Abstract
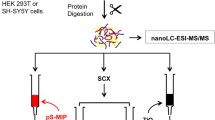
The work describes the development of an enrichment method for the analysis of endogenous phosphopeptides in serum. Endogenous peptides can play significant biological roles, and some of them could be exploited as future biomarkers. In this context, blood is one of the most useful biofluids for screening, but a systematic investigation of the endogenous peptides, especially phosphorylated ones, is still lacking, mainly due to the lack of suitable analytical methods. Thus, in this paper, different phosphopeptide enrichment strategies were pursued, based either on metal oxide affinity chromatography (MOAC, in the form of commercial TiO2 spin columns or magnetic graphitized carbon black-TiO2 composite), or on immobilized metal ion affinity chromatography (IMAC, in the form of Ti4+-IMAC magnetic material or commercial Fe3+-IMAC spin columns). While MOAC strategies proved completely unsuccessful, probably due to interfering phospholipids displacing phosphopeptides, the IMAC materials performed very well. Different sample preparation strategies were tested, comprising direct dilution with the loading buffer, organic solvent precipitation, and lipid removal from the matrix, as well as the addition of phosphatase inhibitors during sample handling for maximized endogenous phosphopeptide enrichment. All data were acquired by a shotgun peptidomics approach, in which peptide samples were separated by reversed-phase nanoHPLC hyphenated with high-resolution tandem mass spectrometry. The devised method allowed the identification of 176 endogenous phosphopeptides in fresh serum added with inhibitors by the direct dilution protocol and the Ti4+-IMAC magnetic material enrichment, but good results could also be obtained from the commercial Fe3+-IMAC spin column adapted to the batch enrichment protocol.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The term “biomarker” is used to indicate a broad subcategory of medical signs, indicators for normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. Biomarkers are mostly connected to disease, but they can also be indicators of environmental exposure [1].
Blood, together with urine and other biofluids, is the sample of choice to investigate for possible biomarkers. Biomarker discovery based on blood has the advantages of the availability of blood, plasma, or serum samples, which are commonly employed also for routine analysis and on the wide range of substances therein present, which comprise proteins [2], lipids [3], metabolites [4], vesicles and cells and cell fragments [5, 6], and nucleic acids [7], whose profile depends on the physiological and pathological states.
Due to their bioactivities [8,9,10,11,12], peptides and proteins are suitable compounds for biomarker investigation, because they can be involved in pathways specific of disease [13,14,15]. In order to perform a comprehensive profile of possible biomarkers, high-throughput methods for molecular analysis are fundamental. Low molecular weight proteins have been considered a source of useful diagnostic biomarkers [16], as they comprise cytokines, peptide hormones, endogenous peptide products, and potentially bioactive fragments derived from the parent proteome. Peptides are small enough to easily move within the body by crossing the vascular permeable barriers, including the blood–brain barrier and tumor barriers. The investigation of the endogenous peptidome is important to provide additional information on the state of the system, as endogenous peptides can originate by a regulated process which would represent a post-translational maturation of proteins [17]. Thus, the peptidome is not simply the result of protein degradation and peptides can also play important roles in the body, for example as signaling molecules [18].
When dealing with the investigation of peptides as biomarkers, peptides with post-translational modifications (PTMs) can also provide valuable information. Protein phosphorylation is biologically significant, as it regulates several cell functions in living organisms; in humans, abnormal phosphorylation has been linked to several diseases, such as cancer [19]. Differently from other PTMs, the analysis of protein phosphorylation can be achieved by shotgun proteomics approaches [20].
Peptidomics is the tool of choice to investigate the endogenous peptides in complex matrices. In fact, peptidomics allows a comprehensive qualitative and quantitative analysis of peptides in a biological sample. Peptidomics can be considered a subfield of proteomics, from which the separation approaches and analytical and computation technologies are borrowed. Peptidomics well applies to biomarker discovery, which represents the main field of peptidomics applications thus far [18].
The complex biological matrices typically examined in peptidomics experiments require systematic peptide extraction to achieve a successful analysis. The analysis of the endogenous peptidome in blood poses some challenges: blood is a very complex matrix, in which many compounds are present [21], and direct analysis can be hindered. Endogenous peptides in blood with no PTM have already been addressed in the literature, and strategies for their selective analysis have been described [22,23,24]. As far as the endogenous phosphopeptide analysis is concerned, blood derivative products can be much better exploited for analysis, in particular serum, which is free from chelating agents used to prepare plasma. In the analysis of phosphorylations in blood, on the one hand, phosphoproteins in human have already been addressed [25], also for searching new biomarker candidates [26], and on the other hand, the investigation of endogenous phosphopeptides is limited to new material applications [23, 27, 28] and application of database search strategies, either by a focused database [29] or a de novo search sequencing-assisted database search approach [30]. As a consequence, the investigation of endogenous phosphopeptides in serum is limited to few peptides [31]. Still, endogenous phosphopeptides represent interesting targets in serum, possible phosphopeptide biomarker candidates have been suggested in comparative studies for breast cancer [26], and differences in the endogenous phosphopeptide profiles were reported for gastric cancer [31], gallbladder carcinoma [32], and hepatocellular carcinoma [33].
Given the different performance of the most common phosphopeptide enrichment strategies [34] and the lack of a suitable comparison for the analysis of the endogenous serum phosphopeptides, in this work, both the immobilized metal ion affinity chromatography (IMAC) and the metal oxide affinity chromatography (MOAC) were exploited for the enrichment of the endogenous phosphopeptides in human serum. A comparison of their performance was accomplished: for MOAC enrichment, commercial TiO2 spin columns and magnetic graphitized carbon black-TiO2 composite [35] were tested to compare spin column and batch enrichment. For IMAC, commercial Fe3+-IMAC spin columns and Ti4+-IMAC magnetic material [36] were used. The sample preparation prior to phosphopeptide enrichment was also systematically addressed, by comparing the performance of serum direct dilution to protein precipitation by organic solvent pretreatment. Experiments were performed on commercial serum, but the effect of phosphatase inhibitors added to fresh serum was also evaluated. The best combination of sample preparation and enrichment was exploited for endogenous serum phosphopeptide analysis by means of a shotgun peptidomics workflow.
Materials and methods
All chemicals, reagents, protein standards, and organic solvents of the highest grade available were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated. Doubled-distilled water (ddH2O) was prepared by arium 611 VF system from Sartorius (Göttingen, Germany).
Preparation of serum samples
The serum samples employed in the comparison experiments were purchased by Sigma-Aldrich. The commercial serum was employed for all comparative experiments, except where clearly stated, which is for assessment of phosphatase inhibitors effect on phosphopeptide identification. Only for these experiments, freshly prepared serum (referred to as “fresh serum added with inhibitors”) was employed and prepared as follows. Fresh serum added with inhibitors was prepared from the human whole blood provided by the Department of Experimental Medicine (Sapienza University of Rome) according to institutional bioethics (informed consent). In this latter case, 5 mL of whole blood was drawn by venipuncture from 20- to 40-year-old healthy donors and pooled. Soon after collection, blood was left to clot for 30 min, then it was centrifuged at 2000×g for 10 min at 4 °C. The supernatant was gently withdrawn to avoid clots and hemolysis. At this point, one tablet of cOmplete™ Mini, EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich), and one tablet of PhosSTOP Phosphatase Inhibitor Cocktail (Sigma-Aldrich) were added to 10 mL of fresh serum. Both the Sigma pool and the fresh sera added with inhibitors were stored in 1-mL aliquots at − 20 °C until use. Before each enrichment, the aliquots were thawed at 4 °C and centrifuged at 12,000×g for 10 min to remove insoluble debris.
Selective enrichment of phosphopeptides from serum by the Ti4+-IMAC magnetic material
For comparative purposes, two serum preparation protocols were followed for the enrichment of endogenous phosphopeptides from serum. Enrichment was performed either directly on 1:10 diluted samples or on the supernatant obtained after serum protein precipitation by organic solvents (see the Electronic Supplementary Material (ESM) for experimental details).
In the final method (see the ESM for experimental details about the commercial TiO2 spin column and magnetic graphitized carbon black-TiO2 composite protocols), 1 mL of the clarified serum sample was diluted with 9 mL of loading buffer (H2O/acetonitrile (ACN), 50:50 (v/v) with 0.1% TFA). After dilution, the sample was shaken for 5 min until clear. After this step, the sample was applied to the enrichment media as later described.
The Ti4+-IMAC magnetic material was prepared by seeded polymerization [37, 38] as previously described [36]. In the final protocol, 10 mg of Ti4+-IMAC magnetic material was conditioned with 200 μL of loading buffer under gentle agitation (Digital Vortex-Genie 2 by Scientific Industries, Bohemia, NY, USA) for 2 min. After this and the following steps, a 30-s centrifugation (14,000×g) was performed to settle drops adhering to the vial and then the magnet (a permanent magnetic disk Nd-Fe-B, 25 mm × 5 mm, by Supermagnete, Gottmadingen, Germany) was applied to retrieve the magnetic phase. The conditioning operation was performed twice. After this step, the conditioned phase was added to the serum sample and gently shaken for 40 min. The supernatant was discarded, and three washing steps with 500 μL of the loading buffer (2-min shaking) were performed. Finally, phosphopeptides were eluted twice with 500 μL of 1.5% NH3 (aq) by gently shaking for 5 min. Combined eluates were acidified with TFA to pH 2.5, desalted, dried down in a Speed-Vac SC250 Express (Thermo Savant, Holbrook, NY, USA), and dissolved with 100 μL of 0.1% formic acid (FA). Three experimental replicates were performed for each condition.
Commercial Fe3+-IMAC spin column enrichment
For comparative purposes and to provide a commercial material for application of the final protocol, the Pierce™ Fe-NTA Phosphopeptide Enrichment Kit (Thermo Fisher Scientific) was purchased from VWR (Radnor, Pennsylvania) and used according to instructions, with some modifications to adapt it to the batch enrichment. Since the kit has volume restrictions due to the spin column size (600 μL ca.), the direct dilution enrichment experiments from serum were performed by dispersing the affinity material in the diluted serum inside 15-mL PP falcon tubes, employed exclusively for the incubation step. After incubation, the tubes were centrifuged at 7000×g for 5 min and the entire volume was stepwise transferred to the spin column. All other operations were performed according to the instruction manual.
NanoHPLC-MS/MS analysis and peptide identification
For each sample, a 20-μL sample was analyzed by nanoHPLC and high-resolution MS/MS. Chromatographic separation was performed on a Dionex Ultimate 3000. Samples were online preconcentrated (Dionex, 300 μm i.d. × 5 mm Acclaim PepMap 100 C18 μ-column, 5 μm particle size, 100 Å pore size), loaded with ddH2O/ACN 99:1 (v/v) containing 0.1% (v/v) TFA at a flow rate of 10 μL min−1. RP separation was performed on an EASY-Spray column (50 cm × 75 μm i.d. PepMap C18, 2-μm particles, 100 Å pore size; Thermo Scientific) operated at 200 nL min−1 and at 40 °C. A 166-min-long multistep gradient was for peptide separation, using ddH2O with 0.1% FA as phase A and ACN with 0.1% FA as phase B. One percent phase B was maintained for 5 min, then it was linearly increased to 5% within 2 min; afterwards, phase B was gradually increased to 35% in 90 min. For column washing, B was brought to 90% in 3 min and kept constant for 20 min and then the column was re-equilibrated for 45 min. Eluting peptides were ionized by an EASY-Spray source and analyzed by high-resolution tandem MS by Orbitrap Elite Hybrid Ion Trap-Orbitrap mass spectrometer (Thermo Scientific) in the m/z range of 380–1400 Da and 30,000 (full width at half maximum at m/z 400) resolution for the full scan. A data-dependent mode acquisition was enabled in top 10 mode, rejecting + 1 and unassigned charge states and fragmenting precursor ions by HCD at 15,000 resolution, normalized collision energy of 35%, and an isolation window of 2 m/z. To minimize redundant spectral acquisitions, dynamic exclusion was enabled with a repeat count of 1 and a repeat duration of 30 s with exclusion duration of 70 s. For each sample, three technical replicates were performed.
The acquired raw MS/MS data files from Xcalibur software (version 2.2 SP1.48, Thermo Fisher Scientific) were searched against UniProt database by Proteome Discoverer software (version 1.3, Thermo Scientific) and the Mascot (v.2.3.2, Matrix Science) search engine, as previously described [39], using Swiss-Prot and human taxonomy (20,284 entries). Finally, physicochemical features of the identified peptides were calculated using the freeware ProPAS [40].
Results and discussion
Serum endogenous phosphopeptide investigation by MOAC systems
Experiments for comparison between the two most important strategies for phosphopeptide enrichment, i.e., IMAC and MOAC, were performed to assess differences between the two, given the well-known partial complementarity [34].
Experiments were initially performed using the commercial TiO2 spin columns, which represent one of the most widespread approaches for phosphopeptide enrichment, within which hand-packed pure TiO2 spin columns can be included. The main problem with spin columns is the strict volume limitation, since they are assembled from 200-μL micropipette tips. Due to volume limitations, the serum sample preparation by dilution was not a compatible procedure. Firstly, the use of large volumes implied an extremely long loading time during enrichment; secondly, column clogging issues could not be bypassed, not even by initial serum clarification. Given the above, a classical acetone precipitation strategy was tested. Acetone precipitation is a pretreatment often employed for the analysis of endogenous peptides, as it allows to remove proteins from the matrix and enrich free peptides. However, as later also observed for the magnetic graphitized carbon black-TiO2 composite and the Ti4+-IMAC magnetic material, the performance of acetone precipitation was disappointing. Coupling acetone precipitation of serum with the commercial TiO2 spin column enrichment allowed to identify a single phosphopeptide, QQYHRALVAVLLSRT, which belonged to the translator activator GCN1. The most abundant fibrinogen peptides were not detected at all. The result was search space independent, as the use of focused database [29] only added three more identifications (KSIIEEKTQELDSITKKLQEINKEISGR, ADSGEGDFLAEGGGVR, VEQTAIKVSLK).
To try improving MOAC performance by bypassing the volume restrictions, a batch enrichment using the magnetic graphitized carbon black-TiO2 composite [35] was tested, with three composite amounts (3, 5, and 10 mg) to exclude saturation phenomena, but also in this case, the detection of endogenous phosphopeptides was limited, with no more than 11 identifications (ESM Table S1). For comparison, the precipitation protocol was tested also for the magnetic graphitized carbon black-TiO2 composite, employing three organic solvents with different polarity: acetone, methanol, and ACN. The aim of testing different solvents for precipitation was to assess any effect on the isolation of the endogenous phosphopeptides, which might be large peptides potentially removed by precipitation. Attempts were all unsuccessful, and no improvement could be accomplished (ESM Table S1).
The poor performance provided by MOAC systems agreed with previous reports using a TiO2-based strategy [41]. At this point, it was clear that a MOAC strategy was not feasible for serum, probably due to interfering compounds which competed for binding to the TiO2 active material and thus displaced the less abundant phosphopeptides. The hypothesis about the presence of compounds competing with phosphopeptides for binding to TiO2 agreed with a previous report about the use of magnetic rutile echinus microspheres for removal of phosphocholines and lysophosphocholines from plasma, as those phospholipids were the main matrix interferences in the detection of low abundance metabolites [42]. It was evident that the use of a MOAC strategy for enrichment of the endogenous phosphopeptides in serum was not feasible under the tested experimental conditions, due to the contemporary presence of interfering species.
Serum endogenous phosphopeptide investigation by IMAC systems
Given the poor results of MOAC materials for the enrichment of the endogenous phosphopeptides in serum, the IMAC approach was tested. IMAC was not chosen simply because it is a common alternative to MOAC in the enrichment of phosphopeptides. In fact, assuming the interaction with phospholipids can hinder phosphopeptide enrichment, the electrostatic nature of the interaction of IMAC materials appeared suitable to improve the selectivity for phosphopeptides over phospholipids. In IMAC, the interaction exploits the affinity of phosphonate groups to the metal center with formation of a complex as for MOAC, but there is also a strong electrostatic interaction. The latter could be exploited to both enrich phosphopeptides and repel zwitterionic phospholipids, namely phosphatidylcholine, phosphoethanolamine, and phosphatidylserine derivatives, which are abundant in serum.
The first IMAC system employed was the Ti4+-IMAC magnetic material previously used to enrich phosphopeptides from yeast protein digests used as a model system [36]. The Ti4+-IMAC magnetic material appeared to be the ideal system for enrichment of phosphopeptides from serum. Due to the magnetic properties, the Ti4+-IMAC magnetic material can be employed in batch format, with no volume restriction or clogging issues. Moreover, from results obtained in the previous work [36], the Ti4+-IMAC magnetic material showed a better interaction with hydrophilic peptides, which proved useful in reducing co-enrichment of non-phosphorylated peptides, due to reduced hydrophobic interactions, thus increasing the selectivity.
Different protocols were applied to find the best conditions (see the ESM for details): to begin, the precipitation protocols were tested; however, the results were still not positive, and a limited number of peptides, both phosphorylated and unmodified peptides, were identified for acetone and ACN precipitation. Results, although improved with respect to MOAC materials, were still unsatisfactory, with up to 6 phosphopeptides for the ACN precipitation and 11 for acetone precipitation (Table 1, ESM Table S2).
Only the methanol precipitation provided slightly better results, with up to 35 phosphopeptides out of 889 total identifications. This result only partially agreed with previous literature reports, in which endogenous peptides were purified by organic solvent precipitation due to their very limited solubility [24, 43]. In these studies, the endogenous peptides were isolated from serum by ACN precipitation and the precipitate was then selectively extracted with water modified by organic solvents and collected over C18 resin. These studies [24, 43] were not aimed at identifying the free phosphopeptides; however, they can explain the low recovery of endogenous phosphopeptides obtained by analyzing the supernatants from organic solvent precipitation experiments, even if the latter procedure was previously exploited for profiling free phosphopeptides in different biofluids, including serum [44]. Thus, given the possibility that some phosphopeptides could be precipitated with the proteins during organic solvent precipitation, the direct dilution protocol was tested. The direct dilution protocol is the simplest among the tested ones, but it also provided the best results both for phosphopeptide characterization and for enrichment performance. Given the improvement with respect to the precipitation protocols, for the direct dilution, the amount of Ti4+-IMAC magnetic material employed for enrichment was optimized; thus, 5, 10, and 15 mg were tested for enrichment. The results indicated that the amount of enrichment phase did affect the performance, as previously observed for other materials [45], and it was significant, indeed. For both the 5- and 15-mg experiments, the total number of phosphopeptide identifications did not exceed 59, with up to 410 total peptide identifications (Table 1, ESM Table S3). The enrichment, calculated as the total number of phosphopeptides divided by the total number of identifications, was higher than the previous experiments and ranged between 12 and 15%. The best results were, however, obtained for the 10-mg direct dilution experiments, which provided up to 114 phosphopeptides out of 488 total peptide identifications, and the enrichment was also significantly improved up to 23% (Table 1, ESM Table S4). The identification of such a large number of endogenous phosphopeptides indicated a problem connected to a large dynamic range in phosphopeptide natural abundance, since in most experiments, no more than ten endogenous phosphopeptides were identified and they probably were the most abundant ones [46]. Due to the large dynamic range and heterogeneity of phosphopeptides in serum, a comprehensive approach, as the shotgun peptidomics one, is the best one for phosphopeptide characterization in serum.
Finally, an attempt of phospholipid removal from serum was performed, to test if any improvement could be accomplished by removing the non-zwitterionic lipids. In fact, it is generally known that they can severely hinder other analyte detection by RP-HPLC-MS, due to their strong retention on hydrophobic columns and the important ion suppression during ionization in the mass spectrometer [47]; thus, they outcompete analyte molecules for ionization and, given that such problems already affect phosphopeptide analysis, this could provide an additional obstacle. Moreover, a competition for interaction with the IMAC phase is also possible and would significantly affect the enrichment capability. Thus, a purification step prior to enrichment was introduced, based on a literature procedure, and DCM was employed to remove phospholipids in serum [48]. DMC pretreatment did not improve the performance than the simple dilution and enrichment protocol (Table 1, ESM Table S5), even though it improved the selectivity with respect to the precipitation methods, from 7 to 16%.
A final issue was considered. Many biological systems (including blood and digestive samples) contain proteases, either endogenous or produced by other organisms (e.g., bacteria). In order to use peptides as biomarkers and guarantee method reproducibility to compare different samples, postsample collection proteolysis should be eliminated (by the addition of protease inhibitors) [18]. This applies to phosphopeptide analysis too, since phosphatases are endogenous in blood as well. Thus, fresh human serum samples were prepared and immediately added with protease and phosphatase inhibitors. The fresh serum samples thus prepared were used with the direct dilution protocol and enrichment with the Ti4+-IMAC magnetic material. Results (Table 1) clearly showed a remarkable improvement in the characterization of endogenous phosphopeptides (ESM Table S6). Up to 176 endogenous phosphopeptides were identified from the analysis of serum fresh samples, and the number of total identifications increased as well, up to 750 total endogenous peptide identifications. The enrichment % did not improve as much and reached 23%. In the case of the present work, a standard commercial serum sample was employed for method development to ensure sample homogeneity and remove irreproducibility due to individual donors and serum preparation. However, these results did not only indicate the need of a standardized protocol in which phosphatase inhibitors are added as soon as possible to the sample but also showed how endogenous peptides, regardless of phosphorylation, can undergo degradation during conservation, also at low temperatures; protease inhibitors are also fundamental for endogenous peptide characterization [43], thus should be considered for biomarker investigation to meet the requirement for clinical research [49].
Finally, the proposed method, based on batch magnetic solid-phase extraction with 10 mg of Ti4+-IMAC magnetic material and the direct dilution protocol, was compared with a commercial Fe3+-IMAC spin column for two main reasons: to compare the performance of different IMAC systems and to provide an established commercial system suitable for the characterization of the endogenous phosphopeptides in serum. As the Fe3+-IMAC spin columns are commercially available, the use of such devices allowed to avoid preparation and characterization of the Ti4+-IMAC material and would thus be accessible virtually to any laboratory. The test was performed on the commercial serum pool and gave overall good results, with up to 83 out of 846 total peptide identifications (Table 1, ESM Table S7). Thus, if on the one hand, the number of identified phosphopeptides was close to the one obtained by the Ti4+-IMAC magnetic material, the selectivity was not as good and the number of co-enriched peptides was significantly larger. Nevertheless, the commercial Fe3+-IMAC spin column was suitable for analysis of the endogenous phosphopeptides in serum and provided a valuable alternative.
General features of serum endogenous phosphopeptides
A general investigation of the physicochemical features of the identified peptides was done. To this purpose, the grand average of hydropathy (GRAVY) index, the molecular weight, the isoelectric point (pI), and the amino acid composition were calculated, pooling the results by the Sigma and fresh serum pool for the dilution experiments. As far as the GRAVY was concerned, the endogenous peptides identified in this work were all extremely hydrophilic and had a negative value, regardless whether they were phosphorylated (95%) or co-enriched peptides (97%, Fig. 1). The large presence of hydrophilic peptides could be expected, since they were extracted from an aqueous matrix. However, some concerns about possible interaction with proteins may arise (larger proteins [50], lipids [51], etc.), despite the employ of organic solvents in the loading buffer. Moreover, although a hydrophilic support is considered to increase the selectivity of the system [52, 53], still the large abundance of co-enriched very hydrophilic peptides with GRAVY value between − 2 and − 1 may be ascribed to hydrophilic interactions with the material and some improvement or complementary information could be obtained by the use of a different material.
As far as the molecular weight distribution was concerned, the endogenous peptides identified in the study were small peptides, mainly in the range 2000–3000 Da (ESM Fig. S1). Additionally, the pI distribution analysis indicated that they were mainly acidic peptides, with a maximum at 5 (ESM Fig. S2). Finally, the amino acid distribution (ESM Fig. S3) showed that the most frequent residues were glutamic acid, for both the phosphopeptides (10.1%) and the co-enriched peptides (14.5%), a phenomenon which can be ascribed to the affinity of the metal cation for acidic peptides together with the phosphorylated ones. Additionally, the second significantly represented residue was serine (13.8% for the phosphopeptides and 10.2% for the co-enriched peptides). Apart from these small differences, all other residues showed a similar distribution for the two groups, and a small bias for acidic peptides in both groups could be observed as well, which could still be attributable to the material.
The residues at the first and last positions of the identified phosphopeptides were considered to find some hint on the proteases which might have produced the identified peptides. The results of the analysis of the distribution of the amino acids in the first and last residues in the identified phosphopeptides showed that glutamic acid was the most frequent residue at the end of such peptides (Fig. 2). However, no enzyme in the human body shows specificity for cleaving after such amino acid but some bacterial ones do, such as the glutamyl endopeptidase. A very represented cleavage was also the one after arginine, which could be attributed to thrombin and factor Xa, both of them being involved in the coagulation cascade of blood. Finally, threonine was frequently the last amino acid in the identified phosphopeptides and can be attributed to the action of chymotrypsin, which is also a marker of acute pancreatitis and in renal failure [54]. The first amino acids (P1′) most represented in the phosphopeptides were serine, lysine, arginine, and alanine, but only the latter could be connected with any enzyme specificity, in particular with thermolysin, which, however, has a bacterial origin.
As far as the phosphorylated proteins related to the endogenous phosphopeptides identified by this comprehensive study were concerned, the 11 most abundant phosphopeptides belonged to fibrinogen alpha chain, which had a coverage of 54.74%. The large presence of peptides from fibrinogen alpha chain agreed with most previous reports, for which at least four phosphopeptides belonging to fibrinogen were always reported. However, it should be noted that such large coverage did not derive entirely from the phosphopeptides; a significant contribution to fibrinogen alpha chain is provided by the co-enriched peptides as well, for which the ten most abundant peptides also belong to fibrinogen alpha chain. Such large dynamic range needs to be considered in case of the quantification of endogenous phosphopeptides between a physiological condition and a pathological one; if the fibrinogen phosphopeptides were not significantly expressed in different amounts between two conditions, suitable depletion systems would greatly help in extending the characterization of endogenous phosphopeptides to the least abundant ones.
Conclusions
In this work, a comparison between MOAC and IMAC enrichment materials for phosphopeptides was performed and applied to the characterization of serum endogenous phosphopeptide. Despite the generally recognized similar performance of IMAC and MOAC strategies, in serum, the enrichment of the endogenous phosphopeptides by MOAC is hindered, probably due to matrix components, whereas IMAC performs significantly better. Sample preparation proved critical, and better results could be obtained using phosphatase inhibitors soon after serum preparation. Moreover, minimal sample pretreatments allowed to avoid depletion of endogenous phosphopeptides. The identified endogenous phosphopeptides were very hydrophilic; thus, common precipitation treatments employed in peptidomics could possibly depleted the endogenous phosphopeptides. From the results, the largest number of phosphopeptide identifications could be obtained by direct dilution of serum combined to an IMAC enrichment. Even if the Ti4+-IMAC magnetic material provided the largest number of identifications, the commercial Fe3+-IMAC spin column could be adapted to the dilution protocol and employed for phosphopeptide enrichment with good results. In both cases, few abundant phosphopeptides, mainly belonging to fibrinogen and to the coagulation cascade involved in serum preparations, may hinder the detection of the least abundant phosphopeptides; thus, the development and the use of suitable depletion media for the most abundant phosphopeptides are desirable. The developed method relies on shotgun technologies; thus, the introduction of quantification strategies typical of shotgun proteomics is completely compatible with this method, especially label-free quantification. Thus, in the end, the work provides an improved method for endogenous phosphopeptide identification in serum and, definitely, the starting point for screening of differently expressed phosphopeptides in disease.
References
Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2011;5:463–6. https://doi.org/10.1097/COH.0b013e32833ed177.
Barbosa AI, Reis NM. A critical insight into the development pipeline of microfluidic immunoassay devices for sensitive quantitation of protein biomarkers at point-of-care. Analyst. 2017;142:858–82. https://doi.org/10.1039/C6AN02445A.
Ishikawa M, Maekawa K, Saito K, Senoo Y, Urata M, Murayama M, et al. Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS One. 2014;9:e91806. https://doi.org/10.1371/journal.pone.0091806.
Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, et al. The human serum metabolome. PLoS One. 2011;6:e16957. https://doi.org/10.1371/journal.pone.0016957.
Capriotti AL, Caruso G, Cavaliere C, Piovesana S, Samperi R, Laganà A. Proteomic characterization of human platelet-derived microparticles. Anal Chim Acta. 2013;776:57–63. https://doi.org/10.1016/j.aca.2013.03.023.
H. Rashed M, Bayraktar E, K. Helal G, Abd-Ellah M, Amero P, Chavez-Reyes A, et al. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci. 2017;18:538. https://doi.org/10.3390/ijms18030538.
Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. https://doi.org/10.1038/nrc3066.
Piovesana S, Capriotti AL. Magnetic materials for the selective analysis of peptide and protein biomarkers. Curr Med Chem. 2017;24:438–53.
Zenezini Chiozzi R, Capriotti AL, Cavaliere C, La Barbera G, Piovesana S, Laganà A. Identification of three novel angiotensin-converting enzyme inhibitory peptides derived from cauliflower by-products by multidimensional liquid chromatography and bioinformatics. J Funct Foods. 2016; https://doi.org/10.1016/j.jff.2016.09.010.
Piovesana S, Capriotti AL, Cavaliere C, La Barbera G, Samperi R, Zenezini Chiozzi R, et al. Peptidome characterization and bioactivity analysis of donkey milk. J Proteome. 2015; https://doi.org/10.1016/j.jprot.2015.01.020.
Zenezini Chiozzi R, Capriotti AL, Cavaliere C, La Barbera G, Piovesana S, Samperi R, et al. Purification and identification of endogenous antioxidant and ACE-inhibitory peptides from donkey milk by multidimensional liquid chromatography and nanoHPLC-high resolution mass spectrometry. Anal Bioanal Chem. 2016; https://doi.org/10.1007/s00216-016-9672-z.
Capriotti AL, Cavaliere C, Foglia P, Piovesana S, Samperi R, Zenezini Chiozzi R, et al. Development of an analytical strategy for the identification of potential bioactive peptides generated by in vitro tryptic digestion of fish muscle proteins. Anal Bioanal Chem. 2015;407:845–54. https://doi.org/10.1007/s00216-014-8094-z.
Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2010;21:228–37. https://doi.org/10.1016/j.tcb.2010.12.002.
Mahendru S, Roy K, Kukreti S. Peptide biomarkers: exploring the diagnostic aspect. Curr Protein Pept Sci. 2016;17:1–1. https://doi.org/10.2174/1389203717666160724203746.
Borrebaeck CAK. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat Publ Group. 2017;17:199–204. https://doi.org/10.1038/nrc.2016.153.
Capriotti AL, Caruso G, Cavaliere C, Piovesana S, Samperi R, Laganà A. Comparison of three different enrichment strategies for serum low molecular weight protein identification using shotgun proteomics approach. Anal Chim Acta. 2012;740:58–65. https://doi.org/10.1016/j.aca.2012.06.033.
Marshall J, Kupchak P, Zhu W, Yantha J, Vrees T, Furesz S, et al. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res. 2003;2:361–72. https://doi.org/10.1021/pr030003l.
Dallas DC, Guerrero A, Parker EA, Robinson RC, Gan J, German JB, et al. Current peptidomics: applications, purification, identification, quantification, and functional analysis. Proteomics. 2015;15:1026–38. https://doi.org/10.1002/pmic.201400310.Current.
Cutillas PR. Role of phosphoproteomics in the development of personalized cancer therapies. Proteomics Clin Appl. 2015;9:383–95. https://doi.org/10.1002/prca.201400104.
Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. https://doi.org/10.1038/35085068.
Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum: recent developments. Ann N Y Acad Sci. 2008;1137:1–6. https://doi.org/10.1196/annals.1448.050.
Hu L, Boos KS, Ye M, Zou H. Analysis of the endogenous human serum peptides by on-line extraction with restricted-access material and HPLC-MS/MS identification. Talanta. 2014;127:191–5. https://doi.org/10.1016/j.talanta.2014.04.011.
Gilar M, Olivova P, Chakraborty AB, Jaworski A, Geromanos SJ, Gebler JC. Comparison of 1-D and 2-D LC MS/MS methods for proteomic analysis of human serum. Electrophoresis. 2009;30:1157–67. https://doi.org/10.1002/elps.200800630.
Williams D, Ackloo S, Zhu P, Bowden P, Evans KR, Addison CL, et al. Precipitation and selective extraction of human serum endogenous peptides with analysis by quadrupole time-of-flight mass spectrometry reveals posttranslational modifications and low-abundance peptides. Anal Bioanal Chem. 2010;396:1223–47. https://doi.org/10.1007/s00216-009-3345-0.
Carrascal M, Gay M, Ovelleiro D, Casas V, Gelpí E, Abian J. Characterization of the human plasma phosphoproteome using linear ion trap mass spectrometry and multiple search engines research articles. J Proteome Res. 2010;9:876–84.
Zawadzka AM, Schilling B, Cusack MP, Sahu AK, Drake P, Fisher SJ, et al. Phosphoprotein secretome of tumor cells as a source of candidates for breast cancer biomarkers in plasma. Mol Cell Proteomics. 2014;13:1034–49. https://doi.org/10.1074/mcp.M113.035485.
Klement E, Raffai T, Medzihradszky KF. Immobilized metal affinity chromatography optimized for the analysis of extracellular phosphorylation. Proteomics. 2016;16:1858–62. https://doi.org/10.1002/pmic.201500520.
Yao J, Sun N, Deng C, Zhang X. Designed synthesis of graphene @titania @mesoporous silica hybrid material as size-exclusive metal oxide affinity chromatography platform for selective enrichment of endogenous phosphopeptides. Talanta. 2016;150:296–301. https://doi.org/10.1016/j.talanta.2015.12.050.
Zhu J, Wang F, Cheng K, Song C, Qin H, Hu L, et al. Analysis of human serum phosphopeptidome by a focused database searching strategy. J Proteome. 2012;78:389–97. https://doi.org/10.1016/j.jprot.2012.10.006.
Dong M, Ye M, Cheng K, Dong J, Zhu J, Qin H, et al. Identification of phosphopeptides with unknown cleavage specificity by a de novo sequencing assisted database search strategy. Proteomics. 2014;14:2410–6. https://doi.org/10.1002/pmic.201400268.
Zhai G, Wu X, Luo Q, Wu K, Zhao Y, Liu J, et al. Evaluation of serum phosphopeptides as potential cancer biomarkers by mass spectrometric absolute quantification. Talanta. 2014;125:411–7. https://doi.org/10.1016/j.talanta.2014.03.025.
Hussain D, Najam-ul-Haq M, Jabeen F, Ashiq MN, Athar M, Rainer M, et al. Functionalized diamond nanopowder for phosphopeptides enrichment from complex biological fluids. Anal Chim Acta. 2013;775:75–84. https://doi.org/10.1016/j.aca.2013.03.007.
Hu L, Zhou H, Li Y, Sun S, Guo L, Ye M, et al. Profiling of endogenous serum phosphorylated peptides by titanium (IV) immobilized mesoporous silica particles enrichment and MALDI-TOFMS detection. Anal Chem. 2009;81:94–104. https://doi.org/10.1021/ac801974f.
Yue X, Schunter A, Hummon AB. Comparing multi-step IMAC and multi-step TiO2 methods for phosphopeptide enrichment. Anal Chem. 2015;87:8837–44. https://doi.org/10.1177/0963721412473755.
Piovesana S, Capriotti AL, Cavaliere C, Ferraris F, Iglesias D, Marchesan S, et al. New magnetic graphitized carbon black TiO2 composite for phosphopeptide selective enrichment in shotgun phosphoproteomics. Anal Chem. 2016;88:12043–50. https://doi.org/10.1021/acs.analchem.6b02345.
Capriotti AL, Cavaliere C, Ferraris F, Gianotti V, Laus M, Piovesana S, et al. New Ti-IMAC magnetic polymeric nanoparticles for phosphopeptide enrichment from complex real samples. Talanta. 2018;178:274–81. https://doi.org/10.1016/j.talanta.2017.09.010.
Sparnacci K, Antonioli D, Deregibus S, Laus M, Poggio T, Kapeliouchko V, et al. PTFE-based core-soft shell nanospheres and soft matrix nanocomposites. Macromolecules. 2009;42:3518–24. https://doi.org/10.1021/ma802871y.
Sparnacci K, Laus M, Tondelli L, Bernardi C, Magnani L, Corticelli F, et al. Core-shell microspheres by dispersion polymerization as promising delivery systems for proteins. J Biomater Sci Polym Ed. 2005;16:1557–74. https://doi.org/10.1163/156856205774576673.
Piovesana S, Capriotti AL, Cavaliere C, Ferraris F, Samperi R, Ventura S, et al. Phosphopeptide enrichment: development of magnetic solid phase extraction method based on polydopamine coating and Ti4+-IMAC. Anal Chim Acta. 2016;909:67–74. https://doi.org/10.1016/j.aca.2016.01.008.
Wu S, Zhu Y. ProPAS: standalone software to analyze protein properties. Bioinformation. 2012;8:167–9.
Li XS, Pan YN, Zhao Y, Yuan BF, Guo L, Feng YQ. Preparation of titanium-grafted magnetic mesoporous silica for the enrichment of endogenous serum phosphopeptides. J Chromatogr A. 2013;1315:61–9. https://doi.org/10.1016/j.chroma.2013.09.057.
Li H, Shi X, Qiao L, Lu X, Xu G. Synthesis of a new type of echinus-like Fe3O4@TiO2 core–shell-structured microspheres and their applications in selectively enriching phosphopeptides and removing phospholipids. J Chromatogr A. 2013;1275:9–16. https://doi.org/10.1016/j.chroma.2012.12.023.
Tucholska M, Florentinus A, Williams D, Marshall JG. The endogenous peptides of normal human serum extracted from the acetonitrile-insoluble precipitate using modified aqueous buffer with analysis by LC–ESI–Paul ion trap and Qq-TOF. J Proteome. 2010;73:1254–69. https://doi.org/10.1016/j.jprot.2010.02.022.
Cirulli C, Chiappetta G, Marino G, Mauri P, Amoresano A. Identification of free phosphopeptides in different biological fluids by a mass spectrometry approach. Anal Bioanal Chem. 2008;392:147–59. https://doi.org/10.1007/s00216-008-2266-7.
Li QR, Ning ZB, Tang JS, Nie S, Zeng R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J Proteome Res. 2009;8:5375–81. https://doi.org/10.1021/pr900659n.
He XM, Chen X, Yuan BF, Feng YQ. Graft modification of cotton with phosphate group and its application to the enrichment of phosphopeptides. J Chromatogr A. 2017;1484:49–57. https://doi.org/10.1016/j.chroma.2017.01.020.
Carmical J, Brown S. The impact of phospholipids and phospholipid removal on bioanalytical method performance. Biomed Chromatogr. 2016;30:710–20. https://doi.org/10.1002/bmc.3686.
Neville D, Houghton R, Garrett S. Efficacy of plasma phospholipid removal during sample preparation and subsequent retention under typical UHPLC conditions. Bioanalysis. 2012;4:795–807.
Iliuk AB, Tao WA. Is phosphoproteomics ready for clinical research? Clin Chim Acta. 2013;420:23–7. https://doi.org/10.1016/j.cca.2012.10.063.
McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med Res Rev. 2002;22:168–203. https://doi.org/10.1002/med.10006.
Sanderson JM. Peptide–lipid interactions: insights and perspectives. Org Biomol Chem. 2005;3:201–12.
Novotna L, Emmerova T, Horak D, Kucerova Z, Ticha M. Iminodiacetic acid-modified magnetic poly(2-hydroxyethyl methacrylate)-based microspheres for phosphopeptide enrichment. J Chromatogr A. 2010;1217:8032–40. https://doi.org/10.1016/j.chroma.2010.08.058.
Zhang L, Zhao Q, Liang Z, Yang K, Sun L, Zhang L, et al. Synthesis of adenosine functionalized metal immobilized magnetic nanoparticles for highly selective and sensitive enrichment of phosphopeptides. Chem Commun. 2012;48:6274–6. https://doi.org/10.1039/c2cc31641b.
Lefkowitz RB, Schmid-Schönbein GW, Heller MJ. Whole blood assay for elastase, chymotrypsin, matrix metalloproteinase-2, and matrix metalloproteinase-9 activity. Anal Chem. 2010;82:8251–8. https://doi.org/10.1021/ac101462c.
Conflict of interest
The authors declare that they have no conflicts of interest.
Funding
This work was supported by PRIN 2015, project number 2015TWP83Z.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
No human patients or animals were involved in this study. Blood samples were provided after informed consent was obtained according to the Ethics Committee of the Umberto I Hospital, (Sapienza University of Rome).
Additional information
Published in the topical collection celebrating ABCs 16th Anniversary.
Rights and permissions
About this article
Cite this article
La Barbera, G., Capriotti, A.L., Cavaliere, C. et al. Development of an enrichment method for endogenous phosphopeptide characterization in human serum. Anal Bioanal Chem 410, 1177–1185 (2018). https://doi.org/10.1007/s00216-017-0822-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0822-8