1640 medium with 10% FBS and 1% penicillin-streptomycin under standard conditions. Prior to the experiments, cell suspensions were centrifuged for 5 min at 1000 rpm, followed by discarding of the supernatant. Subsequently, the cell sediment was washed with PBS and then blocked in 1% BSA solution (w/v) for 30 min at 37 °C. Afterwards, the cells were washed twice with PBS and then centrifuged for 5 min at 1000 rpm, followed by discarding of the supernatant. Finally, the cells were re-suspended in PBS containing 4.5 g L−1 glucose monohydrate, 6 mM magnesium chloride hexahydrate, and 1 mM calcium chloride dihydrate. The desired cell concentrations were prepared through hemocytometer-based quantification.
Analysis of specific recognition between aptamers and K562 cells
Flow cytometry was performed to evaluate the selective recognition and binding ability between aptamers and K562 cells. The oligonucleotides (15 pmol) were labeled with Alexa Fluor 488 at the 5′ end, and further incubated with K562 cells (3 × 105 cells) at 37 °C for 30 min. Nonspecific binding sequences were discarded with centrifugation. The fluorescence intensity and binding efficiency of Alexa Fluor 488–labeled aptamers to K562 cells were analyzed by flow cytometry through quantifying 1 × 104 cells for each sample. Further analysis was operated via Kaluza Analysis software (Beckman Coulter, Inc., USA).
Electrochemical determination procedure
A GE with a diameter of 2 mm was polished manually in an alumina slurry (0.3 and 0.05 μm in turn) to clean the surface, followed by sonication in ethanol and water, separately. Next, the GE was immersed in piranha solution for 10 min to remove impurities, followed by rinsing with ultrapure water. GE was then electrochemically deposited in HAuCl4 solution (30 mM) for 20 s under − 0.2 V to obtain AuNP-modified surface. A mixture of MB-modified aptamers (7.0 μM) and thiolated complementary sequences (7.0 μM), as well as 2.0 mM TCEP were firstly denatured at 95 °C for 5 min and then annealed at room temperature to form double-stranded nucleic acids. Disulfide bonds were eliminated by TCEP for 90 min. Afterwards, the prepared double-stranded complexes (3.0 μL) were fixed on the AuNP-modified GE (AuNPs/GE) overnight in a humidified atmosphere at 37 °C. Next, the GE was blocked with 1.0 mM MCH for 30 min at 4 °C, followed by washing with ultrapure water. The double-stranded complex-modified GE (dsDNAs/AuNPs/GE) was incubated with 0.8 mL of K562 cell suspension for cell-aptamer interaction for 90 min at 37 °C. Subsequently, the electrode was washed by PBS carefully. Jurkat and U937 cells, each at the density of 1 × 106 cells mL−1, were tested under the abovementioned procedures. The quantified response was calculated according to the following equation: relative IMB = (I0−I)/I0, where I and I0 were the ACV peak values in the presence and absence of K562 cells, respectively. All experiments were conducted at least three times.
Results and discussion
Principle of the novel aptasensor for tumor cell determination
As shown in Scheme 1, the stem-loop-like and MB-modified aptamers (at the 5′ end) and thiolated complementary chains (at the 3′ end) hybridized to form double-stranded DNAs, and then were self-assembled on the GE through Au–S bond. The close proximity of MB tags to the electrode surface would introduce a high current peak of MB signal. AuNPs were electrodeposited on the bare GE, which would increase the specific surface area for the purpose of fixing more aptamers on the GE. In the presence of K562 cells, the MB-aptamers would specifically recognize and bind with K562 cells because of the high affinity between aptamers and K562 cells, which would further cause the unwinding of dsDNAs. The MB-modified aptamers fell off from the surface of the GE, decreasing the MB signal. Therefore, changes in MB signal were related to the number of K562 cells.
Specific recognition of aptamers to K562 cells
To obtain an aptasensor with the best performance, the specific recognition and binding efficiency of five candidate aptamers to K562 cells were verified through flow cytometry. Alexa Fluor 488–labeled sequences (15 pmol) were incubated with K562 cells (3 × 105 cells) for 30 min at 37 °C, and then the cells were investigated through flow cytometry. As shown in Fig. 1, the binding of Alexa Fluor 488–labeled aptamers 2–5 (Fig. 1c–f) with K562 cells resulted in a minute shift in fluorescence signals, compared to that with the Alexa Fluor 488–labeled random sequence (Fig. 1a). After incubation of K562 cells with Alexa Fluor 488–labeled aptamer 1 (Fig. 1b), the fluorescence intensity apparently increased, compared with that after incubation with Alexa Fluor 488–labeled random sequence. It was indicated that aptamer 1 had specific recognition and high binding capability with K562 cells, which induced marked increment in fluorescence intensity. Based on the above results, aptamer 1 was chosen as the recognition element to capture K562 cells.
Analysis of specific recognition between aptamers and K562 cells by flow cytometry. a Alexa Fluor 488–labeled random sequence. b Alexa Fluor 488–labeled aptamer 1. c Alexa Fluor 488–labeled aptamer 2. d Alexa Fluor 488–labeled aptamer 3. e Alexa Fluor 488–labeled aptamer 4. f Alexa Fluor 488–labeled aptamer 5. Inset plots show the secondary structures of the sequences
Surface and dimensional analysis of the modified electrode
Functional modification on the electrode is highly important for the performance of a biosensor. The morphologies and microstructures of the bare GE and AuNPs/GE were analyzed by FE-SEM. As indicted in Fig. S1a (see ESM), the bare GE exhibited a nearly smooth surface without any modifiers. Compared with that of the bare GE, large amounts of AuNPs were continuously and homogeneously distributed on the surface of GE owing to the electrochemical deposition (see ESM Fig. S1b). The spherical AuNPs had large surface areas, with an average diameter of 20 nm (see ESM Fig. S1c). Additionally, the morphology and structure of the AuNPs/GE did not change obviously over 7 days (see ESM Fig. S1d). It was demonstrated that AuNPs were successfully decorated on the electrode surface. Moreover, the morphologies of AuNPs at different electrodeposition times (10 s, 20 s, and 30 s) were studied (see ESM Fig. S2). The packing density of AuNPs on the electrode surface increased with time. For the purpose of overcoming steric hindrance and electrostatic repulsion of electrochemical probe on the electrode, electrodeposition time of 20 s was chosen for the further study.
In addition, EDS was conducted to analyze the elemental composition of the modified GE. The weight percentage of Au element was high in the bare GE (see ESM Fig. S3a). The weight percentage of Au element increased in the AuNPs/GE (see ESM Fig. S3b), while the weight percentage of C, O, N, and P elements increased in the dsDNAs/AuNPs/GE (see ESM Fig. S3c). Furthermore, S and Cl elements were observed in the dsDNAs/AuNPs/GE. The results showed the successful deposition of AuNPs and immobilization of dsDNAs on the electrode.
Next, the 3D morphology of the sensing platform was captured by AFM. The roughness of the electrode surface was estimated by root mean square roughness (Rq) and average roughness (Ra). The bare GE had a smooth surface with low Ra and Rq values (see ESM Fig. S4a). The AuNPs/GE showed an obvious rough surface with high Ra and Rq values (see ESM Fig. S4b). The topographic height of AuNPs was about 20 nm. The dsDNAs were immobilized uniformly on the AuNPs/GE, which made the electrode surface rougher. Therefore, Ra and Rq values of dsDNAs/AuNPs/GE increased further (see ESM Fig. S4c). The results of AFM images showed the successful deposition of AuNPs and immobilization of dsDNAs on the electrode, which were consistent with the EDS analysis.
To estimate the active surface area of AuNPs/GE, the electrodes (bare GE and AuNPs/GE) were scanned in 0.5 M H2SO4 solution with the CV method (see ESM Fig. S5). The reduction peak of gold oxides was obtained at + 0.9 V with the current values of 3.147 × 10−5 A (bare GE) and 5.441 × 10−5 A (AuNPs/GE). The active surface area of AuNPs/GE could be calculated according to the charge consumed in the reduction process of gold oxides in H2SO4 solution, whereas the specific value of Au was 386 μC/cm2 [34, 35]. The total charges of bare GE and AuNPs/GE were 1.240 × 10−5 C and 1.882 × 10−5 C. Therefore, the active surface areas of bare GE and AuNPs/GE were 3.21 mm2 and 4.88 mm2, respectively. The above results revealed that AuNPs were successfully deposited on the electrode surface and had the potential to enlarge the specific surface area of GE as well as enhance the electron transfer rate.
Electrochemical characterization of the aptasensor
CV and EIS are effective investigative tools to characterize the modification of electrodes in electrochemical sensors [36]. Figure 2a and Fig. S6a (see ESM) show CV results of the stepwise fabrication of the proposed aptasensor. The bare GE showed small potential differences and large redox peak, reflecting a fast electron transfer rate and high conductivity on the clean surface of GE. When the AuNPs were electrodeposited on the GE, the redox peak increased and the potential differences decreased to a small extent, implying the successful fix of the AuNPs on the electrode and the good electron transfer performance by the AuNPs. After the immobilization of DNA duplexes, the redox peak began to decrease because the negative charge of nucleic acids obstructed the diffusion of the redox probe on the sensing surface. Subsequently, the nonspecific sites were blocked by MCH, which apparently decreased the current peak. Finally, aptamers dissociated from DNA duplexes in the presence of K562 cells due to specific recognition between aptamers and K562 cells, which partly recovered the electron transfer of [Fe(CN)6]3−/4− redox probe.
In the impedance spectra, the straight line (low-frequency domain) and the semicircle (high-frequency domain) represent the electron diffusion and transfer process, respectively [37]. The charge transfer resistance (Rct) on the GE surface was equal to the semicircle diameter. As depicted in Fig. 2b and Fig. S6b (see ESM), the semicircle of bare GE was very small, owing to the fast electron transfer of the [Fe(CN)6]3−/4− redox probe. Consistent with the change in CV, the electrodeposition of AuNPs increased the specific surface area and accelerated the interfacial electron transfer rate, showing nearly a straight line. The semicircle of hybrid duplexes noticeably increased, suggesting that the diffusion of [Fe(CN)6]3−/4− was obstructed by the electrostatic repulsion between nucleic acids and redox probe, followed by blocking with MCH. In the presence of K562 cells, aptamers preferred to recognize and bind with K562 cells, causing the aptamers to detach from the electrode, leading to partial recovery of electron transfer on the sensing surface. All these results showed that the stepwise modification of the aptasensor was successful, as expected.
Optimization of experimental conditions
To acquire the optimal analytical performance of our constructed biosensor, three key factors, namely the concentration of the MB-modified aptamers as well as the temperature and time of reaction between K562 cells and aptasensor, were optimized. The concentration of the MB-aptamers was important in the variations of MB signal. As shown in Fig. 3a, the relative IMB increased as the concentration of MB-aptamers increased from 1.0 to 7.0 μM. When the MB-aptamer concentration further increased, the relative IMB began to decrease. It was indicated that the effect of overcoming steric hindrance and electrostatic repulsion was optimal at the concentration of 7.0 μM. Furthermore, the temperature of the reaction between K562 cells and aptasensor was studied (Fig. 3b). The relative IMB increased gradually as the temperature increased from 4 to 37 °C and leveled off at 45 °C. Because 37 °C was suitable for most biological reaction, and there was no statistical difference in relative IMB between reaction at 37 °C and at 45 °C, 37 °C was set as the best reaction temperature in the subsequent experiments. The reaction time between K562 cells and aptamers was critical in the analytical performance of the aptasensor, and was investigated thoroughly (Fig. 3c). The relative IMB increased from 30 min and reached the peak at 90 min. After 90 min, the relative current value decreased, which was attributed to cell lysis caused by long-term exposure to air. Therefore, the optimal reaction time was determined as 90 min in the subsequent experiments.
Determination performance of the electrochemical aptasensor
The sensitivity of the aptasensor was markedly related to early diagnosis of cancer in the clinical setting. In our fabricated biosensor, relative IMB, which represented K562 cell concentration, was calculated as relative IMB = (I0−I)/I0, where I and I0 were the ACV peak values in the presence and absence of K562 cells, respectively (Fig. 4a). As shown in Fig. 4b, the relative IMB decreased proportionally with decreasing K562 cell concentration under the optimal experimental conditions. The linear equation was Y = 0.0488 logC + 0.0111 (R = 0.9944), where Y presented the relative IMB and logC presented the logarithm of K562 cell concentration, respectively. The limit of detection (LOD) was 23 cells mL−1 according to mean + 3SD, where the mean and standard deviation (SD) were equal to the mean value and SD of the relative IMB in a blank sample, respectively. The linear range of detection was from 1 × 102 to 1 × 106 cells mL−1. The LOD and linear range were comparable or better compared with those in previous studies on CTC determination (see ESM Table S2). To detect the extremely low concentration of CTCs in peripheral blood, the sensitivity of the proposed aptasensor could be improved through optimization of aptamers’ structure to enhance the binding affinity with targets.
Selectivity, reproducibility, and stability of the electrochemical aptasensor
Five nonspecific interferences, namely PBS, FBS, BSA, and two kinds of cancer cells, were used to investigate the selectivity of the fabricated aptasensor. As depicted in Fig. 5 and Fig. S7 (see ESM), U937 and Jurkat cells, which were quantified under the same conditions as K562 cells, showed minimal relative current value compared with that shown by K562 cells. In addition, PBS, 10% FBS, and 1% BSA generated even lower relative current value than that generated by K562 cells, showing the good selectivity of the aptasensor for K562 cells.
The reproducibility of the aptasensor was assessed through determination of K562 cells at each concentration by four prepared aptasensors. The relative standard deviation (RSD) was 9.33%, 8.81%, 11.16%, 6.92%, 5.82%, and 4.01% for K562 cells at 10, 1 × 102, 1 × 103, 1 × 104, 1 × 105, and 1 × 106 cells mL−1, respectively. The above results showed that the reproducibility of the aptasensor was adequate.
Stability is essential for long-term use of a biosensor. The aptasensors, which were stored at 4 °C in ultrapure water, were used to quantify the same amounts of K562 cells every 6 days. The relative IMB decreased gradually from 100% of the initial value to 93.95% on the sixth day, and to 91.76% on the twelfth day with an RSD of 4.38%, 5.06%, and 5.75%, respectively. These results showed that the proposed aptasensor was feasible for long-time use.
Determination of K562 cells in human serum
The potential clinical application of the proposed aptasensor was estimated by a recovery experiment. Different concentrations of K562 cells were spiked in healthy human serum (10-fold dilution) and quantified by the biosensor. As shown in Table S3 (see ESM), the recoveries (1 × 102, 1 × 103, 1 × 104, and 1 × 105 cells mL−1) were 93.00%, 103.48%, 104.28%, and 94.82% with an RSD of 9.19%, 8.06%, 6.84%, and 8.01%, respectively. The results revealed that the fabricated aptasensor had potential clinical applications. Additionally, pre-concentration of CTCs such as depletion of white blood cells and lysis of red blood cells is prone to avoid false negative results during CTC detection.
Conclusions
In summary, we fabricated a “signal-off” electrochemical aptasensing platform for high-sensitive determination of tumor cells. The reaction between K562 cells and aptamers induced marked changes in MB signal owing to the high binding affinity of the aptamers to K562 cells and the target-triggered “signal-off” mode. In addition, the introduction of AuNPs onto the electrode surface resulted in the increase of specific surface area and acceleration of the electron transfer ability, further increasing the amounts of aptamers which were immobilized on the GE surface. The LOD of the aptasensor was 23 cells mL−1, with a linear range from 1 × 102 to 1 × 106 cells mL−1. Moreover, the proposed aptasensor exhibited good selectivity, reproducibility, and stability, as well as potential applications in the clinical setting. This proposed aptasensor can be used as a versatile CTC quantification platform with specific aptamers applied for different tumor cells, showing its promising prospect in cancer management.
References
Temilola DO, Wium M, Coulidiati TH, Adeola HA, Carbone GM, Catapano CV, et al. The prospect and challenges to the flow of liquid biopsy in Africa. Cells. 2019;8(8).
Rossi G, Ignatiadis M. Promises and pitfalls of using liquid biopsy for precision medicine. Cancer Res. 2019;79(11):2798–804.
Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2018;19(1):11–34.
Banko P, Lee SY, Nagygyorgy V, Zrinyi M, Chae CH, Cho DH, et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12(1):48.
Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406.
Chen L, Bode AM, Dong Z. Circulating tumor cells: moving biological insights into detection. Theranostics. 2017;7(10):2606–19.
Buscail E, Chiche L, Laurent C, Vendrely V, Denost Q, Denis J, et al. Tumor-proximal liquid biopsy to improve diagnostic and prognostic performances of circulating tumor cells. Mol Oncol. 2019;13(9):1811–26.
Kamande JW, Hupert ML, Witek MA, Wang H, Torphy RJ, Dharmasiri U, et al. Modular microsystem for the isolation, enumeration, and phenotyping of circulating tumor cells in patients with pancreatic cancer. Anal Chem. 2013;85(19):9092–100.
Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31(18):1827–40.
Jackson JM, Witek MA, Kamande JW, Soper SA. Materials and microfluidics: enabling the efficient isolation and analysis of circulating tumour cells. Chem Soc Rev. 2017;46(14):4245–80.
Brown HK, Tellez-Gabriel M, Cartron PF, Vallette FM, Heymann MF, Heymann D. Characterization of circulating tumor cells as a reflection of the tumor heterogeneity: myth or reality? Drug Discov Today. 2019;24(3):763–72.
Sharma S, Zhuang R, Long M, Pavlovic M, Kang Y, Ilyas A, et al. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol Adv. 2018;36(4):1063–78.
Maduraiveeran G, Sasidharan M, Ganesan V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens Bioelectron. 2018;103:113–29.
Lima HRS, da Silva JS, de Oliveira Farias EA, Teixeira PRS, Eiras C, Nunes LCC. Electrochemical sensors and biosensors for the analysis of antineoplastic drugs. Biosens Bioelectron. 2018;108:27–37.
Yang Y, Fu Y, Su H, Mao L, Chen M. Sensitive detection of MCF-7 human breast cancer cells by using a novel DNA-labeled sandwich electrochemical biosensor. Biosens Bioelectron. 2018;122:175–82.
Ge S, Zhang L, Zhang Y, Liu H, Huang J, Yan M, et al. Electrochemical K-562 cells sensor based on origami paper device for point-of-care testing. Talanta. 2015;145:12–9.
Sun D, Lu J, Chen Z, Yu Y, Mo M. A repeatable assembling and disassembling electrochemical aptamer cytosensor for ultrasensitive and highly selective detection of human liver cancer cells. Anal Chim Acta. 2015;885:166–73.
Chen D, Sun D, Wang Z, Qin W, Chen L, Zhou L, et al. A DNA nanostructured aptasensor for the sensitive electrochemical detection of HepG2 cells based on multibranched hybridization chain reaction amplification strategy. Biosens Bioelectron. 2018;117:416–21.
Wang Y, Chang K, Yang C, Li S, Wang L, Xu H, et al. Highly sensitive electrochemical biosensor for circulating tumor cells detection via dual-aptamer capture and rolling circle amplification strategy. J Biomed Nanotechnol. 2019;15(7):1568–77.
Wang H, Song Y, Chai Y, Yuan R. Highly sensitive biosensor based on target induced dual signal amplification to electrochemiluminescent nanoneedles of Ru(II) complex. Biosens Bioelectron. 2019;140:111344.
Chen Z, Xu Q, Tang G, Liu S, Xu S, Zhang X. A facile electrochemical aptasensor for lysozyme detection based on target-induced turn-off of photosensitization. Biosens Bioelectron. 2019;126:412–7.
Zhu S, Lin X, Ran P, Xia Q, Yang C, Ma J, et al. A novel luminescence-functionalized metal-organic framework nanoflowers electrochemiluminesence sensor via “on-off” system. Biosens Bioelectron. 2017;91:436–40.
Wang H, Zhang B, Zhao F, Zeng B. One-pot synthesis of N-graphene quantum dot-functionalized I-BiOCl Z-scheme cathodic materials for “signal-off” photoelectrochemical sensing of chlorpyrifos. ACS Appl Mater Interfaces. 2018;10(41):35281–8.
Zhou Z, Liu M, Jiang J. The potential of aptamers for cancer research. Anal Biochem. 2018;549:91–5.
Kaur H, Bruno JG, Kumar A, Sharma TK. Aptamers in the therapeutics and diagnostics pipelines. Theranostics. 2018;8(15):4016–32.
Pan Q, Luo F, Liu M, Zhang XL. Oligonucleotide aptamers: promising and powerful diagnostic and therapeutic tools for infectious diseases. The Journal of infection. 2018;77(2):83–98.
Zhou G, Latchoumanin O, Bagdesar M, Hebbard L, Duan W, Liddle C, et al. Aptamer-based therapeutic approaches to target cancer stem cells. Theranostics. 2017;7(16):3948–61.
Cui L, Lu M, Li Y, Tang B, Zhang CY. A reusable ratiometric electrochemical biosensor on the basis of the binding of methylene blue to DNA with alternating AT base sequence for sensitive detection of adenosine. Biosens Bioelectron. 2018;102:87–93.
Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J Am Chem Soc. 2005;127(51):17990–1.
Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem. 2005;44(34):5456–9.
Deng C, Pi X, Qian P, Chen X, Wu W, Xiang J. High-performance ratiometric electrochemical method based on the combination of signal probe and inner reference probe in one hairpin-structured DNA. Anal Chem. 2017;89(1):966–73.
Liu LL, Han YW, Zhu KH, Li ZZ, Han YP, Lu Y, et al. Screening and characterization of aptamers of chronic myelognous leukemia K562 cells. Journal of the fourth military medical university. 2009;30(13):1157–60.
Liu LL. Screening, characterization and structure analysis of aptamers binding to chronic myelognous leukemia K562 cells: Lanzhou University (China); 2009.
Meng X, Xu M, Zhu J, Yin H, Ai S. Fabrication of DNA electrochemical biosensor based on gold nanoparticles, locked nucleic acid modified hairpin DNA and enzymatic signal amplification. Electrochim Acta. 2012;71(none):233–8.
Tian M, Pell WG, Conway BE. Nanogravimetry study of the initial stages of anodic surface oxide film growth at Au in aqueous HClO4 and H2SO4 by means of EQCN. Electrochim Acta. 2003;48(18):2675-89.
Zhang X, Xie G, Gou D, Luo P, Yao Y, Chen H. A novel enzyme-free electrochemical biosensor for rapid detection of Pseudomonas aeruginosa based on high catalytic Cu-ZrMOF and conductive Super P. Biosens Bioelectron. 2019;142:111486.
Ma J, Wu L, Li Z, Lu Z, Yin W, Nie A, et al. Versatile electrochemiluminescence assays for PEDV antibody based on rolling circle amplification and Ru-DNA nanotags. Anal Chem. 2018;90(12):7415–21.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81430053, 81401751, 81972027), Chongqing Health Commission (2018QNXM049, 2019ZDXM025), and Medical pre-research project of the Army Medical University (2018XYY04).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The experiments were approved by the ethics committee of Southwest Hospital (China).
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1.31 mb)
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, W., Tang, X. et al. Target-triggered “signal-off” electrochemical aptasensor assisted by Au nanoparticle–modified sensing platform for high-sensitivity determination of circulating tumor cells. Anal Bioanal Chem 412, 8107–8115 (2020). https://doi.org/10.1007/s00216-020-02940-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02940-x