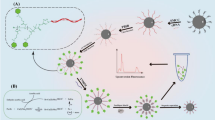
Abstract
An ultrasensitive fluorescence biosensor for detecting cytokeratin fragment antigen 21-1 (CYFRA 21-1) DNA of non-small cell lung carcinoma (NSCLC) is designed using polysaccharide and activator regenerated by electron transfer atom transfer radical polymerization (ARGET ATRP) signal amplification strategy. Thiolated peptide nucleic acid (PNA) is fixed on magnetic nanoparticles (MNPs) by a cross-linking agent and hybridized with CYFRA 21-1 DNA. Hyaluronic acid (HA) is linked to PNA/tDNA heteroduplexes in the form of carboxy-Zr4+-phosphate. Subsequently, multiple 2-bromo-2-methylpropionic acid (BMP) molecules are linked with HA to initiate ARGET ATRP reaction. Finally, a large number of fluorescein o-acrylate (FA) monomers are polymerized on the macro-initiators, and the fluorescence signal is significantly amplified. Under optimal conditions, this biosensor shows a significant linear correlation between the fluorescence intensity and logarithm of CYFRA 21-1 DNA concentration (0.1 fM to 0.1 nM), and the limit of detection is as low as 78 aM. Furthermore, the sensor has a good ability to detect CYFRA 21-1 DNA in serum samples and to recognize mismatched bases. It suggests that the strategy has broad application in early diagnosis by virtue of its high sensitivity and selectivity.

A novel and highly sensitive fluorescence biosensor for quantitatively detecting CYFRA 21-1 DNA via dual signal amplification of hyaluronic acid and ARGET ATRP reaction was developed. This proposed method has a low detection limit, wide detection range, high selectivity, and strong anti-interference.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is a malignant tumor derived from epithelial cells and leads to uncontrolled growth in the tissues of the lung [1]. Statistically, lung cancer is one of the fastest-growing cancers, with the highest morbidity and mortality and the greatest threat to the health of the population [2]. Lung cancer is divided into two main categories: small cell lung carcinoma (SCLC) and non-small cell lung carcinoma (NSCLC) [3, 4], in which an important biomarker of NSCLC is cytokeratin fragment antigen 21-1 (CYFRA 21-1) [5, 6]. Considerable research has shown that CYFRA 21-1 DNA is closely associated with the diagnosis of lung diseases [7, 8]. Despite the rapid development of science and technology in recent decades, the survival rate of patients with lung cancer remains extremely low, so it is urgent to develop sensitive and rapid methods for diagnosing lung cancer.
Over the past decade, multiple strategies for signal amplification have been widely applied to the detection of nucleic acid molecules, such as hybridization chain reaction [9, 10], enzyme-assisted amplification [11], rolling circle amplification [12], loop-mediated isothermal amplification [13], and ligase chain reaction [14]. However, traditional methods of amplification detection have several disadvantages, such as low sensitivity, high cost, and cumbersome operation steps, which hinder their application in biological samples. Therefore, an effective and economical polymerization amplification strategy seems to be an ideal method for improving the performance of biosensors.
Atom transfer radical polymerization (ATRP), a signal amplification method, has extremely broad application and development potential in the detection of trace molecules owing to its mild reaction conditions, moderate reaction temperature, diverse monomers, and easy industrial production [15, 16]. Activator regenerated by electron transfer ATRP (ARGET ATRP), a recently developed type of ATRP, can react in the presence of limited oxygen and require lower concentrations of catalyst due to the presence of excess reducing agents [17, 18], and its use has been gradually increasing in the industrial, biological, pharmaceutical, and environmental fields [17, 19]. In ARGET ATRP, a reversible dynamic equilibrium between dormant species and active species is established by adding reducing agent, in which the growth of the polymer chains mainly relies on atom transfer [20, 21]. As one of the essential requirements in ARGET ATRP reaction, the number of initiation sites is directly related to the quantity of polymer chains. Hence, the signal output can be increased by modifying the initiator structure, by means such as the introduction of functionalized hyaluronic acid (HA). HA is a natural linear polysaccharide, which is widely used as biomaterial in drug design, tissue engineering, and medical devices because of its good hydrophilicity, low toxicity, excellent biodegradability, and biocompatibility [22, 23]. Particularly, the skeleton structure of HA contains numerous oxygen-containing functional groups which can greatly increase the number of modified initiators [24]. Building on these features of HA, the output signal can be further amplified via ARGET ATRP.
At present, developing detection methods for target DNA (tDNA) has become a research hotspot in the field of modern biomedicine, including fluorescence [25], electrochemistry [26], and chemiluminescence methods [27]. Among these, fluorescence spectroscopy stands out because of its high sensitivity, simple operation, and fast response [28,29,30]. Based on these considerations, we designed a novel fluorescent method for DNA detection using polysaccharide-enhanced ARGET ATRP signal amplification.
In this report, we describe a highly sensitive and specific fluorescence detection method for CYFRA 21-1 DNA based on a dual signal amplification strategy using polysaccharide and ARGET ATRP. In this approach, the thiolated peptide nucleic acids (PNAs) are immobilized onto the magnetic nanoparticles (MNPs) via cross-linking and used as probes for specifically recognizing tDNA. After HA is associated with the phosphate groups of heteroduplexes (PNA/tDNA) by Zr4+, multiple 2-bromo-2-methylpropionic acid (BMP) molecules are connected to the HA backbone through an esterification reaction, initiating ARGET ATRP reaction. Plenty of fluorescein o-acrylate (FA) monomers are polymerized on the tDNA by ARGET ATRP reaction, which effectively amplifies the fluorescence signal. The detection results indicate that the biosensor has high selectivity in complex serum and strong anti-interference capability. All in all, this fluorescence method of detecting CYFRA 21-1 DNA has potential applications in the early diagnosis of lung cancer because of its high sensitivity and selectivity.
Materials and methods
Materials
N-hydroxysuccinimide (NHS), hyaluronic acid (HA), fluorescein o-acrylate (FA), 1-ethyl-3-[3-(dimethylamino) propyl] carbodiimide hydrochloride (EDC), and tris [2-(dimethylamino) ethyl] amine (Me6TREN) were acquired from Sigma-Aldrich Trading Co., Ltd. (Shanghai, China). 2-Bromo-2-methylpropionic acid (BMP) and diethylamine (DEA) were acquired from Aladdin Reagent Co., Ltd. (Shanghai, China). 4-(N-Maleimidomethyl) cyclohexane-1-carboxylic acid 3-sulfo-N-hydroxysuccinimide ester sodium salt (SSMCC), zirconium dichloride oxide octahydrate (ZrOCl2∙8H2O), and L-ascorbic acid (AA) were obtained from J&K Scientific Ltd. (Beijing, China). Magnetic nanoparticles (Fe3O4-MNPs, d = 200 nm, 10 mg mL−1) modified with amino groups on the surface were purchased from PuriMag Biotechnology Ltd. (Xiamen, China). Copper bromide (CuBr2) was obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Tetrabutylammonium hydroxide (TBA) was acquired from Shanghai Macklin Biochemical Co., Ltd. Normal human serum was obtained from Yu Duo Co., Ltd. (Shanghai, China). In addition, 0.1 M PBS (pH 7.40) and 0.1 M PBST (0.05% Tween-20, pH 7.40) were prepared and used as reactive buffer and washing solution. Ultrapure water (≥ 18.25 MΩ·cm) was used throughout all experiments. All chemicals used in the experiment were of analytical purity and not further purified.
All the nucleotide sequences used in the experiment were acquired from Sangon Biotech Co., Ltd. (Shanghai, China). Thiolated peptide nucleic acid (PNA) was designed by Panagene Inc. (South Korea). Their sequences are listed in Table 1.
Apparatus
All fluorescence measurements were recorded with an FLS 1000 fluorescence spectrophotometer (Edinburgh, UK). Scanning electron microscopy (SEM) images were obtained using Sigma HD field emission SEM (Zeiss, Germany).
Experimental methods
Functionalization of MNPs
Fe3O4-MNPs (20 μL, 10 mg mL−1) were washed three times and dispersed in PBST solution (160 μL, pH 7.40). Then the SSMCC (20 μL, 0.2 mM) was added for 2 h at 310 K, during which time the SSMCC was immobilized on the Fe3O4-MNPs via amino bonds [31]. The MNPs-SSMCC was then magnetically separated and washed with PBST and distributed in 90 μL PBST. The PNA (10 μL, 0.5 μM) was added and reacted for 2 h under the same conditions, to form the MNPs-SSMCC/PNA combination. The residual adsorbed oligodeoxynucleotides were removed by magnetic separation and washing with ultrapure water, and then MNPs-SSMCC/PNA was recovered in 180 μL PBST buffer.
Hybridization of tDNA with PNA
The tDNA (20 μL, 0.1 nM) was incubated with the prepared MNPs-SSMCC/PNA in PBST buffer for 2 h at 310 K, in which PNA was used as the probes for the specific hybridization of tDNA. Subsequently, the MNPs-SSMCC/PNA/tDNA was washed with PBST buffer and separated magnetically to remove any residual unhybridized tDNA, and then it was resuspended in 180 μL PBST.
Immobilization of initiators
First, Zr4+ (20 μL, 5 mM) was added to the MNPs-SSMCC/PNA/tDNA and reacted for 30 min at 310 K. The resulting MNPs-SSMCC/PNA/tDNA/Zr4+ was resuspended in 180 μL PBST after magnetic separation and washing with 15% ethanol. HA-TBA (20 μL, 5 μM) was mixed with the previously described reaction system for 2 h. For the next step, the MNPs-SSMCC/PNA/tDNA/Zr4+ was washed and magnetically separated with ultrapure water. The BMP-NHS (10 μL, 5 mM) was interacted with 180 μL PBST solution of the MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA for 2 h under the same conditions, with DEA (10 μL, 5 mM) as catalyst [32]. The resulting mixture was washed and magnetically separated with DMSO solution (volume ratio between DMSO and H2O equal to 2). At this point, the HA-TBA/BMP-NHS macro-initiators were connected to the tDNA via Zr4+ to initiate the ARGET ATRP reaction. The MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA/ BMP-NHS was evenly dispersed in 155 μL PBST buffer for use.
The synthesis of 5 μM HA-TBA and 5 μM BMP-NHS mentioned above was performed as follows: 10 μM HA (dissolved in water) and 2.5 mM TBA (dissolved in water) were incubated for 2 h at 310 K to improve the solubility of HA in organic solvents [33]. The carboxyl group of BMP (15 mM) was activated by equivalent EDC (15 mM) and NHS (15 mM) in DMSO at 310 K overnight [32].
ARGET ATRP reaction
The mixture of CuIIBr2/Me6TREN (20 μL, 10 mM, dissolved in DMF, molar ratio between Me6TREN and CuBr2 equal to 1.1), FA (5 μL, 10 mM, dissolved in DMF), and AA (20 μL, 2 mM, dissolved in 70% ethanol) were added to the prepared MNP system and incubated at 310 K for 50 min. A large number of FA fluorescent monomers were polymerized on the surface of MNPs by ARGET ATRP reaction to form MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA/BMP-NHS/FA.
Fluorescence measurement
After magnetically separating and washing with DMF and PBS buffer solution in turn, the prepared functionalized MNPs with tDNA/Zr4+/HA-TBA/BMP-NHS/FA were distributed in 500 μL PBS buffer solution for the fluorescence analysis. The fluorescence signal was scanned from 500 to 600 nm under an excitation wavelength of 490 nm and emission at 511 nm. (The slit width was set at 3 nm.)
Results and discussion
Principle of the strategy
In Scheme 1, the principle of fluorescence CYFRA 21-1 DNA detection is displayed based on signal amplification of polysaccharide and ARGET ATRP reaction. Relative to single-stranded DNA (ssDNA) or RNA, PNA has a higher binding affinity and specificity to its complementary target at low ion concentrations, so it has been widely used as a low-interference nucleic acid probe [34]. First, the carbonyl groups of the SSMCC molecules were connected with the amino groups on the MNPs by amide bonds, and the double bonds in the N-maleimide methyls of the SSMCC reacted with PNA-SH by addition reaction so that PNA probes were connected to the surface of the MNPs. As the PNA sequence was complementary to that of the tDNA, the PNA recognized and hybridized with the tDNA. In order to increase the number of initiators of ARGET ATRP reaction connecting to PNA/DNA heteroduplexes, we attached multiple BMP molecules to the polysaccharide backbone of HA to form the HA-TBA/BMP-NHS macro-initiators. After that, the phosphate groups in tDNA reacted with the carboxy groups in macro-initiators to form “PNA/tDNA/Zr4+/HA-TBA/BMP-NHS” complexes under the coordination of Zr4+. The FA monomers were then continuously polymerized to the tDNA by ARGET ATRP reaction with HA-TBA/BMP-NHS as initiator, Me6TREN as ligand, and Cu(І) as catalyst (Cu(І) was generated from CuBr2 and AA). In this way, the amount of tDNA was measured by detecting the amplified fluorescence intensity emitted by FA.
The mechanism of the ARGET ATRP is illustrated in Scheme. 2. At first, CuIIBr2/Me6TREN was reduced to CuIBr/Me6TREN activator through adding AA. Afterwards, CuIBr/Me6TREN reacted with initiators (Pn-Br) to generate free radicals (Pn˙) and deactivators (CuIIBr2/Me6TREN), the latter of which participated in catalytic cycling once again [35]. The free radicals could be constantly propagated with FA monomers to form a polymerization chain, or returned to the dormant species (Pn-Br) states in the presence of CuIIBr2/Me6TREN deactivators, which were further involved in the reaction. Eventually, long polymer chains with large quantities of FA fluorescence monomers were grafted from the surface of HA, amplifying the detection signal of CYFRA 21-1 DNA [36].
Study of feasibility
To examine the feasibility of the proposed strategy by polysaccharide and ARGET ATRP reaction, the fluorescence signal of different modified MNPs was measured. Obviously, the proposed MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA/BMP-NHS/FA (curve a) generated the strongest fluorescence intensity in Fig. 1, which was attributed to the fact that FA monomers were heavily polymerized onto MNPs by the mechanism of polysaccharide-enhanced ARGET ATRP signal amplification, and the fluorescence signal was observably magnified. The fluorescence signal was at medium intensity in the absence of HA-TBA (curve f), because BMP-NHS molecules similarly contained carboxy groups which were linked to MNPs-SSMCC/PNA/tDNA via Zr4+ for performing the subsequent ARGET ATRP reaction. Under other conditions (in the absence of SSMCC, PNA, tDNA, Zr4+, HA-TBA, and BMP-NHS, respectively), the fluorescence signal was particularly weak. It was proved that the strategy based on polysaccharide-enhanced ARGET ATRP amplification is feasible for use as a signal amplification method in biosensing.
Feasibility of ultrasensitive detection for tDNA assay. Fluorescence spectra of MNPs with different modification, which are a MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA/BMP-NHS/FA (the proposed scheme), b MNPs/PNA/tDNA/Zr4+/HA-TBA/BMP-NHS/FA, c MNPs-SSMCC/tDNA/Zr4+/HA-TBA/BMP-NHS/FA, d MNPs-SSMCC/PNA/Zr4+/HA-TBA/BMP-NHS/FA, e MNPs-SSMCC/PNA/tDNA/HA-TBA/BMP-NHS/FA, f MNPs-SSMCC/PNA/tDNA/Zr4+/BMP-NHS/FA, and g MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA/FA. The inset shows the enlarged view of the fluorescence spectra of different modified MNPs
In addition, the surface morphology of MNPs modified in different modes (MNPs-SSMCC/ PNA/tDNA/Zr4+/HA-TBA/BMP-NHS/FA and MNPs-SSMCC/PNA/Zr4+/HA-TBA/BMP-NHS/ FA) was further characterized by scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDS). As displayed in Fig. 2, SEM investigation revealed that the functional MNPs (Fig. 2b) had a relatively rougher surface than that of the control group without tDNA (Fig. 2a), and floccules were observed on the surface of the functional MNPs. Furthermore, the elemental composition of MNPs-SSMCC/PNA/tDNA/Zr4+/HA-TBA/BMP-NHS/FA could be clearly observed in the EDS images, in which S, P, Zr, and Br (Fig. 2c–f) were characteristic elements of SSMCC (and PNA), tDNA, Zr4+, and BMP, respectively. To sum up, it indicated that the fluorescence biosensor was successfully constructed via polysaccharide and ARGET ATRP signal amplification method.
Optimization of conditions
In order to obtain high-performance fluorescence detection, it was essential to optimize the relevant experimental conditions. The concentration of initiators and reaction time of ARGET ATRP directly determined the total amount of polymer, and affected the signal amplification of the strategy. Therefore, we considered the effects of ARGET ATRP time and BMP-NHS concentration on the polymerization reaction.
Figure 3a shows the impact of BMP-NHS concentration on ARGET ATRP. The fluorescence intensity was obviously enhanced as the concentration of the BMP-NHS increased over the range of 0.05 mM~2.5 mM, and reached a peak at 2.5 mM. The fluorescence intensity then decreased with a further increase in the concentration of BMP-NHS. This was because initiators were indispensable in ARGET ATRP, and with the increase in initiator concentration, more free radicals were polymerized with the FA monomers and the output signal was greatly amplified. At the same time, however, the polymerization rate was lowered [37]. Therefore, 2.5 mM was selected as the most appropriate concentration of BMP-NHS.
Figure 3b shows the fluorescence response of ARGET ATRP reaction time. The fluorescence intensity obviously increased with the prolongation of ARGET ATRP reaction time and reached a maximum of 60,000 a.u. at 50 min. Once the equilibrium was reached, the fluorescence intensity remained stable with the extension of the reaction time, which might be caused by the termination of free radicals. Eventually, 2.5 mM of BMP and 50 min of ARGET ATRP reaction time were selected for this study.
Analytical performance
Under the optimal conditions, we investigated the fluorescence response to different concentrations of tDNA. As illustrated in Fig. 4a, it was observed that the fluorescence intensity increased with increasing concentration of tDNA. A significant linear correlation relationship between the fluorescence intensity and the logarithm of tDNA concentration from 0.1 fM to 0.1 nM is shown in Fig. 4b. The linear regression equation is y = 8838.75 log CtDNA + 18,740.80 with a correlation coefficient (R2) of 0.9983. The limit of detection (LOD) of this method was calculated to be 78 aM based on S/N = 3. This revealed that this DNA biosensor based on amplification from a biocompatible polysaccharide and ARGET ATRP reaction displayed high sensitivity.
The analytical performance for tDNA detection in PBS. a Fluorescence spectra with different tDNA concentrations (1 aM, 10 aM, 0.1 fM, 1 fM, 10 fM, 0.1 pM, 1 pM, 10 pM, and 0.1 nM). b The linear relationship between the fluorescence intensity and the target concentration in the range of 0.1 fM to 0.1 nM at 511 nm, and the dotted horizontal line indicates the fluorescence intensity for LOD estimation. The error bars represent the standard deviation of four measurements
To further confirm the good performance of the fluorescence biosensor, comparison with some different DNA sensors developed in recent years are listed in Table 2. Compared with other DNA sensors, the proposed sensor exhibited a lower LOD and a wider linear range.
Selectivity, anti-interference capability, and reproducibility
The specificity of this strategy for biological detection was appraised by comparing the fluorescence intensity of tDNA with SBM, TBM, and NC under the same experimental conditions. As shown in Fig. 5a, the fluorescence response signal of SBM, which was most likely to cause interference, accounted for only 22.0% of the signal when compared with recognized response of tDNA, and the fluorescence response of TBM was approximately 82.6% lower than that of tDNA. There was no significant difference between NC and background signal. The above results confirmed that the PNA fixed on the surface of MNPs accurately realized complementary pairing recognition with target tDNA, making the method highly specific for quantitative recognition of tDNA. The reason for this phenomenon was that the presence of mismatched base(s) hindered the formation of PNA/DNA heteroduplexes, and thus reduced the fluorescence response intensity due to the high specificity of the neutral PNA probes [42].
a Fluorescence response of this method toward tDNA, SBM, TBM, and NC with a concentration of 100 pM. b The detection capability for tDNA with concentrations of 0.1 nM, 0.1 pM, and 0.1 fM in PBS buffer and 5% normal human serum. The error bars represent the standard deviations of four independent measurements
In order to verify the anti-interference ability of the fluorescence biological analysis, the fluorescence response of 0.1 nM, 0.1 pM, and 0.1 fM tDNA in PBS buffer and 5% normal human serum samples were severally measured. The results of different concentrations of tDNA in normal human serum were compared with those obtained in the 0.1 M PBS (pH 7.4) experimental system in Fig. 5b. The fluorescence intensity of 0.1 nM tDNA in 5% normal human serum was approximately 85.3% of that PBS buffer, and the intensity of 0.1 pM and 0.1 fM in 5% normal human serum was approximately 85.4% and 89.1% of that PBS buffer, respectively. The tDNA in human serum samples still produced an obvious signal. These results showed that this method possessed good anti-interference ability in complex serum samples, indicating that it has great promise for practical clinical application.
In order to further evaluate the practical applicability of the design, five repeated experiments were performed based on intra- and inter-assays at the tDNA concentration of 0.1 nM. The relative standard deviations (RSD) were 3.3% and 3.5%, respectively. The results proved that the biosensor has high precision and reproducibility.
Conclusions
In summary, an ultrasensitive fluorescence biosensor for detection of CYFRA 21-1 DNA was developed based on polysaccharide-enhanced ARGET ATRP signal amplification technology for the first time. Under optimal reaction conditions, the biosensor shows good analytical performance with the LOD as low as 78 aM. Moreover, it displays good detection capability in complex normal human serum and high selectivity for detecting CYFRA 21-1 DNA. Because of its high selectivity, sensitivity, anti-interference capability, and reproducibility, this signal amplification strategy provides a valuable new approach for the early clinical diagnosis and treatment of lung cancer.
References
Kanwal M, Ding X, Cao Y. Familial risk for lung cancer. Oncol Lett. 2017;13(2):535–42.
Zhao G, Li M. Ni-Doped MoS2 biosensor: a promising candidate for early diagnosis of lung cancer by exhaled breathe analysis. Appl Phys A Mater Sci Process. 2018;124(11):751.
Arya SK, Bhansali S. Lung cancer and its early detection using biomarker-based biosensors. Chem Rev. 2011;111(11):6783–809.
Bharti A, Ma PC, Salgia R. Biomarker discovery in lung cancer-promises and challenges of clinical proteomics. Mass Spectrom Rev. 2007;26(3):451–66.
Kammer MN, Kussrow AK, Webster RL, Chen HD, Hoeksema M, Christenson R, et al. Compensated interferometry measures of CYFRA 21-1 improve diagnosis of lung cancer. ACS Comb Sci. 2019;21(6):465–72.
Mizuquchi S, Nishiyama N, Iwata T, Nishida T, Izumi N, Tsukioka T, et al. Serum sialyl Lewis x and cytokeratin 19 fragment as predictive factors for recurrence in patients with stage I non-small cell lung cancer. Lung Cancer. 2007;5(4):374.
Dohmoto K, Hojo S, Fujita J, Ueda Y, Bandoh S, Yamaji Y, et al. Mechanisms of the release of CYFRA21-1 in human lung cancer cell lines. Lung Cancer. 2000;30(1):55–63.
Schmidt B, Beyer J, Dietrich D, Bork I, Liebenberg V, Fleischhacker M. Quantification of cell-free mSHOX2 plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS One. 2015;10(2):e0118195.
Li S, Li G, Du Z, Zhu L, Tian J, Luo Y, et al. The ultra-sensitive visual biosensor based on thermostatic triple step functional nucleic acid cascade amplification for detecting Zn2+. Food Chem. 2019;290:95–100.
Khalil I, Yehye WA, Julkapli NM, Rahmati S, Ibn Sina AA, Basirun WJ, et al. Graphene oxide and gold nanoparticle based dual platform with short DNA probe for the PCR free DNA biosensing using surface-enhanced Raman scattering. Biosens Bioelectron. 2019;131:214–23.
Wang K, Zhai F, He M, Wang J, Yu Y, He R. A simple enzyme-assisted cascade amplification strategy for ultrasensitive and label-free detection of DNA. Anal Bioanal Chem. 2018;411(19):4569–76.
Ding C, Liu H, Wang N, Wang Z. Cascade signal amplification strategy for the detection of cancer cells by rolling circle amplification and nanoparticles tagging. Chem Commun. 2012;48(41):5019–21.
Sun Y, Tian H, Liu C, Sun Y, Li Z. One-step detection of microRNA with high sensitivity and specificity via target-triggered loop-mediated isothermal amplification (TT-LAMP). Chem Commun. 2017;53(80):11040–3.
Yan J, Li Z, Liu C, Cheng Y. Simple and sensitive detection of microRNAs with ligase chain reaction. Chem Commun. 2010;46(14):2432–4.
Chen F, Hou S, Li Q, Fan H, Fan R, Xu Z, et al. Development of atom transfer radical polymer-modified gold nanoparticle-based enzyme-linked immunosorbent assay (ELISA). Anal Chem. 2014;86(20):10021–4.
He P, Lou X, Woody SM, He L. Amplification-by-polymerization in biosensing for human genomic DNA detection. ACS Sens. 2019;4(4):992–1000.
Li X, Wang W, Li B, Zhu S. Kinetics and modeling of solution ARGET ATRP of styrene, butyl acrylate, and methyl methacrylate. Macromol React Eng. 2011;5(9-10):467–78.
Patra S, Roy E, Das R, Karfa P, Kumar S, Madhuri R, et al. Bimetallic magnetic nanoparticle as a new platform for fabrication of pyridoxine and pyridoxal-5′-phosphate imprinted polymer modified high throughput electrochemical sensor. Biosens Bioelectron. 2015;73:234–44.
Zhang Z, Wang X, Tam KC, Sebe G. A comparative study on grafting polymers from cellulose nanocrystals via surface-initiated atom transfer radical polymerization (ATRP) and activator re-generated by electron transfer ATRP. Carbohydr Polym. 2019;205:322–9.
Forbes DC, Creixell M, Frizzell H, Peppas NA. Polycationic nanoparticles synthesized using ARGET ATRP for drug delivery. Eur J Pharm Biopharm. 2013;84(3):472–8.
Ma W, Otsuka H, Takahara A. Poly (methyl methacrylate) grafted imogolite nanotubes prepared through surface-initiated ARGET ATRP. Chem Commun. 2011;47(20):5813–5.
Kwon SS, Kong BJ, Park SN. Physicochemical properties of pH-sensitive hydrogels based on hydroxyethyl cellulose-hyaluronic acid and for applications as transdermal delivery systems for skin lesions. Eur J Pharm Biopharm. 2015;92:146–54.
Hahn SK, Park JK, Tomimatsu T, Shimoboji T. Synthesis and degradation test of hyaluronic acid hydrogels. Int J Biol Macromol. 2007;40(4):374–80.
Fiorica C, Pitarresi G, Palumbo FS, Di Stefano M, Calascibetta F, Giammona G. A new hyaluronic acid pH sensitive derivative obtained by ATRP for potential oral administration of proteins. Int J Pharm. 2013;457(1):150–7.
Wen D, Liu Q, Cui Y, Kong J, Yang H, Liu Q. DNA based click polymerization for ultra-sensitive IFN-γ fluorescent detection. Sensors Actuators B Chem. 2018;276:279–87.
Lazerges M, Bedioui F. Analysis of the evolution of the detection limits of electrochemical DNA biosensors. Anal Bioanal Chem. 2013;405(11):3705–14.
Tanaka T, Matsunaga T. Fully automated chemiluminescence immunoassay of insulin using antibody-protein A-bacterial magnetic particle complexes. Anal Chem. 2000;72(15):3518–22.
Dai N, Kool ET. Fluorescent DNA-based enzyme sensors. Chem Soc Rev. 2011;40(12):5756–70.
Xing X, Liu X, He Y, Lin Y, Zhang C, Tang H, et al. Amplified fluorescent sensing of DNA using graphene oxide and a conjugated cationic polymer. Biomacromolecules. 2013;14(1):117–23.
Song C, Li B, Yang X, Wang K, Wang Q, Liu J, et al. Use of β-cyclodextrin-tethered cationic polymer based fluorescence enhancement of pyrene and hybridization chain reaction for the enzyme-free amplified detection of DNA. Analyst. 2017;142(1):224–8.
de la Torre TZG, Herthnek D, Ramachandraiah H, Svedlindhl P, Nilsson M, Stromme M. Evaluation of the sulfo-succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate coupling chemistry for attachment of oligonucleotides to magnetic nanobeads. J Nanosci Nanotechnol. 2011;11(10):8532–7.
Pitarresi G, Fiorica C, Licciardi M, Palumbo FS, Giammona G. New hyaluronic acid based brush copolymers synthesized by atom transfer radical polymerization. Carbohydr Polym. 2013;92(2):1054–63.
Hu Y, Li Y, Xu F. Versatile functionalization of polysaccharides via polymer grafts: from design to biomedical applications. Acc Chem Res. 2017;50(2):281–92.
Pellestor F, Paulasova P, Hamamah S. Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr Pharm Des. 2008;14(24):2439–44.
Dong H, Tang W, Matyjaszewski K. Well-defined high-molecular-weight polyacrylonitrile via activators regenerated by electron transfer ATRP. Macromolecules. 2007;40(9):2974–7.
Hansson S, Trouillet V, Tischer T, Goldmann AS, Carlmark A, Barner-Kowollik C, et al. Grafting efficiency of synthetic polymers onto biomaterials: A comparative study of grafting-from versus grafting-to. Biomacromolecules. 2012;14(1):64–74.
Audouin F, Larragy R, Fox M, O’Connor B, Heise A. Protein immobilization onto poly(acrylic acid) functional macroporous polyHIPE obtained by surface-initiated ARGET ATRP. Biomacromolecules. 2012;13(11):3787–94.
Karimizefreh A, Mahyari F, VaezJalali M, Mohammadpour R, Sasanpour P. Impedimetic biosensor for the DNA of the human papilloma virus based on the use of gold nanosheets. Microchim Acta. 2017;184(6):1729–37.
Liu Q, Ma K, Wen D, Sun H, Wang Q, Kong J, et al. BisPNA-assisted detection of double-stranded DNA via electrochemical impedance spectroscopy. Electroanalysis. 2018;31(1):160–6.
Liu X, Aizen R, Freeman R, Yehezkeli O, Willner I. Multiplexed aptasensors and amplified DNA sensors using functionalized graphene oxide: application for logic gate operations. ACS Nano. 2012;6(4):3553–63.
Shao K, Wang L, Wen Y, Wang T, Teng Y, Shen Z, et al. Near-infrared carbon dots-based fluorescence turn on aptasensor for determination of carcinoembryonic antigen in pleural effusion. Anal Chim Acta. 2019;1068:52–9.
Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick Hydrogen-bonding rules. Nature. 1993;365(6446):566–8.
Acknowledgments
This work was supported by the project for tackling of key scientific and technical problems in Henan Province (no. 192102310033) and the National Natural Science Foundation of China (no. 21575066).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Human and animal rights
This article does not contain any studies with human or animal subjects.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Zhang, Y., Zhao, L. et al. Polysaccharide-enhanced ARGET ATRP signal amplification for ultrasensitive fluorescent detection of lung cancer CYFRA 21-1 DNA. Anal Bioanal Chem 412, 2413–2421 (2020). https://doi.org/10.1007/s00216-020-02394-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02394-1