Abstract
MS-based metabolite profiling of adherent mammalian cells comprises several challenging steps such as metabolism quenching, cell detachment, cell disruption, metabolome extraction, and metabolite measurement. In LC-MS, the final metabolome coverage is strongly determined by the separation technique and the MS conditions used. Human liver-derived cell line HepG2 was chosen as adherent mammalian cell model to evaluate the performance of several commonly used procedures in both sample processing and LC-MS analysis. In a first phase, metabolite extraction and sample analysis were optimized in a combined manner. To this end, the extraction abilities of five different solvents (or combinations) were assessed by comparing the number and the levels of the metabolites comprised in each extract. Three different chromatographic methods were selected for metabolites separation. A HILIC-based method which was set to specifically separate polar metabolites and two RP-based methods focused on lipidome and wide-ranging metabolite detection, respectively. With regard to metabolite measurement, a Q-ToF instrument operating in both ESI (+) and ESI (−) was used for unbiased extract analysis. Once metabolite extraction and analysis conditions were set up, the influence of cell harvesting on metabolome coverage was also evaluated. Therefore, different protocols for cell detachment (trypsinization or scraping) and metabolism quenching were compared. This study confirmed the inconvenience of trypsinization as a harvesting technique, and the importance of using complementary extraction solvents to extend metabolome coverage, minimizing interferences and maximizing detection, thanks to the use of dedicated analytical conditions through the combination of HILIC and RP separations. The proposed workflow allowed the detection of over 300 identified metabolites from highly polar compounds to a wide range of lipids.

A novel analytical workflow for the LC-MS-based metabolomic analysis of adherent cultured hepatic cells was developed. Three key steps were evaluated: a) cell harvesting, which includes cell detachment and metabolism quenching; b) metabolite extraction; and c) LC-MS analysis, assessing the use of RP and HILIC chromatographies. The final protocol allowed us to extend the metabolome coverage and enabled the detection of a wide range of metabolites from highly polar compounds to a wide range of lipids
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metabolomics aims to measure all the metabolites present in a given biological sample and to use this information to understand the biology or physiology of the system under study for different purposes (discrimination analysis, biomarker discovery, etc.). Its application to mammalian cells in culture in fields like toxicology drug testing [1–3], cell culture monitoring [4, 5], cancer research [6, 7], and foodomics [8] has considerably increased in recent years [9]. The use of in vitro cell models offers advantages over other subject or animal models, which include easily controlled experimental variables, greater reproducibility, lower cost, easily interpretable results, and commercial availability [10]. On the contrary, cell metabolomics involves more complex sample processing procedures (i.e., cell detachment, metabolism quenching, cell disruption, and metabolome extraction) than those used with other matrices as body fluids (serum, plasma, urine, etc.). These critical steps have to be carefully optimized before proceeding to sample analysis as they eventually determine metabolome coverage and therefore the representativeness of the detected metabolite profiles [9, 11]. The optimization of sample processing and analysis of cultured cells by metabolomics are an active field of research. Recently, several studies have explored the influence of different factors on final metabolome coverage, which comprises cell detachment, metabolism quenching, metabolome extraction, and type of analysis [12–17]. A number of protocols have been set up [18–20]; however, no general consensus has yet been reached and new advances are expected in this field. The most common procedure comprises the direct extraction of the metabolites by scrapping the cells using an organic solvent or a mixture of organic solvents with or without water (e.g., methanol, water/methanol (1:3) or water/methanol/chloroform (10:27:3)), whereas the analysis of the extracted metabolites is mainly performed using either generic reversed phase (RP) or hydrophilic interaction chromatography (HILIC) or a combination of both.
The pure metabolomics concept implies the unbiased determination of all the metabolites present in a sample, independently of their chemical nature. However, it is well accepted that no single analytical technique can cover the whole range of metabolites that may be present in a complex biological sample. Among the analytical platforms used to perform an unbiased analysis, nuclear magnetic resonance (NMR) and mass spectrometry (MS) are the most frequently used [9]. The latter is normally coupled to liquid or gas chromatography, which are used as a first step to separate the chemical and physical complexity of the metabolome. In liquid chromatography (LC), metabolite separation and detection are strongly influenced by the chemical nature of the mobile and stationary phases used [21]. Untargeted LC-MS-based metabolite profiling studies are usually performed by using RP chromatography with C8 or C18 columns, in which nonpolar or medium polar compounds can be properly separated [22]. HILIC allows the detection of polar/ionic metabolites that are poorly retained, or not retained at all, by RP chromatography. Recently, the combined use of RP with HILIC has been seen to be a suitable approach to extend metabolome coverage, which provides a more comprehensive metabolomic snapshot of the biological system under study [17, 23, 24].
The aim of the present work was to define a sample processing protocol which focused on maximizing metabolite recovery from cells, together with an LC-MS-based analytical strategy that focused on detecting metabolites which range from polar to highly nonpolar ones. The human hepatoma HepG2 cell line was chosen as a representative model of liver-derived cultured cells because of its widespread use, unlimited life span, stable phenotype, high availability, reproducibility, and easy handling. Furthermore, this cell line is one of the most widely used human cell models for hepatotoxicity evaluations [25].
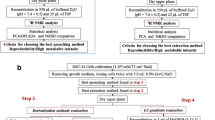
The different variables that may affect metabolome coverage in cells’ metabolite profiling were evaluated and optimized following a two-phase experimental design. First, metabolite extraction and analysis were examined. To this end, five different extraction solvents were chosen for intracellular metabolite recovery. The solvents were selected based on their extraction capabilities and their later compatibility with the chromatography technique used in the sample analysis. Ultra-performance liquid chromatography (UPLC), coupled with quadrupole time of flight (Q-ToF), was the instrument of choice for the untargeted metabolite profiling analysis of the cells extracts. Two complementary separation methods, RP and HILIC, together with three different elution conditions were evaluated in both the positive and negative electrospray ionization (ESI) modes (Fig. 1). Once metabolite extractions and sample analyses were optimized, the influence of cell monolayer detachment and metabolism quenching on metabolome coverage was also evaluated in a second phase. Thus, the three most widely used approaches for cell harvesting were evaluated (see Electronic Supplementary Material (ESM) Fig. S1). The novel workflow integrates sample preparation and analysis by means of compatible extraction solvents and complementary chromatographic techniques, with a special emphasis on detecting both polar and nonpolar metabolites (mainly lipids).
Parameters evaluated during the optimization of metabolome extraction and analysis conditions. Cell harvesting procedure was kept constant for all the different combinations of metabolome extractions and LC-MS analysis. Metabolism quenching was achieved by the addition of liquid N2 over the cells. Then, simultaneous cell detachment and metabolites extraction were performed by scrapping the cells in the different extraction solvents
Materials and methods
Materials
All the LC solvents (i.e., water, methanol, acetonitrile, and isopropanol) were of LC-MS grade and were purchased from Fisher Scientific (Loughborough, UK). All the LC-MS additives (i.e., formic acid and ammonium acetate) and the analytical standards were acquired from Sigma-Aldrich (Madrid, Spain). Deuterium-labeled bile acids were obtained from Steraloids (Newport, USA). Phenylalanine-D5 (Phe-D5) was obtained from Cambridge Isotope Laboratories (Tewksbury, USA).
Cell culture
HepG2 cells were routinely grown in culture grade flasks at 37 °C in a humidified atmosphere, 5 % CO2/95 % air, in Ham’s F-12 / Leibovitz L-15 (1:1, v/v) supplemented with 7 % fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin. The medium was renewed every 2 days. The cells that reached 70–80 % confluence were ready to be used or passaged. For subculturing purposes, cells were detached by treatment with 0.25 % trypsin/0.02 % EDTA at 37 °C [26]. For the metabolomic studies, cells were seeded at a density of 8 × 104 cells/cm2 on 3.5-cm culture dishes [27].
Sample processing and analysis of HepG2 cells
Three major aspects of the unbiased analysis of the metabolome of HepG2 cells were examined: (i) cell harvesting, (ii) metabolome extraction, and (iii) LC-MS analysis. The performance of each procedure was evaluated according to two parameters: (i) the number, class, and intensity of the annotated/identified metabolites and (ii) the total number of detected features. Each experimental condition was tested by three biological replicates. In all cases, amount of biomass was determined by protein quantification using a 96-well-adapted Lowry method [28]. In a first step, the extraction solvent and the metabolite profiling conditions (covering both chromatographic separation and MS polarity) were tested and optimized in a combined manner by a previously described cell harvesting method [14] (Fig. 1). In a second step, three cell harvesting procedures (ESM Fig. S1) were compared and evaluated based on the previously optimized metabolome extraction and analysis conditions. Nevertheless, this strategy did not follow the natural metabolomic workflow (cells harvesting, metabolome extraction, and sample analysis). Instead, it allowed the delineation of a global metabolomic workflow that expanded metabolome coverage by analyzing an affordable number of samples. All the samples were analyzed in the following week after sample collection.
Metabolome extraction
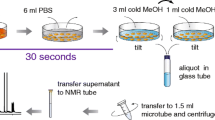
Five different solvents, leading to seven distinct extracts, were examined (Fig. 1). Cell harvesting and processing were fixed in the extraction solvent optimization step [14]. First, the medium was removed by aspiration. Then, the cell monolayer was washed once with 1 mL of cold phosphate buffer saline (PBS) and immediately frozen using liquid N2. Cells were then scraped in the presence of each solvent and submitted to three freeze/thaw (liquid N2/room temperature) cycles to increase cell lysis and metabolite extraction [5]. In all the cases cold solvents, either −20 °C or ice cold, were employed to perform the extractions. Finally, the clarified extracts were evaporated to dryness and reconstituted in the appropriate solvent containing internal standards (IS) selected according to the analytical condition to be employed (see ESM 2). Each extraction solvent was tested by three biological replicates. The extraction procedures were classified as follows:
Monophasic extractions
The following solvents were employed for monophasic extractions: (i) 1 mM Hepes, 1 mM EDTA pH 7.2 in water [5]; (ii) water/methanol/chloroform (10:27:3) [14]; (iii) methanol [29]. In all cases, an initial volume of 750 μL of cold solvent was used to scrape cells. Then a second volume of 250 μL was added to recover possible rests. Then, both extracts were pooled together. After the freeze/thaw cycles, a 75-μL aliquot of cell extract was taken and mixed with 75 μL of 1 M NaOH to carry out protein quantification. Then, the supernatant was separated from the cell pellet by centrifugation (10,000g, 10 min).
Biphasic liquid-liquid extraction
Biphasic liquid-liquid extraction was performed by using a water/methanol/chloroform (1:1:1) mixture [30]. Cells were first scraped with 750 μL of cold methanol/water (1:1) and then with an additional volume of 250 μL. Both extracts were pooled together. After the freeze/thaw cycles, a 75-μL aliquot was taken for protein quantification. Then, 500 μL of chloroform was added to the extract to perform liquid-liquid extraction. After vortexing (3 × 10 s), samples were allowed to rest for 20 min at −20 °C and were centrifuged (10,000g, 10 min). Next, the upper aqueous and the lower organic phases were transferred to separate clean tubes.
Sequential extraction
Sequential extraction was performed by methanol/water (1:1), followed by methanol/chloroform (1:3) [31]. First, 750 μL of cold methanol/water (1:1) was used to scrape cells, and a second volume of 250 μL to recover possible rests was employed and pooled in the tube with the first one. After the freeze/thaw cycles, a 75-μL aliquot was taken for protein quantification. Then, samples were centrifuged (10,000g, 10 min) and supernatants were transferred to clean tubes. Pellets were re-extracted with 1 mL of cold methanol/chloroform (1:3). Subsequently, supernatants were transferred to clean tubes after centrifugation (10,000g, 10 min). Thus, both extracts were collected and analyzed independently.
Cells harvesting
Three different approaches were examined for cell harvesting: (i) method A, quenching, followed by simultaneous detachment and extraction; (ii) method B, simultaneous quenching, extraction, and detachment; (iii) method C, detachment, first followed by quenching, and then by extraction (ESM Fig. S1). To specifically evaluate these issues, the different cell harvesting and processing procedures were examined using the previously optimized conditions for metabolome extraction, chromatographic separation and MS polarity.
The following steps were employed in the three cell harvesting methods (A–C): (i) removing the culture medium by aspiration and washing the cell monolayer once with 1 mL of cold PBS; (ii) freeze/thaw (liquid N2/room temperature) cycles to increase cell lysis and metabolites extraction; (iii) separating cellular debris from supernatants; (iv) evaporation to dryness of clarified extracts; and (v) reconstitution in the suitable solvent used for the LC-MS analysis. The specific steps of each configuration are detailed below:
Method A
After washing, the cell metabolism was immediately quenched by adding liquid N2 to the cell monolayer. Frozen plates were stored at −80 °C until further processing (<7 days). Simultaneous metabolite extraction and cell detachment were performed by adding the extraction solvent to the cell monolayer, followed by cell detachment with a rubber cell scraper.
Method B
Simultaneous quenching, extraction, and detachment were achieved by adding cold extraction solvent on the cell monolayer and using a rubber cell scraper to detach cells from the plate surface.
Method C
Cell harvesting was performed by treatment with 300 μL of 0.25 % trypsin/0.02 % EDTA at 37 °C. Thereafter, cells were recovered in 1 mL of 5 % fetal bovine serum in PBS and were transferred to a clean tube. After centrifugation (3000g, 3 min), the supernatant was removed by aspiration and cells were washed once with 1 mL of cold PBS by repeating centrifugation and removing the supernatant steps. The pellet was frozen in liquid N2 and stored at −80 °C until further processing (<7 days). Metabolome extraction was carried out by adding the cold extraction solvent on the cell pellet.
Ultra performance liquid chromatography
Chromatography was performed with a Waters Acquity UPLC system (Waters, UK). Three different LC analyses were examined: (i) generic-RP analysis, separation was performed using a UPLC HSS T3 (1.7 μm, 2.1 × 100 mm; Waters) column; mobile phase A was 0.1 % formic acid in water, and the mobile phase was B 0.1 % formic acid in acetonitrile [31]; (ii) lipidomic-RP analysis, separation was performed using a UPLC BEH C18 (1.7 μm, 2.1 × 100 mm; Waters) column; mobile phase A consisted in 0.1 % formic acid and ammonium acetate 10 mM in water, and mobile phase B in 0.1 % formic acid and ammonium acetate 10 mM in acetonitrile/isopropanol (5:2) [32]; (iii) HILIC analysis, separation was carried out using a UPLC BEH Amide (1.7 μm, 2.1 × 100 mm; Waters) column; mobile phase A was acetonitrile and mobile phase B was ammonium acetate 20 mM, pH 3, in water [33].
In all the analytical methods, the column and autosampler temperatures were set at 40 and 4 °C, respectively. The sample injection volume was 5 μL, and the flow rate was 0.4 mL/min. Chromatography details of each method are provided in the ESM (Tables S1-S4).
Mass spectrometry
Mass spectrometry was performed with a Waters Synapt HDMS Q-ToF mass spectrometer equipped with an ESI source (Waters, UK). The ESI conditions were as follows: the capillary was set at 3.2 and 2.8 kV in the positive and the negative mode, respectively; cone voltage was set at 40 V; desolvation and source temperatures were set at 380 and 120 °C, respectively; and the flow rates of the cone and nebulization gases were set at 50 and 800 L/h, respectively. A 50–1000 Da range was employed in all cases, except in the combination with lipidomic-RP separation, in which case mass acquisition was performed from 200 to 1200 Da in ESI (+) and from 100 to 1200 Da in ESI (−). The same parameters were applied for the simultaneous MS and MS/MS analyses, with a collision energy ramp from 5 to 60 eV in the MS/MS channel. For both ionization modes, leucine enkephalin (m/z 556.2771 in ESI(+), m/z 554.2615 ESI(−)) prepared at 50 pg/mL in acetonitrile/water (1:1), plus 0.1 % formic acid, was infused at a flow rate of 50 μL/min with an isocratic pump, and was used as a reference compound for lock mass correction.
Quality assurance strategy
Blank samples and a pooled quality control (QC) sample were employed to monitor UPLC-MS system performance. Blank samples were obtained by applying each extraction protocol over empty cultured plates and were employed to identify those background ions that were associated either with the extraction solvents or chromatographic separation (mobile phases plus column bleeding). The pooled QC sample was injected at the beginning of the analysis and intercalated every 10 study samples to assess instrument stability in terms of retention time, peak area, and mass accuracy for each IS added to the QC samples. Study samples were analyzed in randomized order. The quality assurance strategy has been provided in detail elsewhere [23].
MS data preprocessing and metabolite identification
The Masslynx software, version 4.1, was used for data acquisition. Data preprocessing was performed with the MZMine v.2.9.1 free software [34]. Data were normalized in relation to not only the response obtained by the IS added to each sample during the homogenization and preparation processes [23, 35], but also to the total amount of biological sample, represented by the total amount of protein [9] (ESM Fig. S2).
Metabolite identification was performed by the query of the exact mass of the detected features against online databases (Human Metabolome Database (HMDB) [36], LIPID MAPS-Nature Lipidomics Gateway [37], and METLIN Database [38]) within a ±10 ppm mass range. The identity of selected metabolites was confirmed by comparing the MS/MS spectra of the selected features with those of the proposed metabolites in online databases (HMDB [36], Metlin [38], and MassBank [39]) and, whenever possible, by using authentic standards. The degree of confidence in the identification was defined as specified by the Metabolomics Standards Initiative [40].
Statistical analysis
All the statistical analyses and data plots were run with the free R software [41]. The within-replicates coefficient of variation (CV) of each condition was employed as a measure of reproducibility. Only those features that fulfilled the following criteria were considered for further data analysis: (i) an intensity of at least 150 counts (mean value for the three biological replicates analyzed per extraction solvent and analytical condition); (ii) a CV between replicates <30 %; (iii) identification level 3 (as specified by the Metabolomics Standards Initiative) [40]. Prior to the principal component analysis (PCA), data sets were log-transformed, mean-centered, and unit-variance-scaled, while the data for the hierarchical clustering analysis (HCA) were mean-centered and unit-variance-scaled.
Results and discussion
Metabolite extraction and analysis by using RP/HILIC separations
To reliably and comprehensively detect the endo-metabolome, metabolites have to be efficiently extracted from inside cells. The extraction procedure should avoid potential interferences, ensure minimal loss of metabolites, and be compatible with the subsequent analytical procedures [9, 42]. Thus, the extractive capabilities of five different solvent combinations were evaluated by the use of three LC methods, one intended to specifically separate polar metabolites (HILIC) [33], one focused on lipidome detection (lipidomic-RP) [32], and a generic one (generic-RP) [31], whereas detection was performed by untargeted MS in both ESI (+) and ESI (−) modes (Fig. 1). Representative chromatograms that correspond to each analytical method are depicted in the ESM (Fig. S3). Each method (i.e., metabolite extraction coupled to LC-MS analysis) was evaluated by comparing the number, class and intensity of the assigned metabolites (Table 1), and the total number of features (Table 2). To ensure analysis reproducibility and reliability, only those assigned metabolites with a CV between the replicates below 30 % were considered. Confounding factors, regardless of metabolite extraction and sample analysis, were ruled out because the same standard cell harvesting protocol was applied to all the proposed analytical variations (Fig. 1) [14]. LC-MS data quality was assessed by the use of IS and QC samples (ESM Table S5).
Nonsupervised multivariate data analysis techniques were adopted to investigate the relationship among the metabolites comprised in the seven extracts. Three sample groups were observed in the PCA scores plot, such separation was guided by the differences in the physical and chemical properties of the metabolites comprised in each extract (Fig. 2). One cluster included those extracts that efficiently recovered both polar and lipidic compounds (E1, E2, E3, and E6). The second group included those extracts that exclusively covered lipids (E5 and E7). The third one corresponded to extract E4, which comprised mainly polar metabolites (Fig. 2a). As observed in the correlation biplot, extracts separation, specifically along the first principal component, was driven by the eluotropic capability of the solvent (Fig. 2b). To gain more information about the specific classes of metabolites and their relative levels in each extract, an HCA was carried out (ESM, Figs. S4 and S5). This analysis showed two main clusters, which corresponded to the extracts that comprised lipids and polar compounds, respectively. However, more subtle differences were detected in the levels of polar compounds, and in specific groups of lipids. Briefly, E4 was able to extract only polar compounds, E5 and E7 efficiently covered lipid compounds, and extracts E1, E2, E3, and E6 covered both polar and lipidic compounds, but with different specificities, mostly due to the percentage of the organic solvent used (E1 < E6 < E2 < E3) (Table 1, ESM Fig. S4). Regarding lipid metabolites, extract E5 displayed the best performance in general terms, although extract E3 provided similar results for FA, DG, PL, and LysoPL (Table 1, ESM Fig. S4). For the polar metabolites, similar results were obtained for extracts E1, E2, E4, and E6 in general terms. However, differences in individual species or in specific groups were observed among them (Table 1, ESM Fig. S5). For example, E1 outperformed its capability to extract nucleobases, nucleotides, and some amino acids, while E4 and E2 exhibited greater average extraction capabilities for the different classes of polar metabolites (ESM Fig. S5). The tight clustering of the different replicates of each extract, shown by the PCA and HCA analyses, demonstrated the good reproducibility of the in vitro model and the analytical workflow used.
a The PCA scores plot showing the natural interrelation among the different extracts. Each point represents all the metabolites detected in each extract by using the six analytical methods (the columns in Table 1). Lines denote the 95 % confidence interval of Hotelling’s ellipse for the three different identified clusters. b Biplot correlation analysis showing the relationship between extracts (big shaped marks) and the identified metabolites (small dots). Extracts are colored according to their eluotropic capacity (green: low; orange: medium, red: high). Metabolites are colored according to their polarity (green: polar; orange: high to medium polar lipids, red: nonpolar lipids). E1–E7 refer to the different extracts obtained (Fig. 1). E1: extraction with 1 mM Hepes, 1 mM EDTA, pH 7.2, in water; E2: extraction with water/methanol/chloroform (10:27:3); E3: extraction with methanol; E4 (aqueous), and E5 (organic): extracts obtained after a water/methanol/chloroform (1:1:1) biphasic extraction; E6 (aqueous), and E7 (organic): extracts obtained after a water/methanol (1:1), methanol/chloroform (1:3) sequential extraction
Venn diagrams were drawn to examine the overlapping of the different analytical methods in terms of the number of assigned metabolites (Fig. 3). In all, 140 positive assignments were found for the polar metabolites. Of these, 103 were detected by the HILIC analysis and 37 by the generic-RP analysis, and most were detected by ESI (−) (Fig. 3a). Regarding the lipidome (Fig. 3b), 616 features were assigned to known lipids. The generic-RP analysis detected 160 lipids and covered high to medium polar ones, such as fatty acids (FA), phospholipids (PL), lysophospholipids (LysoPL), etc. (Table 1). The lipidomic-RP analysis contributed with 456 lipids, which extended metabolome coverage to highly nonpolar species (e.g., triacylglycerides (TG) and cholesterol esters (CE)) (Table 1, Fig. 3b). Complementary information was obtained for both the lipidomic-RP and generic-RP approaches with the ESI (+) and ESI (−) ionization modes. In general, 542 metabolites were detected, of which 95 matched polar compounds and 447 matched lipids.
Table 1 summarizes detailed information on metabolite class, number, and relative intensities that each extract and analytical method provided. For polar metabolites, the largest number of metabolites with the highest intensity was obtained for extracts E1 and E4 by HILIC chromatography. Extracts E3, E5, and E7, analyzed by lipidomic-RP and ESI (+) provided the broadest coverage of lipids. However, E5 gave improved metabolite extraction yields, particularly for highly nonpolar lipids (i.e., CE and TG). E1, E2, E3, and E6 were composed of a mixture of both polar and lipidic metabolites (ESM Fig. S4). Of these extracts, E1 and E3 achieved the best coverage of polar metabolites and lipidic compounds, respectively. In contrast, extract E2 provided the highest combined coverage of both polar and lipidic metabolites, 21 and 51, respectively, by the generic-RP ESI (−) procedure.
Extraction with methanol alone, or in combination with other solvents (e.g., water or water/chloroform), is usually performed as a first approach for the metabolite profiling of cells because it allows the extraction of metabolites that range from polar to medium nonpolar ones [9, 43]. However, the use of methanol hinders the comprehensive characterization of the lipidome as it lacks the extraction of highly nonpolar lipids [44]. The aim of the present study was to detect from polar to highly nonpolar metabolites, despite this possibly implied harder sample processing, longer analysis times and more resources. Accordingly, a workflow comprising the use of different extracts was set up, which included aqueous and organic fractions, subsequently separated by complementary chromatography techniques and detected by MS. The final selection, which always attempted to avoid redundant information, was: (i) E4, an aqueous extract that resulted from a water/methanol/chloroform (1:1:1) biphasic extraction and HILIC chromatography, which covered polar compounds; (ii) E5, an organic extract that resulted from a water/methanol/chloroform (1:1:1) biphasic extraction, and the lipidomic-RP ESI (+) method, which covered polar (i.e., LysoPL), medium polar (i.e., PL, FA) and nonpolar (i.e., TG, CE) lipids; (iii) E2, extraction with water/methanol/chloroform (10:27:3) and the generic-RP ESI (−) method, which delivered a complementary set of polar metabolites and lipids. As shown in Table 1, a total of 344 metabolites were detected by the use of the selected strategy. This represents only 16 % increase in the number of detected metabolites (mainly lipids) when compared to the same LC-MS detection strategy but based on a more generic extraction protocol (i.e., extract E2, water/methanol/chloroform (10:27:3)), which rendered a total of 297 metabolites, and higher total intensity of several classes of lipids. Besides the increase in both the number and intensity of detected metabolites, in the optimized strategy, each class of metabolites is analyzed separately in the most suitable LC-MS condition, thus minimizing interferences and maximizing detection. Moreover, when compared to the most common analytical approaches, that is, the use generic RP or HILIC separations alone or in combination, E2 rendered the following figures: 131 metabolites in generic-RP conditions, 70 in HILIC, and a total of 201 combining both modes. Thus, the optimized strategy represents increases of approximately 150, 400, and 70 % respectively in the number of detected metabolites. Although the combined use of HILIC and RP separations has been proposed by our group [23, 33] and others [17, 45, 46] as a straightforward strategy to extend metabolome coverage, one of the main advantages of the workflow proposed herein is that special attention is being paid to detect highly nonpolar lipids by including a specific complementary lipidomic analysis.
Cell harvesting optimization
Cell harvesting should provide appropriate cell metabolism quenching, cell detachment, and efficient metabolite extraction and should avoid metabolome changes at the time of sampling. These steps can be carried out simultaneously or sequentially; if performed sequentially, the quenching and/or detachment steps must avoid cell disruption because metabolite leakage would occur [9]. To explore the advantage of using sequential or all-in-one procedures and/or trypsin for cell detachment, the performances of three different methods were compared (ESM Fig. S1). To specifically focus on the influence that cell harvesting has on metabolome coverage, all the samples were processed and analyzed by the metabolite profiling workflow proposed above (Fig. 4). Each method provided similar figures and yields in terms of the number of lipids detected: approximately 220 for lipidomic-RP ESI (+) and approximately 40 for generic-RP ESI (−). However, lower feature intensities were observed for method C, with a mean decrease in intensity of around 20 % compared to methods A and B (Table 3). Based on the total number of the polar metabolites detected by the combination of HILIC and generic-RP ESI (−), methods A and B rendered 93 and 89 polar metabolites, respectively, while only 65 polar metabolites were detected by method C, which also showed a lower mean intensity (an approximate 30 % decrease) (Table 3). A PCA analysis was performed to evaluate the natural interrelation among the different cell harvesting methods, and showed a separate cluster for each method (Fig. 5a). Along the first component, a clear separation was observed between method C and the two other methods. Method C involved using trypsin to detach cells from the plate surface, which implied that metabolism quenching was delayed compared to the other two methods. Trypsinization is a harvesting technique used routinely for subculturing purposes; however, its application to metabolomics is ill-advised. It has been reported to possibly compromise cellular membrane integrity, which could lead to loss of intracellular metabolites [12, 47]. The generic decrease in the intensity of the metabolites observed in Table 3 reinforces this suggestion. Delayed metabolism quenching may also contribute to the enzymatic degradation of some metabolites [9]. Interestingly, a significant decrease for the adenylate energy charge ratio (expressed as (ATP + ADP/2)/(AMP + ADP + ATP)) [48] was observed for method C. That change was also accompanied by an increase in hypoxanthine-to-ATP ratio (indicative of ATP degradation) and an increase in cysteine-glutathione disulfide levels, an oxidative stress marker; however, no increases were observed with regard to oxidized glutathione (Fig. 5b). Although no significant differences were found between methods A and B with respect to the total metabolite coverage or the levels of selected metabolites (Table 3, Fig. 5c), method A is preferred for practical considerations such as samples can be stored at −80 °C after metabolism quenching without having to process them immediately after sample collection. This is particularly useful when a large number of samples is generated at the same time, or when complex extraction procedures are required. Greater metabolite stability has also been reported when samples are stored as intact cells (frozen plates) rather than as frozen cellular extracts [14]. All these facts allowed us to select method A as the optimum strategy to perform sample processing when analyzing the endo-metabolome of cultured adherent cells.
Cell harvesting methods evaluated. Metabolome extraction and LC-MS analyses are performed based on previously optimized conditions. Method A: Metabolism quenching by the addition of liquid N2 followed by the simultaneous detachment of the cells and extraction of the metabolites by scrapping the cells using extraction solvent. Method B: Simultaneous metabolism quenching, cell detachment and metabolites extraction by scrapping the cells in extraction solvent. Method C: Cell detachment by trypsinization followed by metabolism quenching using liquid N2 and metabolites extraction
a The PCA scores plot showing the natural interrelation among the three cell harvesting methods. Each point represents all the metabolites detected using the workflow represented in Fig. 4, and the data presented in Table 3. Lines denote the 95 % confidence interval of Hotelling’s ellipse for the three different cell harvesting methods. b, c Influence of harvesting method in adenylate charge ratio and oxidative stress. Boxplots showing the values for the different ratios of interest b and statistical results of the ANOVA test and the corresponding Tukey HSD post hoc tests c. Method A: Metabolism quenching by the addition of liquid N2 followed by the simultaneous detachment of the cells and extraction of the metabolites by scrapping the cells using extraction solvent. Method B: Simultaneous metabolism quenching, cell detachment and metabolites extraction by scrapping the cells in extraction solvent. Method C: Cell detachment by trypsinization followed by metabolism quenching using liquid N2 and metabolites extraction. GSH reduced glutathione, GSSG oxidized glutathione, CSSG cysteine-glutathione disulfide
Optimized metabolite profiling workflow for cells in adherent culture
The final metabolite profiling workflow, which comprised a combination of cell harvesting, metabolome extractions, extract separations, and MS detection, is represented in Fig. 6. Detailed information about how the selected conditions are combined and assembled into a single straightforward protocol is provided in ESM 8. The combined use of different metabolome extractions to obtain polar and nonpolar fractions, analyzed separately by HILIC and RP liquid chromatographic techniques, allowed the detection of more than 300 metabolites with a CV < 30 % in the determination between biological replicates (ESM Tables S6-S9), with high diversity in classes, including 73 polar (e.g., amino acids, nucleotides, cofactors, etc.) and 243 lipid (e.g., TG, FA, PL, etc.) metabolites. The main advantage of the present work over previously published ones [5, 12–20] lies, on the one hand, in the number of parameters evaluated during the optimization process, where aspects of all the steps involved in the cell sample processing and analysis were evaluated and, on the other hand, in the nature of the target metabolites to be profiled. While most previously published methods have focused on the detection of polar metabolites [12, 14–20], the main goal of the present work was to extend metabolite coverage to a wider polarity range, from highly polar to highly nonpolar, and to place special emphasis on the comprehensive determination of the lipidome. Most previously published studies have assessed their metabolite coverage by examining the number of features detected, which is highly dependent on both the instrument and conditions employed for data preprocessing. Other studies have centered on the targeted determination of a set of representative metabolites, which is more appropriate for comparison purposes, but still presents some bias. In terms of the number of detected metabolites, the results shown herein are comparable [17, 19] (≈100 polar metabolites) and even below the figures obtained by others [18], where a targeted analysis of 258 polar metabolites was developed. However, these approaches focused on extracting and detecting polar compounds, rather than combining the detection of both polar and nonpolar ones. As regards the metabolite yields reported for other biological samples, our results showed comparable or slightly superior yields to those reported for worms (142 mostly polar metabolites) [49], atherosclerotic plaques (226, which comprised both lipids and polar compounds) [50], liver tissue (120 lipids and 210 polar compounds) [51], or serum (185 lipids) [52]. On the other hand, the major weakness of the present protocol is that only a reduced set of IS was used, what may constitute an important issue when looking for subtle differences as it may be in many real studies. Future research has to be conducted to incorporate a more complete and representative set of IS, which may allow for improved signal correction.
Optimized workflow for the untargeted metabolite profiling of adherent cultured cells. a Schematic workflow of the options selected with respect to each of the evaluated steps (i.e., cell harvesting, metabolite extraction and LC-MS analysis). b Schematic representation of how the different selected options are integrated into a single protocol
To test whether the developed metabolite workflow could be easily adapted to other mammalian cells, it was also applied to the metabolite profiling of primary cultured rat hepatocytes, a particularly relevant model, together with HepG2 cells, in different research areas [10, 25]. Interestingly, similar results were obtained for the number and nature of the identified metabolites (ESM Tables S10-S13), which indicates the versatility of the new cells’ metabolite profiling workflow.
Conclusions
Optimization of sample treatment for the analytical platform is essential for obtaining more comprehensive, reliable, and representative metabolite profiles of the biological system under study. The goal of the present study was to develop a straightforward method for sample processing and metabolite profiling of cells that focuses on extending metabolome coverage and on placing special emphasis on the detection of both polar and lipids compounds. To this end, a two-phase optimization strategy was followed. First, metabolome extractions and analyses were optimized in a combined manner (Fig. 1). Then, the influence of different cell harvesting methods was evaluated by using the previously optimized extractions and analyses methods (Fig. 4). This strategy allowed the delineation of a global metabolomic workflow that expands metabolome coverage by integrating sample preparation and analysis (Fig. 6). The specific separation and analysis of metabolites, according to their chemical and physical properties, allowed us to minimize the interferences and maximize metabolome coverage. This constitutes a crucial advantage for the metabolic phenotyping of cells and the possibility of discovering biomarkers. The development of new analytical strategies that increase metabolome coverage reinforces the utility of cell cultures and metabolite profiling as a model to determine altered biochemical pathways or changes associated with pharmacological effects, toxic insults or pathological processes.
References
Bai J, Wang MX, Chowbay B, Ching CB, Chen WN (2011) Metabolic profiling of HepG2 cells incubated with S(−) and R(+) enantiomers of anti-coagulating drug warfarin. Metabolomics 7(3):353–362
Croixmarie V, Umbdenstock T, Cloarec O, Moreau A, Pascussi JM, Boursier-Neyret C et al (2009) Integrated comparison of drug-related and drug-induced ultra performance liquid chromatography/mass spectrometry metabonomic profiles using human hepatocyte cultures. Anal Chem 81(15):6061–6069
Brown MV, Compton SA, Milburn MV, Lawton KA, Cheatham B (2013) Metabolomic signatures in lipid-loaded HepaRGs reveal pathways involved in steatotic progression. Obesity (Silver Spring) 21(12):E561–E570
Paglia G, Hrafnsdottir S, Magnusdottir M, Fleming RM, Thorlacius S, Palsson BO et al (2012) Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS). Anal Bioanal Chem 402(3):1183–1198
Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R et al (2012) The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res 22(1):168–177
Frezza C, Zheng L, Tennant DA, Papkovsky DB, Hedley BA, Kalna G et al (2011) Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS One 6(9):e24411
Yizhak K, Gaude E, Le Devedec S, Waldman YY, Stein GY, van de Water B et al (2014) Phenotype-based cell-specific metabolic modeling reveals metabolic liabilities of cancer. eLife 3:e03641
Ibanez C, Valdes A, Garcia-Canas V, Simo C, Celebier M, Rocamora-Reverte L et al (2012) Global Foodomics strategy to investigate the health benefits of dietary constituents. J Chromatogr A 1248:139–153
Leon Z, Garcia-Canaveras JC, Donato MT, Lahoz A (2013) Mammalian cell metabolomics: experimental design and sample preparation. Electrophoresis 34(19):2762–2775
Gomez-Lechon MJ, Castell JV, Donato MT (2008) An update on metabolism studies using human hepatocytes in primary culture. Expert Opin Drug metab Toxicol 4(7):837–854
Vuckovic D (2012) Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal Bioanal Chem 403(6):1523–1548
Dettmer K, Nurnberger N, Kaspar H, Gruber MA, Almstetter MF, Oefner PJ (2011) Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal Bioanal Chem 399(3):1127–1139
Fei F, Bowdish DM, McCarry BE (2014) Comprehensive and simultaneous coverage of lipid and polar metabolites for endogenous cellular metabolomics using HILIC-TOF-MS. Anal Bioanal Chem 406(15):3723–3733
Lorenz MA, Burant CF, Kennedy RT (2011) Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem 83(9):3406–3414
Bi H, Krausz KW, Manna SK, Li F, Johnson CH, Gonzalez FJ (2013) Optimization of harvesting, extraction, and analytical protocols for UPLC-ESI-MS-based metabolomic analysis of adherent mammalian cancer cells. Anal Bioanal Chem 405(15):5279–5289
Ritter JB, Genzel Y, Reichl U (2008) Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal Biochem 373(2):349–369
Ivanisevic J, Zhu ZJ, Plate L, Tautenhahn R, Chen S, O’Brien PJ et al (2013) Toward ‘omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal Chem 85(14):6876–6884
Yuan M, Breitkopf SB, Yang X, Asara JM (2012) A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 7(5):872–881
Martano G, Delmotte N, Kiefer P, Christen P, Kentner D, Bumann D et al (2015) Fast sampling method for mammalian cell metabolic analyses using liquid chromatography-mass spectrometry. Nat Protoc 10(1):1–11
Sellick CA, Hansen R, Stephens GM, Goodacre R, Dickson AJ (2011) Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat Protoc 6(8):1241–1249
Lahoz A, Gombau L, Donato MT, Castell JV, Gomez-Lechon MJ (2006) In vitro ADME medium/high-throughput screening in drug preclinical development. Mini Rev Med Chem 6(9):1053–1062
Lenz EM, Wilson ID (2007) Analytical strategies in metabonomics. J Proteome Res 6(2):443–458
Garcia-Cañaveras JC, Donato MT, Castell JV, Lahoz A (2011) A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J Proteome Res 10(10):4825–4834
Rojo D, Barbas C, Ruperez FJ (2012) LC-MS metabolomics of polar compounds. Bioanalysis 4(10):1235–1243
Donato MT, Lahoz A, Castell JV, Gomez-Lechon MJ (2008) Cell lines: a tool for in vitro drug metabolism studies. Curr Drug Metab 9(1):1–11
Gomez-Lechon MJ, Donato MT, Martinez-Romero A, Jimenez N, Castell JV, O’Connor JE (2007) A human hepatocellular in vitro model to investigate steatosis. Chem Biol Interact 165(2):106–116
García-Cañaveras JC, Jiménez N, Gómez-Lechón MJ, Castell JV, Donato MT, Lahoz A (2015) LC-MS untargeted metabolomic analysis of drug-induced hepatotoxicity in HepG2 cells. Electrophoresis 36(18):2294–2302
Gomez-Lechon MJ, Ponsoda X, O’Connor E, Donato T, Jover R, Castell JV (2003) Diclofenac induces apoptosis in hepatocytes. Toxicol in Vitro 17(5–6):675–680
Cao B, Aa J, Wang G, Wu X, Liu L, Li M et al (2011) GC–TOFMS analysis of metabolites in adherent MDCK cells and a novel strategy for identifying intracellular metabolic markers for use as cell amount indicators in data normalization. Anal Bioanal Chem 400(9):2983–2993
Wu H, Southam AD, Hines A, Viant MR (2008) High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal Biochem 372(2):204–212
Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS et al (2013) Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc 8(1):17–32
Nygren H, Seppanen-Laakso T, Castillo S, Hyotylainen T, Oresic M (2011) Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics for studies of body fluids and tissues. Methods Mol Biol 708:247–257
Cortes M, Pareja E, Garcia-Canaveras JC, Donato MT, Montero S, Mir J et al (2014) Metabolomics discloses donor liver biomarkers associated with early allograft dysfunction. J Hepatol 61(3):564–574
Pluskal T, Castillo S, Villar-Briones A, Oresic M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf 11:395
Quintás G, Portillo N, García-Cañaveras JC, Castell JV, Ferrer A, Lahoz A (2012) Chemometric approaches to improve PLSDA model outcome for predicting human non-alcoholic fatty liver disease using UPLC-MS as a metabolic profiling tool. Metabolomics 8(1):86–98
Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y et al (2013) HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res 41(Database issue):D801–D807
Fahy E, Sud M, Cotter D, Subramaniam S (2007) LIPID MAPS online tools for lipid research. Nucleic Acids Res 35(Web Server issue):W606–W612
Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR et al (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27(6):747–751
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K et al (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45(7):703–714
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA et al (2007) Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3(3):211–221
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Álvarez-Sánchez B, Priego-Capote F, Castro M (2010) Metabolomics analysis II. Preparation of biological samples prior to detection. TrAC Trends Anal Chem 29(2):120–127
Villas-Boas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J (2005) Mass spectrometry in metabolome analysis. Mass Spectrom Rev 24(5):613–646
Seppanen-Laakso T, Oresic M (2009) How to study lipidomes. J Mol Endocrinol 42(3):185–190
Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N et al (2011) Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 6(7):1060–1083
Chen J, Zhou L, Zhang X, Lu X, Cao R, Xu C et al (2012) Urinary hydrophilic and hydrophobic metabolic profiling based on liquid chromatography-mass spectrometry methods: differential metabolite discovery specific to ovarian cancer. Electrophoresis 33(22):3361–3369
Teng Q, Huang W, Collette TW, Ekman DR, Tan C (2009) A direct cell quenching method for cell-culture based metabolomics. Metabolomics 5(2):199–208
Atkinson DE (1968) The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 7(11):4030–4034
Saric J, Want EJ, Duthaler U, Lewis M, Keiser J, Shockcor JP et al (2012) Systematic evaluation of extraction methods for multiplatform-based metabotyping: application to the Fasciola hepatica metabolome. Anal Chem 84(16):6963–6972
Vorkas PA, Isaac G, Anwar MA, Davies AH, Want EJ, Nicholson JK et al (2015) Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: application to cardiovascular disease. Anal Chem 87(8):4184–4193
Yamazaki M, Miyake M, Sato H, Masutomi N, Tsutsui N, Adam K-P et al (2013) Perturbation of bile acid homeostasis is an early pathogenesis event of drug induced liver injury in rats. Toxicol Appl Pharmacol 268(1):79–89
Sarafian MH, Gaudin M, Lewis MR, Martin FP, Holmes E, Nicholson JK et al (2014) Objective set of criteria for optimization of sample preparation procedures for ultra-high throughput untargeted blood plasma lipid profiling by ultra performance liquid chromatography-mass spectrometry. Anal Chem 86(12):5766–5774
Acknowledgments
This work has been supported by the Instituto de Salud Carlos III of the Spanish Ministry of Science and Innovation (FIS PI14/00026 and FIS PI13/0986). A.L. is grateful for a Miguel Server II contract (CPII14/00004) from the above Ministry/Instituto de Salud Carlos III. J.C. G.-C. is grateful for a pre-doctoral contract from the Vali + d program of the Conselleria d’Educació (Regional Valencian Ministry of Education). S.L. is grateful for a contract (PTA2012-7224-I) from the Spanish Ministry of Economy and Competitiveness.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All the animals received human care and all experimental protocols were approved by the institutional animal ethics committee and performed in accordance with national and institutional regulations.
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2.28 mb)
Rights and permissions
About this article
Cite this article
García-Cañaveras, J.C., López, S., Castell, J.V. et al. Extending metabolome coverage for untargeted metabolite profiling of adherent cultured hepatic cells. Anal Bioanal Chem 408, 1217–1230 (2016). https://doi.org/10.1007/s00216-015-9227-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-9227-8