Abstract
Casein phosphopeptides (CPP) were identified in small amounts in milks heated at various intensities by using matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry. CPP selectively concentrated on hydroxyapatite (HA) were regenerated using phosphoric acid mixed in the matrix. Unphosphorylated peptides not retained by HA were removed by buffer washing. This procedure enhanced the MALDI signals of CPP that are ordinarily suppressed by the co-occurrence of unphosphorylated peptides. CPP, belonging to the β-casein (CN) family, i.e., (f1-29) 4P, (f1-28) 4P, and (f1-27) 4P, and the αs2-CN family, i.e., (f1-21) 4P and (f1-24) 4P, were observed in liquid and powder milk. The lactosylated counterparts were specific to intensely heated milks, but absent in raw and thermized/pasteurized milk. Most CPP with C-terminal lysines probably arose from the activity of plasmin; an enzyme most active in casein hydrolysis. A CPP analogue was used as the internal standard. The raw milk signature peptide β-CN (f1-28) 4P constituted ~4.3% of the total β-CN. Small amounts of lactosylated peptides, which varied with heat treatment intensity, were detected in the milk samples. The limit of detection of ultra-high-temperature milk adjunction in raw or pasteurized milk was ~10%.

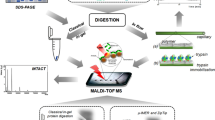
Schematic representation of the procedure for phosphoprotein/phosphopeptide enrichment using hydroxyapatite (HA). Native and lactosylated casein phosphopeptides are captured by HA, while non-phosphorylated peptide was washed out by the loading buffer. Signature peptides of UHT milk are detected through direct analysis by MALDI-TOF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The European Union Directives (92/46 CEE and 94/71 CEE) set specific rules for the production of heat-treated milk and milk-based products such as the use of specific temperature/time combinations for pasteurized and ultra high-temperature (UHT) milk. The use of low value ingredients such as UHT and milk powder in mixtures with raw or pasteurized milks is strictly prohibited. Nowadays, rapid methods are mainly based on the evaluation of furosine, a useful indicator of heat damage in processed milk. Studies of milk adulterations are also focused on lactosylation of milk proteins. Endogenous milk proteinases and those associated with somatic cells in high counts hydrolyze caseins of raw milk. Some heat-stable proteinases may continue to do it during storage of heat-treated milks. The number of somatic cells should not exceed 200,000 cells/mL bulk raw milk [1], although the European Union Directives (92/46 CEE and 94/71 CEE) set a limit of 400,000 cells/mL for SCC for drinking milk. Alternatively, an indirect means to monitor milk quality in relation to SCC would be to know one or more signature peptides. Milk contains colloidal and soluble casein subjected to hydrolysis by proteinases. Casein releases water-soluble peptides under the action of plasmin (EC 3.4.21.7) and lysosomal proteases, such as carboxylprotease and cathepsin D (EC3.4.23.5). While β-casein (CN) and αs2-CN are the preferred substrates of plasmin, αs1-CN [2] and κ-CN [3] are more resistant to enzymatic degradation. Therefore, one would expect to find β- and αs2-CN-derived peptides in milk. Indeed, β-CN is the most hydrolyzed casein fraction partly converted into pH 4.6 insoluble γ-CN and complementary soluble proteose peptones (PP). PP include peptides such as β-CN (f1-105/107) (component PP5), (f1-28) (component PP8-fast), (f29-105/107) 1P (component PP8-slow) [4, 5], and others. Raw milk of good quality has 1–2 mg/mL PP, which can increase during storage. A significant PP5 accumulation has been reported for commercially packaged pasteurized milk [6]. Because as much as 30–40% plasmin survives UHT treatment [7], gelation could be caused for long-stored UHT milk [8].
While the role of proteases in casein proteolysis has been fully clarified, the subsequent lactosylation of water-soluble peptides remains to be clarified. Although determined for whey proteins and casein, lactosylation of milk peptides has not been deeply studied with the objective of distinguishing various types of milk [9]. For heat-treated liquid and powder milk, a reaction could occur between the ε-amino group of protein-bound lysine or amino-termini of peptides and the carbonyl group of lactose. In the initial step of the Maillard reaction, lactosylation occurs both during heating and, to a lesser extent, during milk powder storage [10]. The degree of lactosylation of β-CN increases steadily with temperature between 37 °C and 60 °C [11]. Lactose specifically binds to Lys-34 in αs1-CN and to Lys-107 in β-CN under moderate heat treatment conditions (72–85 °C for 15–30 s). The number of binding sites increases to seven for αs1-CN and five for β-CN after intensive treatment (142–145 °C for 2–5 min) [12]. The extent of lactosylation measured under the same conditions is 25% for β-lactoglobulins (β-Lg) and 35% for β-CN [11]. Especially in drinking milk, but also in milk powders, milk quality is greatly affected by the initial proteolysis of raw milk. Due to the specificity of plasmin, which acts preferentially on Lys-X bonds, milk heating induces lactosylation of the released peptide at basic C-terminal residues (mainly Lys or Arg). Even though the extent of lactosylation affects drinking milks differently, little is known about the impact of Maillard reactions on the formation of CPP markers.
Several milk peptides have been shown to derive from the proteolytic digestion of caseins [13]. Separation of signature peptides from other peptides may often be achieved by chemical methods. Thus, to reduce the complexity of the peptide fraction, the chromatography on hydroxyapatite (HA) has been recently introduced [14]. This method achieves the recovery of casein phosphopeptides (CPP) from complex peptide mixtures [14, 15] as an alternative to immobilized metal affinity chromatography (IMAC)-based methods [16, 17] and other affinity-based methods [18]. While unphosphorylated peptides are washed out, CPP were retained on microgranules of HA. This was useful for isolating CPP free of unphosphorylated peptides. A HA–CPP complex was then spotted onto a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) target plate for analysis [14]. Due to plethora of milk peptides, we focused on signature CPP released in definite number by milk proteinase(s). The efficient separation of the unphosphorylated peptides also enhanced the MALDI signals of CPP. The research results could demonstrate the nature and activity of milk proteinases and provide information about the proteolysis induced thereof. We were interested to detect heated milk and quantify peptides which undergo lactosylated once released by proteinases. By direct MALDI analysis, in situ immobilized CPP, both native and lactosylated, can be identified. Similarly, lactosylation sites can be localized on peptide sequence by post-source decay (PSD) fragmentation. The comparison between the potentially lactosylable and actually lactosylated sites could serve to differentiate processed milks according to heat intensity. The lactosylated CPP could serve to discriminate raw and pasteurized from UHT milk. In this work, we have realized a procedure for detecting UHT milk in amounts not lower than 10% spiking pasteurized milk. We suggest extending this approach to the analysis of any dairy products with suspected addition of UHT milk, milk protein, or milk powder.
Experimental
Chemicals
HA (Macro-Prep Ceramic Hydroxyapatite Type I) was supplied by Bio-Rad (Milan, Italy). Tris(hydroxymethyl)aminomethane hydrochloride (Tris–HCl), potassium chloride (KCl), urea, trifluoroacetic acid (TFA), acetonitrile (ACN) for HPLC, 85% orthophosphoric acid (PA), and AMBIC were from Carlo Erba (Milan, Italy). Dithiothreitol (DTT) was from AppliChem (Darmstadt, Germany). TPCK-treated trypsin from bovine pancreas was from Sigma (St. Louis, MO, USA). Sinapinic acid (SA), 2,5-dihydroxybenzoic acid (DHB), and sodium acetate (AcNa) trihydrate were obtained from Fluka (St. Louis, MO, USA). Acetic acid was purchased from Baker Chemicals B.V. (Deventer, the Netherlands). Water was prepared using a Milli-Q system (Millipore, Bedford, MA, USA). Pooled raw milk was from local dairy farms. The pasteurized UHT milks were purchased from a local store. Milk protein powder and milk powder were supplied by Sacco Industry (Milan, Italy).
Sample preparation
Pasteurized milk was added with UHT milk to a final concentration of 90% to 1%. Then, each sample, including isoelectric casein (sample I), milk protein powder (sample V), or milk powder (sample VI) in solution, raw milk (sample II), pasteurized milk (sample III), and UHT milk (sample IV), was treated with HA as described below. The chemical composition of milk samples as well as the time/temperature combinations are reported in Table 1.
HA-based phosphoprotein/CPP enrichment from various milk samples
HA (100 mg), previously equilibrated with the loading buffer (50 mM Tris–HCl, 0.2 M KCl, 4.5 M urea, and 10 mM DTT, pH 8.0), was put in contact with protein in solution (10 mg) or equivalent amounts of raw and skimmed milk. The HA-bound proteins were incubated for 15 min at room temperature and centrifuged for 5 min at 4,000×g per minute. The resin was successively washed with three different buffers: loading buffer (1 mL), 50 mM Tris–HCl at pH 8.0 (1 mL), and 20 mM Tris–HCl in 20% ACN (v/v) at pH 8.0 (1 mL). The resin was washed with Milli-Q water (1 mL) and freeze-dried with a SpeedVac concentrator system (Thermo Electron, Milford, MA).
In order to confirm the degree of lactosylation, the dried HA-bound CPP (1 mg) were dissolved in a 5% aqueous PA solution (120 μL) and desalted with a ZipTip C18 pipette tip before dephosphorylation with alkaline phosphatase (ALP), carried out according to the procedure previously described [14]. Assays were needed especially for CPP isolated from milk protein and milk powder. However, since HA is known to interact with phosphoprotein and CPP, the aqueous suspension of HA was trypsinized according to the procedure previously described [14]. The HA-bound tryptic CPP were then analyzed by MALDI.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
MALDI-TOF mass spectra were recorded using a Voyager DE-PRO mass spectrometer (Applied Biosystems, Framingham, MA, USA) in positive linear mode. All spectra were acquired in the range of 10–30 kDa (for proteins) and 1–5 kDa (for peptides) with the following settings: an accelerating voltage of 25 kV (for proteins) or 20 kV (for peptides) and a grid voltage of 93% (for proteins) or 95% (for peptides) of the accelerating voltage, a guide wire of 0.15% (for proteins) or 0.05% (for peptides), and a delayed ion extraction time of 485 ns (for proteins) or 175 ns (for peptides). The laser power was set just above the ion generation threshold to obtain peaks with the highest possible signal-to-noise ratio. All spectra were acquired with 200 shots in three replicates. Measured molecular masses, with a ±0.5-Da tolerance, matching the calculated masses, enabled the identification of ion signals. Different MALDI matrix solutions were freshly made for different types of molecules to be analyzed. For phosphoprotein analysis, 10 mg/mL of SA was dissolved in 50% ACN and 0.1% TFA in an ultrasonic bath. For CPP, 10 mg/mL of DHB was dissolved in 1 mL of H2O/ACN/PA (49:50:1) using an ultrasonic bath. Prior to HA enrichment, the samples were analyzed by MALDI. Solid milk samples (1 mg/mL) were solubilized in H2O/ACN (50:50) solution with 0.1% TFA. Liquid milk samples (10 μL) were diluted 1:100 with the above solution. Each protein solution (1 μL) was loaded in one well of a MALDI plate and covered with SA matrix (1 μL) for the preliminary analysis. After enrichment, ~1,000 HA-phosphoprotein microgranules were deposited onto the MALDI plate and covered with the SA matrix (1 μL) to promote analyte/matrix co-crystallization in the presence of 0.1% TFA. For CPP, 1% PA was included in the matrix solution to achieve maximum ionization efficiency by MALDI and minimize binding of CPP to HA [19].
PSD-MALDI-TOF experiments
PSD fragment ion spectra were acquired after isolation of the appropriate precursor ions using timed ion selection. In this manner, the spectra of lactosylated β-CN(f1-28) 4P (m/z 3,803 Da) and its unphosphorylated counterpart (m/z 3,483 Da) were acquired. Acquisition was performed with the following settings: accelerating voltage, 20 kV; grid voltage, 80% of the accelerating voltage; guide wire, 0.02; and delayed time, 100 ns. Each spectrum was the average of 300 shots in three replicates.
Synthesis of peptide analogues
The peptides were synthesized by solid-phase methods using the 9-fluorenylmethyloxycarbonyl (Fmoc) strategy on a Pioneer peptide synthesizer (Synthesis System 9050 instrument; PE-Biosystems, Framingham, MA, USA). The level of the β-CN (1-28) 4P plasmin-derived fragment was chosen as the index for quantification of casein proteolysis. The method was applied to all of the milks to select a marker specific for intensely heated milks. The natural and the analogue-synthesized peptides (Table 2) were used to generate a calibration curve.
A constant concentration (10 μg/μL) of synthesized modified peptides was used to spike different solutions of synthesized natural peptides to create a calibration curve, with the natural/modified area ratio as a function of the concentration ratio of the corresponding peptides. The amount of β-CN (f1-28) 4P was obtained by spiking 10 μL raw milk with 10 μg/μL internal standard (IS) and calculating the quantity of this peptide according to the equation of the calibration plot.
Results and discussion
Strategy of HA-based enrichment of casein and CPP
Isolation of casein/CPP is one of the most difficult tasks because of the heterogeneity in the number of phosphate groups (P), varying from 1 to 13, attached to specific serine residues of the four casein fractions [20]. Phosphoproteins/peptides compete for the C sites on ceramic HA over a wide pH range [21], and both were eluted for MALDI analysis [15, 22]. Our procedure did not use elution because the HA-phosphocaseins/CPP complex was directly solubilized by spotting on the MALDI plate matrix mixed to TFA (for casein) or PA (for CPP) [14]. In addition to this, PA enhances the detection of phosphopeptide ions by MALDI-TOF [19]. The main objective of our work was to determine whether the HA-based procedure is suitable for characterizing signature peptides of heat-treated milk samples. Because the affinity of CPP for HA could decrease with increasing nonspecific binding of the unphosphorylated peptides, a preliminary study measured the bound and unbound proteins/peptides of raw, pasteurized, UHT, milk protein powder (casein + whey), and milk powder samples. Our results indicate that MALDI is useful in identifying lactosylation, one of the most common posttranslational protein modifications that occur during heating or spray drying. MALDI analysis provided molecular mass value of peptides and information on the presence/absence of signature peptides. Thus, severe heating induced changes of lactosylation in whey proteins of samples I to VI (Electronic supplementary material (ESM) Table S1). The signals of non-enzymatic lactosylation of β-Lg A and B and α-lactalbumin (α-La) B dominated the MALDI-TOF spectra of different samples (ESM Table S1 and Figs. S4 and S6). The molecular masses of β-Lg B (unmodified m/z 18,280.4) and β-Lg A (unmodified m/z 18,365.9) increased by 314.2 Da per lactose molecule or multiple integrals up to 6 (ESM Table S1). The lactosylation of β-Lg A and B compared with control raw milk was higher in powdered milk (sample VI, six lactose residues/molecule) than in protein milk powder (sample V, four lactose residues/molecule) and UHT milk (sample IV, one lactose residue/molecule). Unmodified β-Lg A and B were still present in all samples except milk powder (ESM Table S1 and Fig. S6). α-La B was less lactosylated in the milk protein powder (sample V, three lactose residues/molecule) than in powdered milk (sample VI, four lactose residues/molecule). In UHT milk (sample IV), α-La B (unmodified m/z 1,4187.2) was lactosylated as β-Lg (one lactose residue/α-La molecule). This seems to contrast at least partially with previous findings on casein lactosylation which occurs at similar levels in UHT and in-bottle sterilized milk [12]. Indeed, the extent of lactosylated proteins in pasteurized milk does not differ from that measured in raw milk. In contrast, the different levels of lactosylation of proteins in the powder samples were probably due to the lower lactose content of ultrafiltered milk or whey. It seems that heating extensively enhanced lactosylation by exposing lysine residues on the surface of the proteins. However, Maillard reaction was incomplete even in the powdered milk preparations. In recent studies, Lys-47, Lys-138, and Lys-141 of β-Lg were lactosylated, while Lys-98, Lys-114, or Lys-122 of α-La were lactosylated in UHT milk [23]. In general, the degree of lactosylation increases steadily with temperature, progressively from pasteurization to spray drying passing through to UHT. However, the Amadori compound, the first intermediate in the Maillard reactions, is probably degraded by heat [24] via the Strecker degradation [25]. Assessment of the whey protein powder quality in infant formula on the basis of the number of lactose residues per protein molecule can detect five lactoses/α-La and ten lactoses/β-Lg [26]. In the solid state, all 15 lysine residues of β-Lg and the N-terminal leucine were lactosylated [27], in addition to Arg-124, for a total of 17 lactosylated amino acid residues [28].
While the effect of heat treatment on the whey protein denaturation has been well established by many decades of research, little is known about casein lactosylation. By applying our procedure, lactosylated casein would be detected in heated milk (ESM Fig. S4 and S6). All of the MALDI spectra for the milk samples actually contained the signals of (1) unmodified κ-CN or the mono-lactosylated component but milk powder (ESM Table S1 and Fig. S6); (2) multi-phosphorylated αs2-CN after enrichment of isoelectric casein on HA; (3) lactosylated β-CN A1 and A2, which differed based on His67(β-CN A1) for Pro67(β-CN A2); and (4) diffusely lactosylated β-CN and αs1-CN in milk protein (V) and milk powder (VI) samples due to the low resolution of MALDI (ESM Table S1 and Figs. S4 and S6). However, monomer κ-CN was missing in the milk powder sample because of its possible association with whey proteins. On the whole, HA procedure provides a system for the selective enrichment of caseins/CPP while whey proteins are washed away (ESM Table S1 and Fig. S8 and S9 and S10).
Identification of HA-bound CPP and unphosphorylated peptides
To follow the evolution of the peptide profile according to the heating intensity, the MALDI-TOF spectra were explored in the 10- to 14-kDa range. After enrichment on HA, raw (Fig. 1b and ESM Table S2) and pasteurized milk (ESM Fig. S11) had dominant signals of β-CN (f1-97) 5P (11,362.3 Da) and αs2-CN(f53-150) 6P (11,860.4 Da). The impact of UHT treatment and of milk and milk protein drying seemed higher than in pasteurized milk (ESM Fig. S11).
MALDI-TOF spectra of raw milk peptides in the 10- to 14-kDa mass range before (a) and after enrichment (b) using HA as concentrating probe. Component identification is reported in ESM Table S2
The MALDI signals of unphosphorylated peptides significantly decreased, while those of CPP considerably increased. The results indicate that (1) a selective enrichment occurred as a result of displacement of CPP bound to HA by the addition of TFA; (2) the CPP enrichment reduced the sample complexity; and (3) low-abundance CPP in mixtures with unphosphorylated peptides can be detected directly “on-beads.” This is confirmed by unphosphorylated γ3- and γ2-CN dominating the spectra of the raw milk (Fig. 1a) whose signals were lost during the repeated buffer washes, while β-CN (1-105) 5P and β-CN (1-107) 5P (complementary to γ2-CN and γ3-CN, respectively; Fig. 1b) were detected in HA-bound CPP. Taken together, these results demonstrate the outstanding enrichment capacity of HA for MS analysis.
MALDI-TOF in the 1.2- to 5-kDa range
Raw, pasteurized, UHT, milk protein, and milk powder samples were examined by MALDI-TOF in the 1.2- to 5-kDa range. As an example, the MALDI spectrum for raw milk is shown in Fig. 2a (ESM Table S3). Here was registered a substantial increase of peptides belonging to the β-CN, αs2-CN and αs1-CN family. This occurred probably because of the presence of active plasmin and somatic cell proteinases. After enrichment on HA, only phosphorylated peptides were detected, as shown in Fig. 2b (ESM Table S3).
MALDI-TOF spectra of raw milk peptides in the 1- to 5-kDa mass range before (a) and after enrichment (b) using HA as concentrating probe. Component identification is reported in ESM Table S3
Similarly, the major CPP were identified in the other commercial milk samples (ESM Fig. S12). In all the milk samples, both native and lactosylated forms of CPP were detected, except in raw and pasteurized milk. β-CN (f1-28) 4P, (f1-27) 4P, and αs2-CN (f1-24) 4P were the main components of CPP (Fig. 3 and ESM Table S4). The native and lactosylated αs2-CN (f1-21) 4P co-existed in milk powder. In this manner, we identified the lactosylated β-CN (f1-29) 4P as a signature peptide of milk proteins and powdered milk preparations because it was missing in other types of milk. It was missing in raw, pasteurized (71.7 °C for 15 s; Fig. 3a), and intensely pasteurized milk (121 °C for 2–4 s, spectrum not shown). αs1-CN-derived CPP failed to form due to the internal location of the phosphorylation cluster. In contrast, they are released in long-ripened cheese [29].
Partial view of MALDI spectra for peptides of pasteurized milk (a), UHT milk (b), milk protein powder (c), and milk powder (d) in the 1- to 5-kDa mass range after HA enrichment. Lactosylated β-CN-derived casein phosphopeptides (CPP) mass signals were magnified and circled in red. Component identification is reported in ESM Table S4
To screen lactosylated CPP, in vitro dephosphorylation was carried out using ALP. The difference between the native and dephosphorylated peptide mass values allowed us to assign the phosphate groups correctly (one phosphate group = 80 Da). The results of CPP dephosphorylation are shown in Table 3 (Fig. 4). Each peptide was a mono-lactosylated component. The degrees of phosphorylation and lactosylation of β-CN (f1-28) 4P were further confirmed by PSD MS experiments (Fig. 5).
An example of determination of phosphorylation stoichiometry of lactosylated CPP by combining alkaline phosphatase-based dephosphorylation with peptide identification by mass spectrometry. By comparing the MALDI spectra recorded for the phosphatase-treated to the untreated sample in Fig. 3, the candidate CPP were identified. Because of 320 Da mass shift (1 phosphate group=80 Da) each peptide, indicated with a m/z value in the MALDI spectra, had four phosphorylation sites
Post-source decay (PSD)-MALDI spectrum of a HA-bound CPP occurring in the sample of milk powder. The red circled peak represents a peptide exhibiting a neutral loss of 340 Da (a lactosyl group) that was one of the most intense ions, and 392 Da for phosphoric acid (1P= 98 Da) from the precursor ion at m/z 3,802.9 Da. These structural informations allowed to assign four phosphates and one lactose group unequivocally to the peptide β-CN (f1-28) at m/z 3,803 Da that was present in the milk powder sample
The PSD spectrum in Fig. 5 exhibited a prominent neutral loss of 340 Da (one lactosyl group) from the precursor ion at m/z 3,802.9 Da. Moreover, the PSD spectrum of lactosylated β-CN (f1-28) 4P yielded a neutral loss of 98 Da (H3PO4), consistent with a peptide containing four phosphoserine residues (Fig. 5). The PSD spectrum of lactosylated β-CN (f1-28) 0P confirmed the loss of one lactose group (m/z 3,140.2 Da) from the peptide (Fig. 6).
In vitro trypsinolysis of HA-bound CPP or caseins from milk protein powder
Trypsinolysis of the HA-bound CPP in addition to the native β-CN (f1-25) 4P and β-CN (f1-25) 3P (3,123.0 and 3.043.6 Da) released mono-lactosylated counterparts (3,447.2 and 3,367.3 Da).
Notwithstanding the fact that Arg25-X and Lys28-X were evenly susceptible to trypsin, Lys28-X was split by plasmin because of the strict specificity of enzyme towards lysine [30]. It is likely that lactosylation especially affects plasmin-mediated peptides [31]. Heating produced a consistent amount of lactosylated peptides. To observe this effect, a shift of +324 Da was expected for peptides containing a reactive NH2 group, yielding the so-called Amadori product. If Arg25 were lactosylated, Arg25-X as well as Lys28-X would be trypsin- and plasmin-resistant. Indeed, an amount of N-terminally mono-lactosylated β-CN (f1-28) 4P corresponding to ~3% total β-CN (f1-28) 4P was found in the milk protein powder sample. In contrast, a higher quantity of mono-lactosylated β-CN (f1-25) 4P, ~17% of the total β-CN, was released by trypsinolysis of HA-bound casein/CPP. This means that both lactosylated caseins and lactosylated CPP combine to determine the levels of signature peptides.
Such peptides were found in dried milks before digestion with trypsin.
Quantification of β-CN (f1-28) 4P in raw milk by MALDI-TOF-MS
Based on a set of naturally occurring peptides, β-CN (f1-28) 4P was chosen because it is a phosphorylated member of a plasmin-mediated CPP family that undergoes glycosylation during heat treatment. Specifically, for the absolute quantification of the mono-lactosylated β-CN (f1-28) 4P signature peptide, a synthetic 25-residue analogue that was identical to the N-terminal 25-mer peptide except for the amino acid substitution Leu3→Trp3 was used as IS. The natural proteotypic peptide was mixed with IS in known concentrations, and the MALDI intensity of the two co-occurring signals was measured (ESM Figs. S14 and S15). MALDI measurements performed on four binary solutions containing natural peptide/IS ratios of 0.22:1, 0.11:1, 0.055:1, and 0.0275:1 produced a line defined by the equation y = 6.2451x + 0.3635 (R 2 = 0.9855, mean of ten replicates; ESM Fig. S16). Using this approach, raw milk β-CN was found to have released ~4.3% of the β-CN (f1-28) 4P (ESM Fig. S17).
Specific detection of lactosylated milk CPP
Raw milk is rejected by dairy processing plants if the sample is found to be contaminated with UHT or other heat-treated milk. The HA-based method could ensure that UHT milk and sterilized milk are not used for the production of pasteurized milk. To demonstrate this, aliquots of pasteurized milk were artificially spiked with 90%, 70%, 50%, 30%, 10%, 5%, or 1% UHT milk. The limit of detection (LOD) was determined by MALDI searching for the lactosylated β-CN (f1-28) 4P, assumed as the signature peptide of UHT milk. The method did not discriminate UHT milk in amounts below 10% (Fig. 7).
Using improved MS equipment, in terms of mass resolution and sensitivity, we think that the LOD could be substantially lowered. Trypsinolysis of HA-bound CPP, enhancing the signal of lactosylated β-CN (f1-25) 4P, could further lower the LOD value.
Sequences of cleaved peptides
To verify the role played by proteolytic enzymes in casein hydrolysis, the amino acid sequence of CPP was investigated (Fig. 2b). The in vivo and in vitro plasmin-mediated peptides that arose from raw milk were compared (ESM Fig. S18 and Table S5).
The choice of plasmin as the proteolytic enzyme was dictated by the role played by this enzyme during milk storage. Because peptide bonds containing a C-terminal Lys or Arg can be hydrolyzed by plasmin, the majority of β- and αs2-CN CPP reported in ESM Table S3 must have been released by plasmin. This agrees with the known activities of plasmin and other proteases associated with milk somatic cells. CPP differed only in the level of lactosylation, which was absent in raw milk but extensive in milk powder. Thus, lactosylated CPP may serve as suitable chemical markers of the intensity of heat treatment. When CPP levels are higher than normal, milk is of poor quality because it is rich in proteolytic enzymes. It seems that the level of lactosylation in milk is related to the intensity of the heating process.
Disadvantages and advantages of the HA procedure
Current techniques use gel electrophoresis and immunoblotting for separating and detecting specifically proteins. Antibodies developed against the Amadori compound containing lactose as a glycating agent were used as a surrogate for measuring furosine, distinguishing the lactosylated from non-lactosylated caseins [32]. In general, anti-peptide or monoclonal antibodies are specific reagents for measuring lactosylated peptides [32]. The drawbacks of polyclonal anti-peptide antibodies are mainly (1) the amino acid sequences of the peptides would be known in advance to target their lactosylated proteins; (2) the antibodies might not recognize native proteins; and (3) a cross-reaction with other members of the protein families would not be excluded a priori. On other hand, concentration of low-abundance peptides/proteins by gel electrophoresis carries the risk that water-soluble peptides escape to the staining procedure with Coomassie Blue. Separation of proteins by 2DE coupled with identification of tryptic peptides through offline MALDI-TOF-MS is one of the most commonly used techniques in proteomic analysis [33]. Cumbersome 2D electrophoresis for protein separations requires extensive manual manipulation for mostly qualitative experiments. Online HPLC coupled with mass spectrometry have been widely used for concentrating/identifying low-abundance peptides/proteins. However, one sample at a time is evaluated by HPLC, so samples are analyzed multiple times during a multi-sample sequence. In addition, low-abundance lactosylated CPP have to be concentrated before injection into the HPLC apparatus. The chromatographic techniques using IMAC and titanium dioxide (TiO2) require both a preliminary purification [34, 35] and elution step [36, 37] for isolating phosphopeptides prior to MALDI analysis. The HA-based procedure has the advantage of capturing distinctly the plasmin-derived CPP and the counterparts that have undergone lactosylation under different heat treatments. The chemical changes induced by processing milk to heating should be confirmed by MS through identification of the amino acid sites modified by lactose. The advantages of using lactosylated signature peptides as analytical surrogates of the proteins are that (1) it is easier to separate and detect peptides than proteins, (2) structure of the lactosylated CPP does not alter during the analysis, (3) native CPP do not interfere with lactosylated counterparts, and (4) putative peptides suggested from databases can be easily recognized and eventually synthesized for external standards. Another advantage of the procedure is that different mass range acquiring analysis can be explored to detect co-existing HA-bound phosphoproteins and CPP in three separated range masses of the MALDI spectra. Once a lactosylated peptide is discriminative for the heat treatment type, the relative signature peptide could be detected and quantified within a restricted mass range.
Conclusion
The lactosylated phosphopeptides we detected were indicative of the casein quality. The ability to dynamically characterize lactosylation as a distinguished signature modification of caseins could help monitor the heat-induced modifications of casein micelles. CPP lactosylation can be regarded as a model for studying the accessibility of lactose to casein micelles. The major drawback encountered in concentrating CPP was resolved by capturing CPP on HA. In addition to this, the quality of MALDI signals of lactosylated CPP improved. These peculiarities make the new method able to detect also low-abundance signature peptides by MALDI.
The results of our experiments have demonstrated that (1) native and lactosylated CPP are equally bound to HA; (2) CPP have higher affinity for HA than unphosphorylated peptides co-purifying with them using other methods; (3) as expected, the extent of the casein lactosylation is a function of the heating intensity; and (4) mono-lactosylated β-CN (f1-28) 4P represents a distinct signature peptide that is detected distinctly from the native counterpart, making possible the detection of not less than 10% UHT milk spiking raw or pasteurized milk. However, the present procedure needs further signature peptides to distinguish thermized and pasteurized milks. Even with this limitation, the method could be applied in the design and control of any dairy product at the molecular level.
Abbreviations
- ACN:
-
Acetonitrile
- ALP:
-
Alkaline phosphatase
- AMBIC:
-
Ammonium bicarbonate buffer
- α-La:
-
alpha-Lactalbumin
- β-Lg:
-
beta-Lactoglobulin
- CN:
-
Casein
- CPP:
-
Casein phosphopeptides
- DHB:
-
5-Dihydroxybenzoic acid
- DTT:
-
Dithiothreitol
- HA:
-
Hydroxyapatite
- IMAC:
-
Immobilized metal affinity chromatography
- IS:
-
Internal standard
- LOD:
-
Limit of detection
- MALDI-TOF-MS:
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- PA:
-
Phosphoric acid
- PP:
-
Proteose peptones
- PSD:
-
Post-source decay
- SA:
-
Sinapinic acid
- SCC:
-
Somatic cell count
- TFA:
-
Trifluoroacetic acid
- UHT:
-
Ultra high temperature
References
International Dairy Federation (1999) Suggested interpretation of mastitis terminology. IDF Bulletin 338, Brussels, Belgium, pp 3–26
Le Bars D, Gripon JC (1993) Hydrolysis of αs1-casein by bovine plasmin. Lait 73:337–344
Diaz O, Gouldsworthy AM, Leaver J (1996) Identification of peptides released from casein micelles by limited trypsinolysis. J Agric Food Chem 44:2517–2522
Andrews AT, Williams RJH, Brownsell VL, Isgrove FH, Jenkins K, Kanakanian AD (2006) β-CN-5P and β-CN-4P components of bovine milk proteose-peptone: large scale preparation and influence on the growth of cariogenic microorganisms. Food Chem 96:234–241
Farrell HM Jr, Jimenez-Flores R, Bleck GT, Brown EM, Butler JE, Creamer LK, Hicks CL, Hollar CM, Ng-Kwai-Hang KF, Swaisgood HE (2004) Nomenclature of the proteins of cows' milk—sixth revision. J Dairy Sci 87:1641–1674
De Noni I, Pellegrino L, Cattaneo S, Resmini P (2007) HPLC of proteose peptones for evaluating the ageing of packaged pasteurized milk during refrigerated storage. J Int Dairy 17:12–19
Alichanidis E, Wrathall JHM, Andrews AT (1986) Heat stability of plasmin (milk protease) and plasminogen. J Dairy Res 53:259–269
Kohlmann KL, Nielsen SS, Ladisch MR (1991) Effects of a low concentration of added plasmin on ultra-high temperature processed milk. J Dairy Sci 74:1151–1156
Arena S, Renzone G, Novi G, Scaloni A (2011) Redox proteomics of fat globules unveils broad protein lactosylation and compositional changes in milk samples subjected to various technological procedures. J Proteomics 74:2453–2475
Guyomarc’h F, Warina F, Muirb DD, Leaverb J (2000) Lactosylation of milk proteins during the manufacture and storage of skim milk powders. J Int Dairy 10:12863–12872
Groubet R, Chobert JM, Haertle T (1999) Functional properties of milk proteins glycated in mild conditions. Sci Aliment 19:423–438
Scaloni A, Perillo V, Franco P, Fedele E, Froio R, Ferrara L, Bergamo P (2002) Characterization of heat-induced lactosylation products in caseins by immunoenzymatic and mass spectrometric methodologies. Biochim Biophys Acta 1598:30–39
Phelan M, Aherne A, FitzGerald RJ, O'Brien NM (2009) Casein-derived bioactive peptides: biological effects, industrial uses, safety aspects and regulatory status. J Int Dairy 19:643–654
Pinto G, Caira S, Cuollo M, Lilla S, Fierro O, Addeo F (2010) Hydroxyapatite as a concentrating probe for phosphoproteomic analyses. J Chromatogr B 878:2669–2678
Mamone G, Picariello G, Ferranti P, Addeo F (2010) Hydroxyapatite affinity chromatography for the highly selective enrichment of mono- and multi-phosphorylated peptides in phosphoproteome analysis. Proteomics 10:380–393
Kokubu M, Ishihama Y, Sato T, Nagasu T, Oda Y (2005) Specificity of immobilized metal affinity-based IMAC/C18 tip enrichment of phosphopeptides for protein phosphorylation analysis. Anal Chem 77:5144–5154
Zhang L, Wang H, Liang Z, Yang K, Zhang L, Zhang Y (2011) Facile preparation of monolithic immobilized metal affinity chromatography capillary columns for selective enrichment of phosphopeptides. J Sep Sci 34:2122–2130
Thingholm TE, Jensen ON, Larsen MR (2009) Analytical strategies for phosphoproteomics. Proteomics 9:1451–1468
Kjellstrom S, Jensen ON (2004) Phosphoric acid as a matrix additive for MALDI MS analysis of phosphopeptides and phosphoproteins. Anal Chem 76:5109–5117
Mercier JC (1981) Phosphorylation of caseins, present evidence for an amino acid triplet code posttranslationally recognized by specific kinases. Biochimie 63:1–17
Schmidt SR, Schweikart F, Andersson ME (2007) Current methods for phosphoprotein isolation and enrichment. J Chromatogr B 849:154–162
Addeo F, Chobert JM, Ribadeau-Dumas B (1977) Fractionation of whole casein on hydroxyapatite. Application to a study of buffalo κ-casein. J Dairy Res 44:63–68
Siciliano R, Rega B, Amoresano A, Pucci P (2000) Modern mass spectrometric methodologies in monitoring milk quality. Anal Chem 72:408–415
Ledl F, Schleicher E (1990) New aspects of the Maillard reaction in foods and in the human body. Angew Chem Int Ed Engl 29:565–594
Yaylayan VA, Huyghues-Despointes A (1994) Chemistry of Amadori rearrangement products: analysis, synthesis, kinetics, reaction, and spectroscopic properties. Crit Rev Food Sci Nutr 34:321–369
Marvin LF, Parisod V, Fay LB, Guy PA (2002) Characterization of lactosylated proteins of infant formula powders using two-dimensional gel electrophoresis and nanoelectrospray mass spectrometry. Electrophoresis 23:2505–2512
Fogliano V, Monti SM, Visconti A, Randazzo G, Facchiano AM, Colonna G, Ritieni A (1998) Identification of a beta-lactoglobulin lactosylation site. Biochim Biophys Acta 1388:295–304
Fenaille F, Morgan F, Parisod V, Tabet JC, Guy PA (2004) Solid-state glycation of β-lactoglobulin by lactose and galactose: localization of the modified amino acids using mass spectrometric techniques. J Mass Spectrom 39:16–28
Ferranti P, Barone F, Chianese L, Addeo F, Scaloni A, Pellegrino L, Resmini P (1997) Phosphopeptides from Grana Padano cheese: nature, origin and changes during ripening. J Dairy Res 64:601–615
Weinstein MJ, Doolittle RF (1972) Differential specificities of the thrombin, plasmin and trypsin with regard to synthetic and natural substrates and inhibitors. Biochim Biophys Acta 258:577–590
Dalsgaard TK, Nielsen JH, Larsen LB (2007) Proteolysis of milk proteins lactosylated in model systems. Mol Nutr Food Res 51:404–414
Pizzano R, Nicolai MA, Siciliano R, Addeo F (1998) Specific detection of the Amadori compounds in milk by using polyclonal antibodies raised against a lactosylated peptide. J Agric Food Chem 46:5373–5379
Mamone G, Caira S, Garro P, Nicolai A, Ferranti P, Picariello G, Malorni A, Chianese L, Addeo F (2003) Casein phosphoproteome: identification of phosphoproteins by combined mass spectrometry and two-dimensional gel electrophoresis. Electrophoresis 24:2824–2837
McNulty DE, Annan RS (2008) Hydrophilic-interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol Cell Proteomics 7:971–980
Nie S, Dai J, Ning ZB, Cao XJ, Sheng QH, Zeng R (2010) Comprehensive profiling of phosphopeptides based on anion exchange followed by flow-through enrichment with titanium dioxide (AFET). J Proteome Res 9:4585–4594
Imanishi SY, Kochin V, Eriksson JE (2007) Optimization of phosphopeptide elution conditions in immobilized Fe(III) affinity chromatography. Proteomics 7:174–176
Aryal UK, Ross ARS (2010) Enrichment and analysis of phosphopeptides under different experimental conditions using titanium dioxide affinity chromatography and mass spectrometry. Rapid Commun Mass Spectrom 24:219–231
Acknowledgments
Publication in partial fulfillment of the requirements for PhD in Sciences and Technologies of Food Productions -XXV Cycle, University of Naples ‘Federico II’.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 437 kb)
Rights and permissions
About this article
Cite this article
Pinto, G., Caira, S., Cuollo, M. et al. Lactosylated casein phosphopeptides as specific indicators of heated milks. Anal Bioanal Chem 402, 1961–1972 (2012). https://doi.org/10.1007/s00216-011-5627-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-011-5627-6