Abstract
A minimally invasive and repeatable approach for real-time epidermal growth factor receptor (EGFR) mutation surveillance would be highly beneficial for individualized therapy of late stage lung cancer patients whose surgical specimens are often not available. We aim to develop a viable method to detect EGFR mutations in single circulating tumor cells (CTCs). Using a model CTC system of spiked tumor cells in whole blood, we evaluated EGFR mutation determination in single tumor cells enriched from blood. We used magnetic beads labeled with antibody against leukocyte surface antigens to deplete leukocytes and enrich native CTCs independent of epithelial marker expression level. We then used laser cell microdissection (LCM) to isolate individual CTCs, followed by whole-genome amplification of the DNA for exon 19 microdeletion, L858R and T790M mutation detection by PCR sequencing. EGFR mutations were successfully measured in individual spiked tumor cells enriched from 7.5 ml whole blood. Whole-genome amplification provided sufficient DNA for mutation determination at multiple sites. Ninety-five percent of the single CTCs microdissected by LCM (19/20) yielded PCR amplicons for at least one of the three mutation sites. The amplification success rates were 55 % (11/20) for exon 19 deletion, 45 % (9/20) for T790M, and 85 % (17/20) for L858R. Sequencing of the amplicons showed allele dropout in the amplification reactions, but mutations were correctly identified in 80 % of the amplicons. EGFR mutation determination from single captured tumor cells from blood is feasible with the approach described here. However, to overcome allele dropout and to obtain reliable information about the tumor's EGFR status, multiple individual tumor cells should be assayed.

Captured CTC from patient blood. CTC (green arrow) and WBCs (red arrow) enriched from 7.5 ml peripheral blood of a NSCLC patient were immunostained with anti-Cytokeratin 18 (green) and anti-CD45 (red) antibodies. Cell nuclei were stained with DAPI (blue). Compared with H1975 (Fig. 1), CTC from the NSCLC patient appeared to have reduced CK18 expression
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Lung cancer has become the leading cause of cancer-related death in the world. Multiple clinical studies have shown that non-small cell lung cancer (NSCLC) patients with activating epidermal growth factor receptor (EGFR) mutations, i.e., a short deletion in exon 19 and L858R point mutation in exon 21, respond favorably to EGFR tyrosine kinase inhibitor (TKI) therapy, while a TKI-resistant point mutation T790M in exon 20 is found present in a majority of relapsed tumors after an initial response to EGFR-TKIs [1, 2]. Since EGFR mutations have significant value for tailoring the individualized EGFR-TKI therapy, continuous monitoring of EGFR mutational status before and during TKI therapy is highly desirable. Currently, tumor surgery specimens are used for most EGFR mutation detections. However, the availability of tumor specimens in the clinical setting is often limited, especially if repeated tests for monitoring are considered. In a prospective study, surgical specimens were only available for less than 50 % of the patients [3]. Alternatives such as bronchoscopic biopsy can be used to obtain tissue samples for late stage patients not suitable for surgery [4], yet they have varying performances and remain invasive, costly, and difficult to repeat for real-time surveillance of the tumor's genetic status. It is therefore highly desirable to establish a minimally invasive method to detect EGFR mutations without tissue biopsy. Detecting mutations in circulating cell-free DNA is one approach. However, since only a small portion of cell-free DNA is of tumor origin, this approach is hindered by the technical challenge of detecting a mutant allele in a background of wild-type alleles in the cell-free DNA population. Circulating tumor cells (CTCs), which are disseminated from tumor tissues [5], offer the potential to change our approach to biomarker evaluation, especially for late stage patients, by providing a source of tumor material that is easily accessible through a blood draw. Molecular characterization of CTCs in NSCLC, including EGFR or KRAS mutational status, has recently been described for a population of cells isolated by either commercial CTC isolation kit or custom-made microfluidic devices [6, 7]. However, they all sampled a population of cells including non-tumor cells as contaminants and were biased towards cells with strong expression of the epithelial marker EpCAM targeted by the CTC isolation scheme. These assays, therefore, may not accurately reflect tumor mass. Here, we describe a single CTC-based EGFR mutation detection approach that employs an epithelial marker-independent enrichment strategy and laser capture microdissection (LCM) for single CTCs, followed by whole-genome amplification (WGA) and PCR sequencing to detect the three EGFR mutations.
Materials and methods
To develop and optimize the multistep method, we spiked NSCLC cell line H1975 (ATCC) into normal peripheral blood. H1975 cells harbor heterozygous L858R and T790M mutations and wild-type exon 19 [8]. Blood from healthy volunteers or NSCLC patients from Peking Union Medical College Hospital were collected into 7.5-ml ACD tubes after an initial 2-ml collection into a precollection tube. Written consent was obtained for each sample under IRB-approved protocol. Each blood sample was mixed with a defined number of cells from H1975 cell suspension to mimic peripheral blood from NSCLC patients. After red cell lysis, spiked tumor cells were recovered by removing most of the white blood cells with immunomagnetic beads conjugated with antibodies against human leukocyte surface antigens as previously reported [9]. The remaining cell suspension were spread onto a Leica PEN slide and dried overnight at room temperature for immediate use or storage at −20 °C. The slide was immunostained with a cocktail of Alexa 594-conjugated anti-CD45 and Alexa 488-conjugated anti-CK18 for 1 h in the dark, and tumor cells were identified as CK18+ and CD45− with intact cell nuclei as visualized by DAPI (Fig. 1). The cancer identity of these cells was confirmed by FISH as aneuploid or tetraploid cells using chromosomal 7 and 8 probes (data not shown).
Identification of enriched H1975 cells. Cells enriched from H1975-spiked 7.5-ml peripheral blood were subjected to immunofluorescent staining with both anti-Cytokeratin 18 and anti-CD45 antibodies. Nuclei of cells were stained with 4-6-diamidino-2-phenylindole (DAPI). Only tumor cells were found CK18 positive but CD45 negative with intact nuclei
For LCM, tumor cells on the slide were counted and photographed, and individual tumor cell coordinates were recorded to facilitate subsequent target cell identification. The slide coverslip was next removed by dipping the slide into 100 % ethanol, and after drying, the sample was loaded onto the stage of a Leica LMD6000 system under a ×40 objective. After microdissection with a 337-nm laser beam, the target cell was dropped by gravity into a 10-μl lysis solution (40 μg/ml proteinase K in PBS, 0.5 % Triton X-100) on the inside surface of a 200-μl microcentrifuge tube lid (Fig. 2a–c). The success of the procedure was verified by in situ examination of the lid (Fig. 2d). After lid closure, the cell was collected to the bottom of the tube with brief centrifugation and digested by proteinase K at 55 °C for 1 h in a sealed tube. Following heat inactivation of proteinase K at 95 °C for 10 min, the DNA in the resulting lysate was whole-genome amplified using a REPLI-g Mini WGA kit (Qiagen) according to the manufacturer's protocol (or for comparison, used directly for PCR sequencing). Nested PCR was used to separately amplify EGFR exons 19, 20, and 21 fragments containing the mutation sites (see Table 1 for primer sequences). Each 20-μl PCR contained 2 μl of WGA DNA, 0.5 μM primers, and 1× GoTaq Colorless Master Mix (Promega). After initial denaturation at 95 °C for 5 min, the DNA was amplified with external primers after 40 cycles of 95 °C for 30 s, annealing (61 °C for exon 19 and 21, 55 °C for exon 20) for 30 s and 72 °C for 30 s. Two microliters of the product was amplified with internal primers under the same condition except that the total reaction volume was 50 μl and the annealing temperatures were 55 °C for exon 19, 58 °C for exon 20, and 56 °C for exon 21. A mitochondrial gene, Cytb, was amplified as an internal positive control for amplification. PCR products were analyzed by agarose gel electrophoresis and bi-directional Sanger sequencing.
Laser capture microdissection. The identified tumor cell (encircled) was first marked by the user on the laser microdissection system (a), dissected with a UV beam, and collected by gravity onto a collection lid (schematic in b). Dissection was verified with bright-field view after LCM (c), and the collected cell was visualized on the lid (d)
Results
We performed 25 runs of enrichment of 7.5-ml whole blood spiked with ten H1975 cells, with an average recovery rate of 77 %. For LCM of enriched H1975 cells on the slide, we compared the performance of Leica PEN slide with that of the low-autofluorescence DIRECTOR slide (Expression Pathology, USA) and chose PEN slide for its ability to preserve the integrity of the dissected cell and to consistently allow the dissected cell to drop by gravity to the collection cap.
The DNA yield of the 50-μl whole-genome amplification reaction was about 5 μg. From 20 samples of whole-genome amplified DNA, nested PCR amplifications for EGFR exons 19, 20, and 21 fragments had success rates of 55 % (11/20), 45 % (9/20), and 85 % (17/20), respectively, as determined by gel electrophoresis (Fig. 3). Interestingly, the same sample could give successful amplification of one exon while failing another exon (Fig. 3). Notably, PCR for the control gene Cytb, a high-copy number gene, had a sequence-verified success rate of 100 % (data not shown), suggesting that the amplification procedure was effective. To investigate whether insufficient template quantity may be the cause of PCR failures, we attempted nested PCR of isolated single H1975 without prior WGA. The entire DNA of a single H1975 was used for nested PCR of exon 21, and the success rate for amplification was 60 % (24/40), not significantly different from that using WGA-preamplified DNA. This would indicate that the PCR failure was likely not primarily due to insufficient template, but possibly due to template damages prior to WGA, for example due to a UV-induced DNA break as a result of UV exposure of the cell during tumor cell identification and LCM.
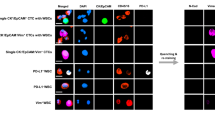
Single-cell nested PCR after WGA for detection of exon 19, 20, and 21 mutations. Exon fragments were separately amplified with nested PCR from whole-genome-amplified DNA of single H1975 cells (nos. 60–80) and analyzed by agarose electrophoresis. M size marker, no. 00 negative control. Positive amplifications were identified for the following cells: for exon 19, nos. 62, 63, 64, 66, 67, 68, 70, 71, 73, 75, and 76; for exon 20, nos. 62, 63, 64, 68, 70, 73, 74, 78, and 80; and for exon 21, all except nos. 65, 75, and 80. The only cell that failed to amplify in all three exons was no. 65
Of the 17 samples positive for exon 21 amplification, ten yielded sufficient DNA for sequencing. Results showed eight of them with L858R mutation including five homozygous and three heterozygous, and two of them were wild type (Fig. 4). For the nine samples positive for exon 20 amplification, sequencing showed seven homozygous T790M mutation and two wild type. All 11 exon 19 amplification-positive samples showed no deletion mutation as expected. Since H1975 harbors heterozygous H858R and T790M mutations [8], the presence of wild-type and homozygous mutant amplicons suggests allele dropout, a common phenomenon seen in single-cell amplification [10]. Despite this, mutation was correctly identified in 80 % (15/19) of the amplicons, likely owing to the preferential copy gain of the mutant EGFR alleles in the genome of H1975 as well as in NSCLC tumors [11]. If exon 19 results were included, our method achieved 87 % (26/30) accuracy for mutation status determination. These results indicate the approach is feasible for mutation detection when multiple isolated tumor cells are individually analyzed. Clinical validation of the approach has been initiated using blood samples from NSCLC patients (Fig. 5). Initial results obtained from the ongoing validation study revealed that among the first five recruited NSCLC patients, two of them showed EGFR L858R mutation on exon 21 which was successfully identified on the microdissected single CTC enriched from 7.5 ml of patients' peripheral blood, indicating feasibility of the described strategy for lung cancer study. Further clinical validation on a large number of clinical samples is underway.
Sequencing chromatograms of nested PCR amplicons for exon 21. Ten exon 21 amplicons from Fig. 3 were sequenced for L858R mutation. a Homozygous mutant sequence found for cell nos. 62, 63, 68, 76, and 78. b Homozygous wild-type sequence found for cell nos. 61 and 74. c Heterozygous mutant sequence found for cells nos. 64 and 70. d Heterozygous mutant sequence found for cell no. 66
CTC from patient blood. CTC (green arrow) and WBCs (red arrow) enriched from 7.5 ml peripheral blood of a NSCLC patient were immunostained with anti-Cytokeratin 18 (green) and anti-CD45 (red) antibodies. Cell nuclei were stained with DAPI (blue). Compared with H1975 (Fig. 1), CTC from the NSCLC patient appeared to have reduced CK18 expression
Discussion
To explore potential alternatives to the use of a surgical specimen for mutation monitoring, we have isolated native single tumor cells from blood as the source of tumor tissue. Our CTC enrichment strategy does not involve recognition of CTC surface antigens, thereby avoiding the pitfalls of conventional antibody-capture methods, such as the potential loss of CTCs having reduced EpCAM expression [9, 11, 12]. We have applied this approach for CTC enumeration for 144 stage III–IV NSCLC patients, with a positive rate of 65 %, and CTC numbers ranging from 2 to 2,630 with a median value of 10 [13]. A recent independent study using a similar enrichment strategy reported a comparable positive rate in metastatic carcinoma patients [14], supporting the validity of our enrichment approach.
In the present study, we explored the possibility of extending the utility of circulating tumor cells from enumeration to mutation detection. We have shown that a microdissected target individual tumor cell, upon whole-genome amplification, can indeed provide a sufficient amount of pure tumor DNA, not contaminated by DNA of any other tumor or non-tumor cells, for EGFR mutation analysis and possibly other genomic characterizations. Our study demonstrated technical feasibility of EGFR mutation determination from single captured tumor cells. Although UV exposure of the cells, which is necessary in the current protocol for tumor cell identification under a fluorescence microscope, may lead to DNA breaks which probably resulted in some of the low PCR success rates observed in this study, the UV exposure time can be minimized upon accumulated experience in experimental handling. Alternatively, primer designs for shorter PCR amplicons may help reduce the impact of DNA break. Additionally, the use of digital PCR technology, which employs multiple miniaturized PCRs to amplify a target, has been shown to improve PCR sensitivity to detect EGFR mutations [15] and should increase the PCR success rate on single CTCs. This technology can be tested as it becomes more accessible to the clinical laboratories. Lastly, DNA damage can be minimized by the use of alternative dyes with longer excitation wavelength and lower-powered light source in LCM, as demonstrated recently with microdissected breast cells by Analtonen at al [16]. At present, a maximal number of individual CTCs are interrogated to overcome this limitation. We anticipate in the near future that the amplification success rates can be improved to a level that makes this technology even more attractive for clinical applications.
Our results suggested that allele dropout, which is stochastic amplification of only one of the alleles in a single cell, was associated with single-cell molecular analysis, leading to about 20 % error rate in mutation determination if only one CTC is sequenced for the conclusion. One potential approach to mitigate this is to use digital PCR technology [15], which by multiple miniaturized PCR amplifications should in theory increase the probability that all alleles are amplified, leading to reduced error rate and increased sensitivity of the method. It is necessary at present to assay multiple individual CTCs in order to overcome this intrinsic limitation for accurate diagnosis, especially when the sequencing result from a CTC is wild type. Despite this complication, the approach is still preferable since CTCs can be obtained from a routine blood draw with minimal risk and inconvenience to the patient, compared to a fresh biopsy. While our current study established the technical feasibility of the approach, requirement characteristics such as the minimal number of CTCs to be assayed obviously depend on the characteristics of the clinical CTC and are to be determined in large-scale clinical trials of this approach. Such trials are warranted as the approach can potentially provide real-time information about the patient's current disease state. Analysis of EGFR mutation status in CTCs collected prior to treatment could potentially be used to select an appropriate targeted therapy, while repeated longitudinal sampling during treatment could potentially be used to detect appearance of resistance markers and enable switching to a more appropriate therapy.
Conclusions
Single-cell genomic analysis, especially next-generation sequencing of single cells, can provide a wealth of information that was not possible with conventional bulk analysis [17]. The diagnostic implications of single-cell analysis, however, were seldom discussed. Our data demonstrate that tumor cells in blood can indeed be analyzed individually. Our present study, although not whole-genome analysis but targeted analysis of diagnostic relevance, points to the possibility that circulating tumor cells may provide the first examples of single-cell whole-genome sequencing for cancer diagnostics. However, our data suggest many single cells should be sequenced before any conclusion can be made regarding the tumor, and the stochastic nature of single-cell DNA amplification dictates that any analysis of intratumor genetic heterogeneity or clonal evolution will not be at single-cell resolution as might be originally imagined.
Abbreviations
- CTC:
-
Circulating tumor cells
- EGFR :
-
Epidermal growth factor receptor
- LCM:
-
Laser capture microdissection
- NSCLC:
-
Non-small cell lung cancer
- TKI:
-
Tyrosine kinase inhibitor
- WGA:
-
Whole-genome amplification
References
Lynch TJ, Bell DW, Sordella R et al (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350:2129–2139
Paez JG, Janne PA, Lee JC et al (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304:1497–1500
Costa DB, Kobayashi S, Tenen DG et al (2007) Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer 58:95–103
Horiike A, Kimura H, Nishio K et al (2007) Detection of epidermal growth factor receptor mutation in transbronchial needle aspirates of non-small cell lung cancer. Chest 131:1628–1634
Allard WJ, Matera J, Miller MC et al (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 10:6897–6904
Punnoose EA, Atwal SK, Spoerke JM et al (2010) Molecular biomarker analyses using circulating tumor cells. PLoS One 5:e12517
Maheswaran S, Sequist LV, Nagrath S et al (2008) Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359:366–377
Gandhi J, Zhang J, Xie Y et al (2009) Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One 4:e4576
Wu C, Hao H, Li L et al (2009) Preliminary investigation of the clinical significance of detecting circulating tumor cells enriched from lung cancer patients. J Thorac Oncol 4:30–36
Ray PF, Winston RM, Handyside AH (1996) Reduced allele dropout in single-cell analysis for preimplantation genetic diagnosis of cystic fibrosis. J Assist Reprod Genet 13:104–106
Tewes M, Aktas B, Welt A et al (2009) Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat 115:581–590
Krebs MG, Hou JM, Sloane R et al (2012) Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 7:306–315
Zhou X, Li L, Hao H et al (2010) Circulating tumor cells as a prognostic and predictive indicator in advanced lung cancer patients. Oncol Prog 8:484–490
Liu Z, Fusi A, Klopocki E et al (2011) Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med 9:70
Yung TKF, Chan KCA, Mok TSK et al (2009) Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 15:2076–2084
Aaltonen KE, Ebbesson A, Wigerup C et al (2011) Laser capture microdissection (LCM) and whole genome amplification (WGA) of DNA from normal breast tissue—optimization for genome wide array analyses. BMC Res Notes 4:69
Pennisi E (2012) The biology of genomes. Single-cell sequencing tackles basic and biomedical questions. Science 336:976–977
Acknowledgments
We thank Zhibin Cheng for help with manuscript preparation. The study was supported by a fund from the Wu Jieping Foundation of China (to L.L).
Conflict of interest
None is declared.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ran, R., Li, L., Wang, M. et al. Determination of EGFR mutations in single cells microdissected from enriched lung tumor cells in peripheral blood. Anal Bioanal Chem 405, 7377–7382 (2013). https://doi.org/10.1007/s00216-013-7156-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-013-7156-y