Abstract
Exercise modulates the metabolome in urine or blood as demonstrated previously for humans and animal models. Using nuclear magnetic resonance (NMR) metabolomics, the present study compares the metabolic consequences of an exhaustive exercise at peak velocity (Vp) and at critical velocity (Vc) on mice. Since small-volume samples (blood and urine) were collected, dilution was necessary to acquire NMR spectra. Consequently, specific processing methods were applied before statistical analysis. According to the type of exercise (control group, Vp group and Vc group), 26 male mice were divided into three groups. Mice were sacrificed 2 h after the end of exercise, and urine and blood samples were drawn from each mouse. Proton NMR spectra were acquired with urine and deproteinized blood. The NMR data were aligned with the icoshift method and normalised using the probabilistic quotient method. Finally, data were analysed with the orthogonal projection of latent-structure analysis. The spectra obtained with deproteinized blood can neither discriminate the control mice from exercised mice nor discriminate according to the duration of the exercise. With urine samples, a significant statistical model can be estimated when comparing the control mice to both groups, Vc and Vp. The best model is obtained according to the exercise duration with all mice. Taking into account the spectral regions having the highest correlations, the discriminant metabolites are allantoin, inosine and branched-chain amino acids. In conclusion, metabolomic profiles assessed with NMR are highly dependent on the exercise. These results show that urine samples are more informative than blood samples and that the duration of the exercise is a more important parameter to influence the metabolomic status than the exercise velocity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, the metabolic pathways involved during physical exercise were investigated by the metabolomic methods. Metabolomics is highly sensitive to environmental factors including age, pharmacology and toxicology (for review, see [1]). Therefore, the phenotypic responses of a subject to exercise may be shown using this method.
Among the analytic techniques implemented for metabolomics, mass spectrometry and nuclear magnetic resonance (NMR) spectroscopy are the most common ones and provide values for a high number of parameters. Through mass spectrometry, a highly sensitive technique, metabolites present at low concentration may be detected. On the other hand, the method needs a complex sample preparation protocol. For several reasons, including the chromatographic and the ionization methods, a low reproducibility of the measurements is the main pitfall of mass spectrometry. With proton NMR spectroscopy, the metabolites detected must be at relatively high concentration in biofluids (more than 10 μM). This limitation is balanced by the high reproducibility of the technique associated to the linear analytical response. Moreover, proton NMR sensitivity does not depend upon the physical and chemical properties of the molecules [2]. For most biofluids, no sample pre-processing is needed. Consequently, metabolomic with NMR is fast and reliable and may be fully or partially automated.
Both spectroscopy methods have advantages and disadvantages and have been used to investigate exercise effects mainly for humans [3–6] and less frequently in animal models [7, 8]. However, most studies concerned plasma or serum. Urine has been rarely used despite the fact that this biofluid can be non-invasively collected. This positive argument applies especially for humans. Most previous studies have compared trained subjects and control ones [5] or pathological cases with controls [9]. In all cases, exercise modulates the metabolome, demonstrating that exercise is one of the major metabolism modulators.
Several protocols have been used to analyse and understand the physiological and biochemical consequences of physical exercises [10, 11]. Quantification of exercise was approached, in the first place, with the determination of peak velocity (Vp) as the maximal speed reached by a subject during an incremental test [12]. This parameter can be measured for humans as well as in animal models [13, 14]. More recently, the concept of critical velocity (Vc) was defined as the asymptotic value of the speed as a function of run time before exhaustion [15, 16]. The individual value for Vc is more informative about the endurance capacity of the subject than Vp. The Vc measurement needs the implementation of four different velocities for each subject [17]. It has been reported that Vc may be the threshold velocity for the contribution of the anaerobic metabolism and the involvement of a larger proportion of the type II muscle fibres [10, 16].
For humans, physiological parameters during exercise at Vc and Vp have been compared. It showed that the two types of exercise have distinct physiological effects. However, exercises at Vc and Vp have not been investigated from the metabolomic point of view. Studies involving animal models exercised at Vp or Vc are less frequent. One reason may be that the physiological data are more difficult to assess for animals than for humans. Conversely, in animal models, the metabolome after exercise can be investigated in urine and blood, and the model can easily be used for bio-molecular investigation in tissues and organs.
For pre-clinical animal studies, including mice, the sample volumes that may be collected are frequently lower than the 500 to 600 μL needed to fill the 5-mm-diameter NMR tubes. The low sensitivity of NMR in addition to reduced sample volume may be a barrier to use NMR spectroscopy in animal studies. Overall, urine samples are more difficult to collect for animals than in the case of humans.
We report here the methods we have used to process to a metabolomic investigation with NMR spectroscopy with samples drawn from mice. Considering the small volumes of blood, it was necessary to enhance the sensitivity to detect metabolites by elimination of proteins and lipids with trichloracetic acid. Urine was collected directly from the bladder. In both cases, the dilution in deuterium dioxide was necessary to complete the minimal volume to be introduced in NMR tubes. After spectral acquisition, the realignment [18] of spectra compensates the chemical variations that may be produced by pH and dilution heterogeneity, and the “Probabilistic Quotient Normalization” [19] took into account the dilution parameter.
Using these processing methods, the present study determines whether an acute and exhaustive exercise at Vc and Vp produces metabolic modulation and whether this discrimination can be obtained using urine or blood metabolome. Moreover, it shows that the metabolome is sensitive to the duration of the exhaustive exercise.
Materials and method
Animals and measurements of Vc and Vp
The mice strain is the FVB strain, selected for its high level of endurance performance on a treadmill, as identified previously [20]. Females were excluded to avoid possible effects of gender. The mice were kept in an animal facility (CERFE, Genopole, Evry, France) in a specific and opportunist pathogen-free environment and at a temperature of 22 °C with 12/12-h light–dark cycles. The animals were supplied with water and food ad libitum. The 30 male mice were separated into six cages of five mice. They were 2 months old for the first tests and 4 months old when sacrificed. The exercise tests were carried out in the facility. All protocols were approved by our institution’s Animal Care and Use Committee and comply with the Council of Europe’s European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes.
For this study, mice were divided into three groups defined according to the type of exercise performed in a treadmill (Columbus Instruments, OH, USA), on the test day. The control group contained ten mice, which performed no exercise throughout the duration of the experiment. The critical velocity (Vc) group included seven mice performing an exhaustive test at their individual Vc, 2 h before sacrifice. The velocity at peak (Vp) group included 13 mice performing an exhaustive exercise at maximal speed (Vp) 2 h before sacrifice. The time to attain exhaustion was measured and reported as the time limit (in minutes) used further on in the statistical analyses.
Vp was determined for each mouse by an exhaustive incremental test. After a 10-min rest, the initial speed was set at 10 m/min and increased by steps of 3 m/min every 3 min, until exhaustion.
Vc was determined with four tests during 4 days (one test each day). The mice ran at constant speed during a limited time of 60 min or less. An exercise was performed with a different speed each day, respectively at 100, 95, 88 and 80 % of the maximal speed reached during the exhaustive incremental test. The duration of the course for each test was noted. The Vc was determined with length and duration of the course, varying as a linear function. The slope of the linear regression provides the value of Vc.
Sample collection procedure
After exercise and before sacrifice, mice were housed alone in a cage. Mice were anaesthetized with an intra-peritoneal injection of pentobarbital (70 mg kg−1) and sacrificed by cervical dislocation. Before sacrifice, blood retrobulbar sampling was performed from anaesthetized mice. This sampling method was preferred from jugular sampling because it is faster and does not necessitate surgical incision. The whole blood was frozen at −20 °C for preservation. Before NMR analysis, the total blood was deproteinised using trichloracetic acid (TCA). Therefore, after blood thawing, samples were centrifuged to remove the unbroken cells for 10 min at 1,500 rpm. The supernatant (variable volumes) was added with TCA (10 μL of 0.9 g/L TCA for 100 μL supernatant). The precipitated proteins were eliminated by centrifugation (15 min, 1,500 rpm). The final supernatant was frozen at −20 °C. For NMR analysis, the thawed samples (50 to 150 μL) were introduced into NMR 5-mm tubes and added with deuterium dioxide to reach a final volume of 700 μL. The blood samples collected and analysed were n = 10 controls, n = 6 Vc and n = 13 Vp. One sample is missing because a too small volume was obtained after the process.
The urine samples were drawn from the bladder after animal sacrifice. This procedure was preferred to metabolic cages in order to obtain the whole volume and cleaner urine samples. Nevertheless, when the bladder was empty, no sample could be collected. For this reason, three samples from the control group and one sample from the Vc group are missing. Urine samples were frozen at −20 °C. For NMR analysis, samples (50 to 400 μL) were placed into 5-mm NMR tubes and added with deuterium dioxide to reach a final volume of 700 μL. The urine samples analysed are n = 7 controls, n = 6 Vc and n = 13 Vp.
NMR spectroscopy analysis
Spectra were acquired at 25 °C in Varian® Unity Inova spectrometer at 500 MHz. The pulse sequence was a single 90° pulse acquisition sequence including residual water signal suppression with a presaturation pulse (0.03 mW, 2.5 s) during the relaxation delay (4.5 s). The free induction decays (FIDs) were collected on 32-K data points after 128 transients for 5,000-Hz spectral width.
The FIDs were processed with the MestReNOVA software. The Fourier transform was performed with an exponential function producing a 1-Hz line broadening. Spectra were phased, and the baseline correction was performed with three points at 0, 5 and 9 ppm. Each spectrum was calibrated using allantoin signal 5.4 ppm in urine and acetic acid signal (1.92 ppm) in blood spectra. As pH, ionic strength and dilution may vary among samples, the spectra were also aligned using the icoshift method [18].
The spectral region between 0 and 9 ppm was divided into 9,000 spectral regions of 0.001-ppm width called buckets. Each bucket is labelled with its median chemical shift value. Water region, between 4.6 and 5 ppm, was excluded. For blood samples, the restricted region from 0.1 to 4.6 ppm was divided into 4,500 buckets. Considering the variation of the sample concentration, the spectra were normalised according to the “Probabilistic Quotient Normalization” method [19].
Statistical analyses
A principal component analysis (PCA) was performed using the statistics SIMCA P+ software (ver. 12, Umetrics AB, Umea, Sweden) to check any separation of samples into groups based on NMR signal variability. This method provides the possibility to detect and to exclude any outliers, defined as observations situated outside the 95 % confidence region of the model. The PCA analysis of the 29 spectra from blood samples shows two outlier samples. As no rational reason shows (problem during exercise or sampling, poor quality spectrum), these samples were not withdrawn from further analyses. The PCA analysis of urine shows no outlier.
The buckets' integration values were analysed in relation to the exercise velocity or to the duration of exercise by orthogonal projection of latent-structure analysis (OPLS) using in-house Matlab code. Compared to the classical projection of latent-structure analysis, this method allows improved interpretation of the spectroscopic variations between discriminated groups, by removing information of the NMR signal that has no impact on discrimination. The ability of the model to explain data and to predict new data is expressed by the value of the parameters R2 and Q2, respectively. The value R2 = 1 indicates a perfect description of the data by the model, while Q2 = 1 indicates a perfect prediction of new data.
The OPLS models were validated using the in-house Matlab code with a permutation testing (26 permutations). The aim is to evaluate whether our OPLS models built with two groups or the time limit are significantly better than any other models built with random permutation of the original groups or time limit values.
To take into account the two possible repartitions of the mice (control/exercised and time limit), two OPLS analyses were performed. The first one was computed with a Y matrix corresponding to the two groups, the control group and the exercised group at Vc and Vp. The second model was computed with a Y matrix corresponding to the time limit expressed in minutes. Results are visualised by score and loading plots. Scores are represented as a projection of the different sample spectra on the predictive (tpred) and the orthogonal (torth) components of the model. For easier reading, colours are used depending on the sample group or the time limit. The loading plot represents the co-variance between the Y-response matrix and the signal intensity of the various buckets. Colours are also used in the loading plots depending on the r, correlation coefficient, between the corresponding bucket and the Y variable. The buckets with a correlation coefficient r > 0.5 are assigned to the most probable metabolites, in reference to the spectral assignment previously performed in the literature [3, 21]. This was done through analysis of the original spectra at chemical shift region corresponding to the buckets taking into account the detailed appearance (multiplet, high-resolution peaks and broad signals) and comparison to values of adjacent buckets. On selected urine samples, a 2D correlation spectroscopy (COSY) spectrum was acquired to confirm spectral attributions.
Results
Physiological and exercise performance of mice
The performances of the Vc and Vp group mice are reported in Table 1. Both Vc and Vp groups are identical regarding their weight, oxygen uptake at maximal velocity and their velocity at Vc and Vp. The time limit sustained for the exhaustive exercise shows a wider spread for mice exercising at Vp than for mice exercising at Vc. The blood lactate reported here was measured 2 min after exercise, and those values are significantly different from the mean blood lactate concentration before exercise.
Metabolome of control mice and exercised mice
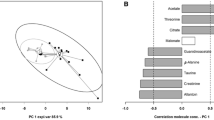
With the blood samples, the OPLS model achieved poor correlation and predictability coefficients when computed for discriminating the control samples from those taken after exercise. With the urine samples, the OPLS model can be achieved with R2 = 0.884 and Q2 = 0.584 as shown in Fig. 1A. Figure 1B indicates the spectral regions involved in the calculation of the OPLS model (intensity of the ordinate) and coloured according to their correlation. The model is validated by the permutation test. The spectral regions with correlation over 0.5 are listed in Table 2 with their possible assignments to metabolites. Those regions correspond mainly to fumarate (6.53 ppm), allantoin (5.4 ppm) and several peaks in the region where inosine (3.85 ppm) and glycerol (3.75 and 4.10 ppm correlated in COSY spectrum) are usually assigned, which are positively correlated to exercise, whereas trimethylamine (2.91 ppm) is negatively correlated to exercise. One resonance at 2.18 ppm could not be assigned and showed no correlation in the 2D COSY spectrum.
a Score plot of the OPLS model obtained with urine from control mice (blue dots) and exercised mice (red dots). b OPLS loading plot. Variations of metabolites are represented using a line plot between 0.5 and 7 ppm. Positive signals correspond to metabolites at increased concentrations in urine of mice after exercise. Conversely, negative signals correspond to metabolites present at increased concentrations in urine of control mice. The colours correspond to the r correlation coefficient. The buckets are labelled according to metabolite assignments presented in Table 2
Metabolome as determined by time limit of the exhaustive exercise
The OPLS models were computed with the time limit taken as the Y matrix. In the case of blood samples, the model cannot significantly discriminate the different times. Whereas, with urine samples, this Y matrix with time limit achieved the best model with R2 = 0.9 and Q2 = 0.625. The score plot is shown in Fig. 2A. The model is validated by the permutation test. The regions of spectra with the highest correlations are coloured in yellow to red in the loading plot of Fig. 2B.
a Score plot of the OPLS model obtained with the time limit (in minutes, coloured scale). b OPLS loading plot. Variations of metabolites are represented using a line plot between 0.5 and 6 ppm. Positive signals correspond to metabolites at increased concentrations in urine of mice with the longest time limit. Conversely, negative signals correspond to metabolites present at increased concentrations in urine of control mice and short time limit. The colours correspond to the r correlation coefficient. The buckets are labelled according to metabolite assignments presented in Table 3
Several regions are highly correlated to the model (Table 3) corresponding to metabolites previously reported in human and murine urine samples such as allantoin (5.40 ppm), glutamine (2.30 ppm with corresponding correlated signals in 2D spectrum) or branched-chain amino acids and their metabolites (below 1 ppm). At 3.31 ppm, a triplet cannot be attributed. It may correspond to inositol, but the correlations cannot be retrieved in 2D COSY spectrum.
Discussion
This study was conducted to discriminate the metabolomics consequences of different types of acute exercise. The exercise load was adapted by measuring the Vc and Vp for each animal. The exercise was performed at a velocity chosen according to individual performances. The results show that exercise modulates the urine metabolome when control mice are compared to exercised ones. As mice ran until exhaustion, the time limit is also an individual parameter which is related to urine metabolome.
To obtain these discriminations with a mouse model, the metabolomic investigation was performed with urine and with deproteinized blood. The statistical analyses show that the discrimination is obtained with urine and not with the blood-derived samples. Urine is the biofluid directly issued from kidney functioning (filtration, secretion and reabsorption). Consequently, urine contains water-soluble metabolites produced in excess during physiological or pathological processes. Therefore, it may be that high amounts of metabolites produced during exercise are detected preferentially in urine rather than in blood, considering the high homeostasis in the latter biofluid.
The use of deproteinized blood presents the advantage of eliminating the broad signals of large molecules in NMR spectra. The large humps in spectra produced by bulk proteins and lipoproteins decrease the detection threshold of signals arising from low concentration species. This is particularly critical when signals are split into several multiplets in cases such as glutamine or leucine and isoleucine. Nevertheless, it has to be considered that in these samples, the metabolites may arise from plasma as well as from the intracellular compartment of blood. Deproteinized blood was previously used in metabolomic studies in relation with physical exercise [22]. The ultrafiltration method used in this previous study could not be implemented with the reduced sample volume collected in mice (less than 1 mL for whole blood, most of the time). The TCA precipitation method allowed the recovery of a larger sample volume than the ultracentrifugation technique with the cost of a 10 % dilution with the TCA solution. NMR tubes have to be completed to 700 μL, introducing another dilution of samples. For this reason, the probabilistic quotient normalisation [19] was preferred to the overall integral normalisation most frequently used. The method was described as more robust to compensate the dilution effects in urine. Consequently, probabilistic quotient normalisation is the most appropriated method to normalise the dilution effect produced by volume compensation with deuterium oxide.
This normalisation was also necessary for urine samples. As all urine samples, the metabolite concentration varies according to the renal function and/or water uptake. In addition, in the present study, the small volume of urine collected had to be compensated with deuterium oxide addition. Moreover, several urine samples contain high amounts of 3-hydroxybutyrate which resonances at 1.18 ppm and 4.01 ppm dominate the spectra. The relative normalisation of the buckets through the total integration ratio produces a relative emphasis of this ketone body metabolite for the samples containing a high level of 3-hydroxybutyrate. For this reason, the probabilistic quotient normalisation was also preferred for urine.
In NMR spectra, the chemical shifts of metabolite resonances are modulated by variations of pH, concentrations of salts and ions with magnetic effects. In the samples used here, these effects are also enhanced by the different dilution with varying amounts of deuterium oxide. In the urine sample, the chemical shift variations required the peaks' realignment. Therefore, the icoshift method was used. Whereas, OPLS models were also calculated before the peak alignment, and similar correlation and predictive coefficients were obtained, the loading plots presented unphased loads, misleading the interpretation of the variation. These models were finally not used. With aligned spectra, the buckets belonging to identical multiplets are clearly phased in the loading plot. In deproteinized blood, the chemical shifts were not varying as much as in urine, probably because the acidic treatment imposed a low pH to all samples. Nevertheless, a similar realignment process was used to maintain identical processing methods for both types of samples. Finally, with these post-acquisition processing, the data were easier to interpret for blood and urine spectra.
Those spectral post-acquisition manipulations produce straightforward interpretable loading plots as presented in Figs. 1 and 2. The intensity of the loads is the co-variance of the two matrixes, and this shows that the highest signals in spectra are those used for model calculation such as 3-hydroxybutyrate, oxo-glutarate, citrate and taurine. Nevertheless, those metabolites are not those with the highest correlations when groups or time limit are compared. The peaks correlated with the Y matrix correspond to lower-intensity signals in the spectra. It is also noteworthy that opposite to what is frequently described for humans, lactate signal is not a predominant resonance in the urine and blood spectra. Moreover, this metabolite is not found increased after exercise. This absence of lactate accumulation (in blood) or elimination (in urine) signifies the divergent metabolic pathways involved when acute exhaustive exercise is imposed to mice compared to what occurs in humans. Nevertheless, the delay (2 h) between the end of the exhaustive exercise and the blood sampling may be another reason.
The computed OPLS models obtained here showed that the determinant parameter to modulate the metabolome is rather the duration of exercise than the velocity itself. This phenomenon may be related to the variability of physical performance in both groups as reported in Table 1. The velocities imposed to the mice, Vc and Vp, are, on the whole, rather close as Vp is 120 % of Vc. Therefore, both exercises may be considered as acute exercise at exhaustion.
Some of the metabolites were correlated in both computed OPLS models. This is the case of allantoin and inosine, which are increased in exercising animal urine when compared to control ones, and this increase is related to the time limit. Allantoin is a product of the purine catabolism formed in the liver, lung, spleen or kidney. It has been reported to be increased after exercise in urine of animals and humans as an effect of oxidative metabolism of adenine nucleotides. Allantoin excretion is increased by physical exercise in human blood [23, 24]. The murine models are able to produce this catabolite, while in human urine, this catabolite has been investigated in relation with other markers of oxidative stress [24]. In the exercise until exhaustion used here with mice, this metabolite elimination is correlated to the time limit, suggesting that the oxidative metabolism is involved in the exhaustion attained by mice. Inosine is also participating in the purine metabolic pathway and is also elevated in urine with increased length of exercise. The increase of this metabolite could be found in the region around 3.9 ppm in the spectra, but the other resonances of aromatic proton (8.2 and 8.3 ppm) are not correlated with time limit, neither are those of aromatic protons of hypoxanthine. In other studies concerning human urine after acute exercise [3, 21, 25], hypoxanthine was a more sensitive metabolite to exercise than inosine.
For both OPLS models, the presence of glutarate is also correlated with exercise and the time limit. In studies of physical exercise, glutarate is not usually reported as a metabolic signature whereas its oxidised metabolite, oxo-glutarate, participating in the Krebs cycle is frequently modulated with exercise [26, 27]. In the case of the animal model used here, despite a high level of oxo-glutarate in urine, the reduced metabolite, glutarate, is found higher after exercise, most likely showing a different turnover of the oxidative metabolism in mice. Malonic acid is another carboxylic acid found increased in both models under the effect of exercise. Malonic acid in urine is usually described in human urine in the case of genetic metabolic disease [28] with an impaired energy metabolism.
When comparing the exercised mice to control ones, the elimination of fumarate was increased. Fumarate is participating in the oxidative metabolism as included in the Krebs cycle. A higher elimination shows the modulation of this metabolic pathway with exercise but without effect of the time limit. In relation to the oxidative metabolism, the time limit was correlated with the increased elimination of the branched-chain amino acids (BCAA: valine, leucine and isoleucine) and their oxidised metabolites such as oxo-isovalerate. The BCAA provide substrate for the Krebs cycle through the oxidation of glutamate into oxo-glutarate. These compounds are produced by proteolysis and especially from the muscle proteins. Intermediate metabolites in this metabolic pathway are oxo-glutarate and glutamate, the latter being increased with exercise duration. Their increase with the duration of exercise proves their accumulation by protein breakdown during exercise. The involvement of protein catabolism in relation with exercise duration highlights the protein metabolism involved with long-term exercise in mice while the lipid catabolism markers are not increased after exercise. The lipid metabolites namely the ketone bodies found in urine, 3-hydroxybutyrate, acetate and acetoacetate, were not discriminating variables. The glycerol resonances identified in spectra at 3. 75 and 4.1 ppm may be those included in several lipid metabolites. This is the only lipid metabolite increased after exercise.
Most of the metabolomic investigations about exercise effects are performed on plasma or serum. Two studies were performed with human subject urine, showing that interval training protocols with different resting intervals [21] or with trained or untrained subjects [3] produced distinct metabolomic profiles. This is an important matter in transferring the urine metabolomic investigation to humans considering that sampling urine is less invasive than sampling blood. In this animal model study, both biofluids could be sampled. The results confirm that NMR metabolomics performed on urine samples is sensitive to metabolic modulation after exercise.
Physical exercise was suggested as a therapeutic tool for several chronic pathologies including cardiac pathology, obesity or diabetes. In these cases, the intensity of exercise to performed is a major point to assess the efficiency without risk or inconvenience for the patients [9]. The specificity of the physical activity may be crucial to modify the physiological and biological profiles of patients. On the other hand, the metabolomic effects of exercise revealed metabolic dysfunctioning in muscle tissue for patients with chronic obstructive pulmonary disease (COPD) [29]. These results were obtained in clinical studies. The investigation of the biological process underlying the effects and/or consequences of a physical exercise necessitates an integrative approach. Integrative biology supposes that several types of biofluids and tissues are available which is easier in animal models than for humans for ethical reason. As an example, with mice undergoing physical exercise, blood and urine may be collected for metabolomic studies as well as tissue samples such as skeletal muscles for molecular biology and biochemistry assays.
On the whole, the metabolites detected in mice urine are very similar to those detected in human urine by Pechlivanis et al. [21]. However, the modulation of the metabolite levels in urine found in humans is different from those found for our mice model. Moreover, the type of exercise imposed in Pechlivanis’ study was an intermittent acute training, and the length of intervals between acute exercise sessions had consequences on the urine metabolome of subjects. In the case of the present study, we prove that the duration of one acute exercise sustained by mice can affect urine metabolome. The metabolic pathways used in this animal model obviously differ from those for humans. Nevertheless, animal models are necessary to investigate the biological process of physical training. For this reason, the pre-clinical studies may be widely informative about the biological consequences of exercise, in particular with an integrative point of view.
Conclusion
The use of an animal model shows that an acute physical exercise sustained until exhaustion is able to modify the metabolome when compared to control mice. This metabolomic study proves that analysis of urine is more discriminating than the analysis of deproteinized blood.
Moreover, the exercise velocity sustained by each animal is adapted to its own capacity. With this individual-adapted exercise protocol, metabolomics demonstrated that metabolic adaptations are depending on the exercise duration rather than the velocity.
These results were obtained with a specific processing method including spectral alignment and probabilistic quotient normalisation before the multivariate statistical analysis. Consequently, our report shows that mice models may be important for investigation of exercise protocols designed to overcome chronic pathologies. The pre-clinical models allow a complete integrative investigation from physiological to metabolic experimentation before the transfer in human clinical cases. The results obtained on urine samples are encouraging to pursue in this direction as urine samples are non-invasively and easily obtainable.
References
Nicholson JK, Lindon JC (2008) Systems biology: metabonomics. Nature 455(7216):1054–1056. doi:10.1038/4551054a
Pan Z, Raftery D (2007) Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem 387(2):525–527. doi:10.1007/s00216-006-0687-8
Enea C, Seguin F, Petitpas-Mulliez J, Boildieu N, Boisseau N, Delpech N, Diaz V, Eugene M, Dugue B (2010) (1)H NMR-based metabolomics approach for exploring urinary metabolome modifications after acute and chronic physical exercise. Anal Bioanal Chem 396(3):1167–1176. doi:10.1007/s00216-009-3289-4
Lehmann R, Zhao X, Weigert C, Simon P, Fehrenbach E, Fritsche J, Machann J, Schick F, Wang J, Hoene M, Schleicher ED, Haring HU, Xu G, Niess AM (2010) Medium chain acylcarnitines dominate the metabolite pattern in humans under moderate intensity exercise and support lipid oxidation. PLoS One 5(7):e11519. doi:10.1371/journal.pone.0011519
Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE (2010) Metabolic signatures of exercise in human plasma. Sci Transl Med 2(33):33ra37. doi:10.1126/scitranslmed.3001006
Yan B, Jiye A, Wang G, Lu H, Huang X, Liu Y, Zha W, Hao H, Zhang Y, Liu L, Gu S, Huang Q, Zheng Y, Sun J (2009) Metabolomic investigation into variation of endogenous metabolites in professional athletes subject to strength-endurance training. J Appl Physiol 106(2):531–538. doi:10.1152/japplphysiol.90816.2008
Duggan GE, Hittel DS, Sensen CW, Weljie AM, Vogel HJ, Shearer J (2011) Metabolomic response to exercise training in lean and diet-induced obese mice. J Appl Physiol 110(5):1311–1318. doi:10.1152/japplphysiol.00701.2010
Huang CC, Lin WT, Hsu FL, Tsai PW, Hou CC (2010) Metabolomics investigation of exercise-modulated changes in metabolism in rat liver after exhaustive and endurance exercises. Eur J Appl Physiol 108(3):557–566. doi:10.1007/s00421-009-1247-7
Barba I, de Leon G, Martin E, Cuevas A, Aguade S, Candell-Riera J, Barrabes JA, Garcia-Dorado D (2008) Nuclear magnetic resonance-based metabolomics predicts exercise-induced ischemia in patients with suspected coronary artery disease. Magn Reson Med 60(1):27–32. doi:10.1002/mrm.21632
Hill DW, Poole DC, Smith JC (2002) The relationship between power and the time to achieve.VO(2max). Med Sci Sports Exerc 34(4):709–714
Froelicher VF Jr, Brammell H, Davis G, Noguera I, Stewart A, Lancaster MC (1974) A comparison of the reproducibility and physiologic response to three maximal treadmill exercise protocols. Chest 65(5):512–517
Taylor HL, Buskirk E, Henschel A (1955) Maximal oxygen intake as an objective measure of cardio-respiratory performance. J Appl Physiol 8(1):73–80
Day JR, Rossiter HB, Coats EM, Skasick A, Whipp BJ (2003) The maximally attainable VO2 during exercise in humans: the peak vs. maximum issue. J Appl Physiol 95(5):1901–1907. doi:10.1152/japplphysiol.00024.2003
Ferreira JC, Rolim NP, Bartholomeu JB, Gobatto CA, Kokubun E, Brum PC (2007) Maximal lactate steady state in running mice: effect of exercise training. Clin Exp Pharmacol Physiol 34(8):760–765. doi:10.1111/j.1440-1681.2007.04635.x
Moritani TA, Nagata HA, deVries HA, Muro M (1981) Critical power as measure of physical work capacity and anaerobic threshold. Ergonomics 24:339–350
Jones AM, Vanhatalo A, Burnley M, Morton RH, Poole DC (2010) Critical power: implications for determination of VO2max and exercise tolerance. Med Sci Sports Exerc 42(10):1876–1890. doi:10.1249/MSS.0b013e3181d9cf7f
Billat VL, Mouisel E, Roblot N, Melki J (2005) Inter- and intrastrain variation in mouse critical running speed. J Appl Physiol 98(4):1258–1263. doi:10.1152/japplphysiol.00991.2004
Savorani F, Tomasi G, Engelsen SB (2010) icoshift: a versatile tool for the rapid alignment of 1D NMR spectra. J Magn Reson 202(2):190–202. doi:10.1016/j.jmr.2009.11.012
Dieterle F, Ross A, Schlotterbeck G, Senn H (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 78(13):4281–4290. doi:10.1021/ac051632c
Lerman I, Harrison BC, Freeman K, Hewett TE, Allen DL, Robbins J, Leinwand LA (2002) Genetic variability in forced and voluntary endurance exercise performance in seven inbred mouse strains. J Appl Physiol 92(6):2245–2255. doi:10.1152/japplphysiol.01045.2001
Pechlivanis A, Kostidis S, Saraslanidis P, Petridou A, Tsalis G, Mougios V, Gika HG, Mikros E, Theodoridis GA (2010) (1)H NMR-based metabonomic investigation of the effect of two different exercise sessions on the metabolic fingerprint of human urine. J Proteome Res 9(12):6405–6416. doi:10.1021/pr100684t
Miccheli A, Marini F, Capuani G, Miccheli AT, Delfini M, Di Cocco ME, Puccetti C, Paci M, Rizzo M, Spataro A (2009) The influence of a sports drink on the postexercise metabolism of elite athletes as investigated by NMR-based metabolomics. J Am Coll Nutr 28(5):553–564
Mikami T, Kitagawa J (2006) Intense exercise induces the degradation of adenine nucleotide and purine nucleotide synthesis via de novo pathway in the rat liver. Eur J Appl Physiol 96(5):543–550. doi:10.1007/s00421-005-0106-4
Shing CM, Peake JM, Ahern SM, Strobel NA, Wilson G, Jenkins DG, Coombes JS (2007) The effect of consecutive days of exercise on markers of oxidative stress. Appl Physiol Nutr Metab 32(4):677–685. doi:10.1139/H07-051
Stathis CG, Carey MF, Hayes A, Garnham AP, Snow RJ (2006) Sprint training reduces urinary purine loss following intense exercise in humans. Appl Physiol Nutr Metab 31(6):702–708. doi:10.1139/h06-074
Almstetter MF, Appel IJ, Gruber MA, Lottaz C, Timischl B, Spang R, Dettmer K, Oefner PJ (2009) Integrative normalization and comparative analysis for metabolic fingerprinting by comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. Anal Chem 81(14):5731–5739. doi:10.1021/ac900528b
Yuan J, Doucette CD, Fowler WU, Feng XJ, Piazza M, Rabitz HA, Wingreen NS, Rabinowitz JD (2009) Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol Syst Biol 5:302. doi:10.1038/msb.2009.60
Wightman PJ, Santer R, Ribes A, Dougherty F, McGill N, Thorburn DR, FitzPatrick DR (2003) MLYCD mutation analysis: evidence for protein mistargeting as a cause of MLYCD deficiency. Hum Mutat 22(4):288–300. doi:10.1002/humu.10264
Rodríguez D, Alcarraz-Vizán G, Díaz-Moralli S, Reed M, Gómez F, Falciani F, Günther U, Roca J, Cascante M (2011) Plasma metabolic profile in COPD patients: effects of exercise and endurance training. Metabolomics 8:508–516. doi:10.1007/s11306-011-0336-x
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Le Moyec, L., Mille-Hamard, L., Triba, M.N. et al. NMR metabolomics for assessment of exercise effects with mouse biofluids. Anal Bioanal Chem 404, 593–602 (2012). https://doi.org/10.1007/s00216-012-6165-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-012-6165-6