Abstract
d-Allose, a rare, naturally occurring monosaccharide, is known to exert anti-proliferative effects on cancer cells. The effects of d-allose on the cellular membranes of hormone-refractory prostate cancer cell line (DU145), hormone-sensitive prostate cancer cell line (LNCaP), and normal prostate epithelial cells (PrEC) were studied at the molecular level by phospholipid (PL) profiling using a shotgun lipidomic method. The molecular structures of 85 PL species including 23 phosphatidylcholines, 12 phosphatidylethanolamines (PEs), 11 phosphatidylserines (PSs), 16 phosphatidylinositols, 9 phosphatidic acids (PAs), and 14 phosphatidylglycerols (PGs) were identified by data-dependent collision-induced dissociation of nanoflow liquid chromatography–tandem mass spectrometry, and the PL amounts were quantified. The addition of d-allose to prostate cancer cell lines during their growth phases had negligible or decreased effects on the relative regulation of PL species, but several new PS molecules (two for DU145 and three for LNCaP) emerged. In contrast, experiments on the PrEC cell line revealed that some high abundant species (14:0/14:0-PE, 16:2/16:0-PG, and 20:6/18:1-PA) showed significant increases in concentration. These findings support a mechanism for the anti-proliferative effect of d-allose on prostate cancer cell lines that involves the induction of programmed cell death since PS molecules are known to induce apoptosis. Principal component analysis was carried out to examine differences in PL distributions among the three cell lines promoted by d-allose.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phospholipids (PLs), the major group of lipids that constitute cellular membranes, are involved in signal transduction, cell proliferation, and cell death [1, 2]. Analyzing PLs in biological fluids, tissues, and cells is of growing interest for the development of candidate biomarkers of diseases such as breast cancer [3, 4], ovarian cancer [5, 6], and prostate cancer [7, 8]. PLs are differentiated by type of polar head group, acyl chain length, number of acyl chains, and the degree of unsaturation in the acyl chain. The polar head groups are derived from choline, ethanolamine, serine, inositol, glycerol, or hydrogen attached to a phosphate group on a glycerol backbone and are phosphatidylcholine (PC), phosphatidyethanolamine (PE), phosphotidylserine (PS), phosphatidylinositol (PI), phosophatidyglycerol (PG), and phosphatidic acid (PA), respectively. In addition to head group variation, acyl chains with different lengths and numbers of double bonds result in complicated mixtures of PLs. Due to the complexity of PL molecules, comprehensive quantitative and qualitative analyses of PLs are often challenging for lipidomic studies of changes in PL content depending on biological or clinical status.
Prostate cancer is now the second leading cause of cancer-related death in men in the USA and Europe, and its incidence has been increasing significantly [9–11]. In particular, androgen-independent hormone-refractory prostate cancer (HRPC) occurs at a certain stage during therapy, which leads to a poor response to anticancer treatments such as hormone therapy, radiotherapy, chemotherapy, and prostectomy [12, 13]. For the development of efficient HRPC treatments, a number of efforts have focused on discovering target molecules and unveiling signaling pathways [14–17].
d-Allose, the C-3 epimer of d-glucose, is a rare, naturally occurring monosaccharide. The biological effects of d-allose are known to include inhibiting neutrophil production and lowering of platelet count without side effects as an immunosuppressant [18], reducing the amount of reactive oxygen species from neutrophils [19] and limiting the proliferation of various cancer [20] and leukemia cell lines [21]. In a recent study [13], d-allose was found to inhibit the proliferation of human HRPC cell lines, DU145 and PC-3, while it did not have any significant effect on the normal prostate epithelial cell line prostate epithelial cells (PrEC). That study showed that the anti-proliferative effect of d-allose arises from the induction of programmed cell death through the modulation of members of the Bcl-2 family of proteins (Bax and Bcl-2 which are pro- and anti-apoptotic proteins, respectively); the release of cytochrome C, which is a mitochondria-mediated apoptotic pathway member; and the cleavage of caspase 3 and poly(ADP-ribose) polymerase. Further investigation of the effect of d-allose on cellular membrane structures at the molecular level to determine how the composition and distribution of PLs in these prostate cancer cell lines are influenced is of interest for further therapeutic validation.
Analyses of PLs in their intact condition can be carried out by direct infusion to an electrospray ionization–mass spectrometry (ESI-MS) for the high-speed screening of PLs coupled with structural identification [22, 23]. While a sophisticated MS method using ESI provides rapid and accurate analysis of PL molecules, physical separation of PL molecules is commonly required for the analysis of complicated PL mixtures. Typical chromatographic methods are thin layer chromatography in combination with matrix-assisted laser desorption and ionization/time-of-flight mass spectrometry [24] and high-performance liquid chromatography (HPLC) coupled to ESI-MS [25–27]. Nanoflow LC with ESI–tandem mass spectrometry (nLC-ESI-MS-MS) has been shown to be capable of separating PL species from tissues and human urine samples with detection limits down to 2.2 fmol for the simultaneous characterization of molecular structures [28–30]. The shotgun lipidomics method has been applied to the identification of candidate markers of PL species from breast cancer [31, 32] and prostate cancer [8] patient urine samples.
In the present study, the shotgun lipidomic method was applied to investigate the effects of d-allose on the distribution and compositions of PL species from human HRPC and hormone-sensitive prostate cancer (HSPC) cell lines, DU145 and LNCaP, respectively, in comparison to those from normal PrEC. Cellular PL mixtures were extracted from each cell line that had been treated with or without d-allose and were analyzed by nLC-ESI-MS-MS. PLs were identified by collision-induced dissociation (CID) in both positive (PCs and PEs) and negative ion modes (PIs, PSs, PGs, and PAs), and the relative concentrations of each component were examined upon d-allose treatment. Principal component analysis (PCA) was carried out to examine the effects of d-allose on PL distribution among the three cell lines.
Experimental
Cell culture and extraction of phospholipids
The prostate cell lines examined in this study—DU 145 (human HRPC cell line), LNCaP (human HSPC cell line), and PrEC (normal prostate epithelial cell line)—were obtained from the Korea Cell Line Bank (KCLB, Seoul, South Korea). Details of the cell culture process may be found in a previous report [13], while a brief summary is as follows. Each cell line was grown in five Petri dishes (5 cm in diameter) containing RPMI-1640 from HyClone Laboratories Inc. (Logan, UT, USA) at 37 °C in a humidified atmosphere containing 5% CO2 for 48 h. Each dish contained cells covering more than 90% of dish area. Then, 1.5 g/L of sodium bicarbonate, 10 mM of HEPES, 1.0 mM of sodium pyruvate, 10% fetal bovine serum, and 1% penicillin–streptomycin solution followed by 40 mM of d-allose obtained from Sigma-Aldrich (St. Louis, MO, USA), dissolved in phosphate-buffered saline solution, were added to the media, and cells were collected after 48 h of treatment.
Extraction of PLs from each cell line began with the sonication of cells (500 μL) using a tip sonicator from Cole-Parmer model CP 130 (Vernon Hills, IL, USA) in pulse mode with an amplitude of 2.5 W for 10 s, twice. The mixture was centrifuged for 1 h at 100,000×g using an ultracentrifuge, model Optima L-100xp from Beckman Coulter Inc. (La Brea, CA, USA), and the sediment was collected for extraction of PLs using the Folch method [33]. The collected pellets were mixed with 0.1 mL each of chloroform and methanol and were vortexed for 5 min. This process was repeated three times for the complete dissolution of the pellets, resulting in a total volume of 0.6 mL. Then, 0.3 mL of chloroform was added and the mixture was vortexed for 20 min. For the phase separation, 0.18 mL of water was added and the mixture was vortexed for 20 min, followed by centrifugation at 15,700×g for 5 min at room temperature. The lower phase containing lipids was recovered and dried using a SpeedVac, model SC 100A SpeedVac Plus from Thermo Savant (Farmingdale, NY, USA). The remaining solid powder was weighed and then redissolved in methanol/acetonitrile (1:1) to yield a final volume of 100 μL for storage (4 °C). For the nLC-ESI-MS-MS analysis, the sample stock solution was diluted with CH3OH/CH3CN (9:1) to give a concentration of 5.0 μg/μL. nLC-ESI-MS-MS analysis was made within a month from PL extraction.
Nanoflow LC with ESI–tandem mass spectrometry
The reverse phase LC column was prepared in our laboratory by packing with Magic C18, 5 μm–100 Å (Michrom Bioresources Inc., Auburn, CA, USA), RP resin into the 75-μm i.d. × 17-cm-long capillary tube under 1,000 psi of He. Silica capillary tubes (Polymicro Technology LLC, Phoenix, AZ, USA) with inner diameters (i.d.) of 20, 50, and 75 μm (360-μm outer diameter for all) were used for the nanoflow LC. Before column packing, one end of the capillary tube was pulled after heating in a flame to produce a sharp needle (tip i.d. ~10 μm) for direct ESI. The pulled tip capillary column prepared in our laboratory was directly connected between a model 1200 capillary pump system from Agilent Technologies (Palo Alto, CA, USA) equipped with an autosampler and an LCQ Deca XP MAX ion trap mass spectrometer (Thermo Finnigan, San Jose, CA, USA) for nLC-ESI-MS-MS. To provide ESI voltage, a Pt wire was inserted into a PEEK micro-cross (Upchurch Scientific, Oak Harbor, WA, USA) in which capillary tubes from the pump, capillary column, and vent tube were all connected. The vent capillary tube (20-μm i.d., 360-μm o.d.) was used to split the pump flow (7 μL/min) such that the required flow rate (300 nL/min) was delivered to the analytical column. During sample loading, the vent tube was closed by an on–off valve located at the end of the tube. Another purpose of using the vent tube was to reduce the dwell time during gradient elution. During gradient elution, the pump flow was set at 7 μL/min with the on–off valve open to minimize the delay in gradient change from the pump to the column and to allow the pump flow to be split just before the column inlet.
Mobile phase solutions were 50:50 (v/v) acetonitrile/water for A and 90:10 isopropanol/acetonitrile for B with 0.1% (v/v) formic acid added for positive ion mode and 0.05% sodium hydroxide added for negative ion mode. All solvents used for the LC mobile phase in this study (acetonitrile, isopropanol, methanol, water (Avantor Performance Materials, Phillipsburg, NJ, USA), chloroform, ammonium hydroxide, and formic acid (Sigma, St. Louis, MO, USA)) were HPLC grade. Gradient elution conditions were as follows. Mobile phase B was increased from 0% to 55% over 1 min and then was ramped to 90% during 90 min for the positive ion mode or during 60 min for the negative ion mode. Injection amounts for each PL sample were 1.5 and 0.3 μg for the positive and negative ion modes, respectively. Two internal standards for both ion modes of MS were added to PL samples to compensate for possible fluctuations in the MS analysis: 14:0/14:0-PC at a concentration of 0.2 pmol/μL for positive ion mode and 16:0/16:0-PI at 0.05 pmol/μL for negative ion mode, both from Avanti Polar Lipids Inc. (Alabaster, AL, USA). ESI voltage for both ion modes was 3 kV, and the normalized collision energy for data-dependent MS-MS analysis was set at 45% (positive ion mode) and 40% (negative ion mode). MS ranges for the first MS scan were m/z 550–950 (positive ion) and 600–900 (negative ion). Maximum injection time was set as 150 ms for both ion modes. Data-dependent MS-MS analysis was carried out for the first three prominent ions in sequence for each MS scan. Characteristic fragment ions were examined manually for the identification of each PL species. For the semiquantitative analysis, the peak area of individual PL species was divided by the peak area of the spiked internal standard for each ion mode. For each extracted sample, measurements in both ion modes were made in triplicate. For PCA analysis, Minitab 16 software (http://www.minitab.co.kr) was utilized.
Results and discussion
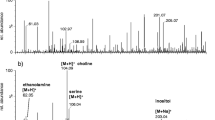
PL mixture samples extracted from the three cell lines before and after treatment with d-allose were analyzed by nLC-ESI-MS-MS in both positive and negative ion modes using gradient elution for structural identification of PL species. Figure 1 shows the simple scheme used to characterize the molecular structures of PL species in this study. Figure 1a shows the base peak chromatograms (BPCs) of PL mixtures from DU145 cells before (top chromatogram) and after treatment with d-allose (bottom) in the MS negative ion mode. Eluted peaks in both BPCs appear similar with only slight variations. Figure 1b exhibits the first MS scan spectra taken at the retention time slice of 21.69 min which has prominent ions [M + H]+ at m/z 737.9, 772.6, and 835.7; the CID fragment ion spectra of the parent ion m/z 835.7 is shown in Fig. 1c. The fragment ion peaks in the CID spectra of Fig. 1c were analyzed to determine the molecular structure of the parent ion as follows. A fragment ion at m/z 673.2 corresponds to loss of the inositol head group (C6H10O5, 162 Da) as [M + H-162]+ from the molecular ion, and fragment ions at m/z 578.8 and 552.8 correspond to the loss of acyl chains in the form of carboxylic acids from the sn-1 and sn-2 carbons of the glycerol backbone, respectively. Fragment ions m/z 596.9 and 570.8 correspond to the loss of fatty acid in the form of a ketene, \( {\left[ {{\text{M}} + {\text{H}} - {{\text{R}}_1}\prime \,{\text{CH}} = {\text{C}} = {\text{O}}} \right]^{ + }} \) and \( {\left[ {{\text{M}} + {\text{H}} - {{\text{R}}_2}\prime \,{\text{CH}} = {\text{C}} = {\text{O}}} \right]^{ + }} \), respectively. The CID spectra in Fig. 1c also show fragment ions dissociated from the parent ion without the inositol head group (\( {\left[ {{\text{M}} + {\text{H}} - 162} \right]^{ + }} \)), at m/z 417.3, 391.3, 435.2, and 409.3, of which the first two and the last two ions result from the dissociation of acyl chains in the form of carboxylic acid and ketene, respectively. In addition, ions at m/z 255.6 and 281.6 correspond to the free carboxylate anions of the fatty acids as [R1COO]− and [R2COO]−, respectively. Overall, the molecular ion observed at m/z 835.7 resulted in the identification of 16:0/18:1-PI based on the characteristic fragment ions. Similarly, eight PIs, seven PSs, four PGs, and four PAs were identified in the negative ion mode for the same sample. In the positive ion mode of the nLC-ESI-MS-MS, 13 PCs and 7 PEs were identified from the DU145 cell line. Characterized PL species for the three cell lines (PrEC, DU145, and LNCaP) are listed in Table 1. From the qualitative analysis of these three cell lines, a total of 85 PL species (23 PCs, 12 PEs, 11 PSs, 16 PIs, 14 PGs, and 9 PAs) were identified, and the relative PL concentrations were obtained from the peak area ratio of each species to the internal standard. For quantitative analysis of these identified molecules, MS scan was obtained without CID in triplicate measurements. Phospholipids in Table 1 are sorted by PL species commonly found in all of the cell lines, in at least two cell lines, unique species for each cell line, and unique species which were observed after treatment with d-allose. Each category of PL was further subdivided into two groups (PCs and PEs from positive ion mode and the four other PL types from negative ion mode), indicated in the table by a horizontal bar, since the two ion modes used different internal standards in different concentrations.
a BPCs of cellular PL mixtures from DU145 cells before and after treatment with d-allose obtained by nLC- ESI-MS-MS. b MS spectra at 21.69 min during the first MS scan in negative ion mode. c MS-MS spectra of the ion m/z = 835.7 showing the typical fragment ion peaks resulting in the identification of the molecular structure as 16:0/18:1-PI. NL represents for normalized limit (arbitrary unit) of signal intensity recorded from MS
The number of identified PL molecules from the three cell lines before and after treatment with d-allose is compared in Fig. 2. Each cell line originally displayed different PL distributions except for a small number of common species. For instance, 14 PL species were found in all three cell lines and 11 PL species were found in only two cell lines. These are listed in Table 1, respectively. The number of commonly found PL species appeared to be smaller than the total number of identified PL molecules; however, some species (14:0/14:0-PE, 16:2/16:0-PG, and 20:0/20:0-PS) among the commonly found PL species in Table 1 were highly abundant compared with the relative peak area of the other species. Since the current experimental data show the relative peak area compared with the internal standard, an accurate calculation of the concentration of each species is needed by including the effect of acyl chain length on MS intensity, but this was not included in this study. After treatment with d-allose, the number of commonly found PL species from all cell lines did not change significantly; however, the total number of PL species increased approximately 10% (from 73 to 80). This increase in the total number of PL species was largely due to the emergence of 12 new PL species after treatment (see Table 1). In particular, six species among the 12 new PLs in Table 1 were PS molecules, which are further discussed below. Besides the new PL species, notable changes were observed for some PL species after d-allose treatment. For instance, the relative concentration of 14:0/14:0-PE in PrEC cells before treatment was 16.67 ± 1.91 (peak area ratio against IS, Table 1), which was considerably greater than other PC or PE species. However, the relative concentration of this species increased to 49.44 ± 7.54 after treatment, while concentrations of the same species in DU 145 and LNCaP cells did not change significantly. Similar trends were found for 22:0/16:0-PC and 20:2/16:0-PE. These results are plotted in Fig. 3a–c. After treatment, two abundant PL species (with relatively large concentrations) such as 16:2/16:0-PG and 20:6/18:1-PA (see Table 1 and Fig. 3d, e) showed a significant concentration increase in PrEC cells, but a considerable decrease (two- to fivefold) in LNCaP cells. These are highly abundant species compared with the other PL molecules. The increase in the concentrations of these molecules in PrEC cells as well as the negligible change or significant decrease in cancer cells can be explained by d-allose not influencing the programmed cell death in normal cells (PrEC), therefore allowing these cells to grow during the period of d-allose treatment. In contrast, for cancer cells, d-allose inhibited cell growth, and thus, the major PL components (the five species mentioned above) had relatively small changes or big decreases in concentration. It was previously reported [13] that proliferation of human prostate cancer cells was inhibited by d-allose: MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell proliferation assay of the 48-h treated cells showed a significant dose-dependent inhibition of cell growth down to 8.7% of DU145and 49.5% for LNCaP compared with untreated cells, while a similar treatment to PrEC cells had no significant effect, showing 98.1%. Another interesting finding in Table 1 was that for 20:0/20:0-PS, in Table 1 (plotted in Fig. 3f), which was found in all cell lines before treatment but was not detected after treatment with d-allose. It is known that PS is closely associated with apoptosis [34–36]. PS molecules, which are minor in number and are mainly located at the cytoplasmic side of the plasma membrane, transit rapidly toward the outer membrane leaflet and are recognized by phagocytes as the cells undergo programmed cell death. It is also known that PS is synthesized from PC or PE by enzymes before externalization of PS molecules [37, 38]. In Table 1, six PS species were newly found after treatment. PS species identified from cancer cells after d-allose treatment were 18:4/18:4-PS and 18:1/18:1-PS for DU145 and 18:3/18:0-PS, 16:1/18:1-PS, and 18:1/18:0-PS for LNCaP cells. It is interesting to note that these five PS molecules contain acyl chains of 18 carbons with a single or no unsaturation in most cases. Although these data are not sufficient to deduce which PS could be related to apoptotic signaling, it is expected that PS molecules that emerged during d-allose treatment could be candidates for the “eat me” signal in programmed cell death.
PCA was carried out for PL species to examine significant differences during d-allose treatment. First, species that exhibited a more than 2.5-fold change in concentration after d-allose treatment were selected; Table 2 is a compilation of concentration ratios for these species. Figure 4a illustrates the statistical differences in the profiles of the selected PL species among the three cell lines before and after d-allose treatment. Data points for each cell line in Fig. 4a are represented by open symbols (before d-allose treatment) and filled symbols (after treatment): circles for PrEC, squares for DU145, and triangles for LNCaP cells. Clustered data points represent the reproducibility of repeated measurements (n = 3) for each sample. The data points for the treated cell lines (filled symbols) in Fig. 4a are individually clustered away from the center of the plot (for untreated samples) apart from each other. This suggests that some species show a clear difference in relative concentration after d-allose treatment, as shown in the accompanying plot in Fig. 4b. Correlations of the two plots (Fig. 4a, b) can be established by matching the same geometrical locations in the two plots. Each component appearing at a specific location in Fig. 4b represents a relative abundance before or after treatment depending on its equivalent location in Fig. 4a. For instance, PL species from PrEC that demonstrated significant increases in concentration can be found in the lower right area of Fig. 4b matched to the ellipsoidal region of Fig. 4a. The PL species labeled C6, E4, and S2, (molecular structure of each label can be found in Table 2) demonstrated significant increases in concentration (more than a threefold increase) in PrEC after d-allose treatment. These three species emerged after d-allose treatment; therefore, the concentration ratio cannot be calculated (expressed with upper arrow in Table 2). Moreover, species E2, G1, C2, E1, and E3, which had a more than threefold increase in concentration, are clustered into two nearby groups. On the other hand, data points clustered toward the center of the plot within the same ellipsoid represent concentrations that were significantly higher prior to treatment, which implies a significant decrease in concentration as a result of treatment. These species were I2, C3, C4, and S1, in order of decreased concentration (the concentration ratios of these species in Table 2 are 0.35, 0.14, 0.05, and almost 0, respectively). For DU145 cells, C1 and I1 increased significantly (6.8- and 4.3-fold, respectively) after treatment along with the four new species (E5, I3, S3, and S4), which overlap in Fig. 4b. Likewise, for LNCaP cells, S1 was shown to significantly decrease after treatment along with the four new species (C7, S5, S6, and S7) that are clustered at the upper left corner of the plot.
PCA plots showing the statistical differences of selected PL molecule concentrations (more than 2.5-fold) from each cell line before (open symbols) and after d-allose treatment (filled symbols) (a) and the distribution of individual components labeled in Table 2 (b). Each component at a specific location of plot b represents its significant abundance before or after treatment depending on its equivalent location in plot a
Conclusions
In this study, a shotgun lipidomic approach with nLC-ESI-MS-MS was utilized to determine the relative concentrations and distributions of PLs from the two prostate cancer cell lines and a normal prostate cell line after treatment with d-allose during cell growth. While d-allose treatment was found to induce significant increases in concentration of some highly abundant PL species (14:0/14:0-PE, 16:2/16:0-PG, and 20:6/18:1-PA) from the normal prostate epithelial cell line (PrEC), it had a negligible or negative effect on the same PL species from the prostate cancer cell lines (DU145 and LNCaP). This observation may reflect the fact that PrEC grew continuously without inhibition from d-allose, but proliferation of cancer cells was inhibited. This result agrees with the experimental findings of previous studies [13]. When d-allose was added during the growth phases of the three cell lines, six new PS molecules were identified, which is noteworthy since PS is known to be a recognition element for phagocytes during programmed cell death. Five of the six new PSs expressed in both prostate cancer cell lines mostly contain acyl chains of 18 carbons with a single or no unsaturation in most cases. The current study deals with the relative changes in only 85 PL species; however, it demonstrates the feasibility of investigating the roles of PL molecules in the proliferation and programmed death of cells using an agent that controls cell growth. A more systematic study on qualitative and quantitative analyses can be obtained, provided with the development of a sophisticated search engine and an improved separation of complicated PL species which can reduce the ionic suppression of low abundant species during ESI.
References
Brouwers JFHM, Vernooji EAAM, Tielens AGM, van Golde LMG (1999) J Lipid Res 40:164–169
Wright MM, Howe AG, Zaremberg V (2004) Biochem Cell Biol 82:18–26
Leach MO, Verrill M, Glaholm J, Smith TAD, Collins DJ, Payne GS, Sharp JC, Ronen SM, McCready VR, Powles TJ, Smith IE (1998) NMR Biomed 11:314–340
Bougnoux P, Chajes V, Lanson M, Hacene K, Body G, Couet C, Folch OL (1992) Breast Cancer Res Treat 20:185–194
Xu Y, Shen Z, Wiper DW, Wu M, Morton RE, Elson P, Kennedy AW, Belinson J, Markman M, Casey G (1998) JAMA 280:719–723
Sutphen R, Xu Y, Wilbanks GD, Fiorica J, Grendys EC Jr, LaPolla JP, Arango H, Hoffman MS, Martino M, Wakeley K, Griffin D, Blanco RW, Cantor AB, Xiao YJ, Krischer JP (2004) Cancer Epidemiol Biomark Prev 13:1185–1191
Swanson MG, Vigneron DB, Tabatabai ZL, Males RG, Schmitt L, Carroll PR, James JK, Hurd RE, Kurhanewicz J (2003) Magn Reson Med 50:944–954
Min HK, Lim S, Chung BC, Moon MH (2011) Anal Bioanal Chem 399:823–830
Gronberg H (2003) Lancet 163:859–864
Clarke RA, Schirra HJ, Catto JW, Lavin MF, Gardiner RA (2010) Cancers 2:1125–1154
Hsing AW, Devesa SS (2001) Epidemiol Rev 23:3–13
Tamura K, Furihata M, Tsunoda T, Ashida S, Takata R, Obara W et al (2007) Cancer Res 67:5117–5125
Naha N, Lee HY, Jo MJ, Chung BC, Kim SH, Kim MO (2008) Apoptosis 13:1121–1134
Knudson CM, Korsmeyer SJ (1997) Nat Genet 16:358–363
Scher HI, Sawyers CL (2006) J Clin Oncol 23:8253–8261
Luck RM, Bossy-Wetzel E, Green DR, Newmeyer DD (1997) Science 275:1132–1136
Lee T, Malathi K, Lohyama S, Silane M, Berenstein A, Jayarama T (2004) Cell Biochem Funct 22:35–40
Arnold EC, Siladay PJ (1997) US Patent No. 5,620,960
Murata A, Sekiya K, Watanabe Y (2003) J Biosci Bioeng 96:89–91
Sui L, Dong Y, Watanabe Y et al (2005) Int J Oncol 27:907–912
Hirata Y, Saito M, Tsukamoto I, Yamaguchi F et al (2009) J Biosci Bioeng 107:562–568
Kim HY, Wang TCL, Ma YC (1994) Anal Chem 66:3977–3982
Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharaju P (2001) J Lipid Res 42:663–672
Fuchs B, Schiller J, Süβ R, Schürenburg M, Suckau D (2007) Anal Bioanal Chem 389:827–834
Taguchi R, Hayakawa J, Takeuchi Y, Ishida M (2000) J Mass Spectrom 35:953–966
Isaac G, Bylund D, Mansson J-E, Markides KE, Bergquist J (2003) J Neurosci Meth 128:111–119
Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, Shimizu T (2005) J Chromatogr B 823:26–36
Bang DY, Kang D, Moon MH (2006) J Chromatogr A 1104:222–229
Bang DY, Ahn E, Moon MH (2007) J Chromatogr B 852:268–277
Ahn E, Kim H, Chung BC, Moon MH (2007) J Sep Sci 30:2598–2604
Kim H, Min HK, Kong G, Moon MH (2009) Anal Bioanal Chem 393:1649–1656
Min HK, Kong G, Moon MH (2010) Anal Bioanal Chem 396:1273–1280
Folch J, Less M, Stanley GHS (1957) J Biol Chem 226:497–509
Schroit AJ, Zwaal RF (1991) Biochim Biophys Acta 1071:313–3129
Iguchi K, Hirano K, Ishida RM (2010) Apoptosis 6:263–268
Chaurio RA, Janko C, Munoz LE, Frey B, Herrmann M, Gaipl US (2009) Molecules 14:4892–4914
Aussel C, Pelassy C, Breittmayer JP (1998) FEBS Lett 431:195–199
Uchida K, Emoto K, Daleke DL, Inoue K, Umeda M (1998) J Biochem 123:1073–1078
Acknowledgments
This study was supported by grant NRF-2010-0014046 and in part by grant NRF-2008-2003136 from the National Research Foundation of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jeong, R.U., Lim, S., Kim, M.O. et al. Effect of d-allose on prostate cancer cell lines: phospholipid profiling by nanoflow liquid chromatography–tandem mass spectrometry. Anal Bioanal Chem 401, 689–698 (2011). https://doi.org/10.1007/s00216-011-5113-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-011-5113-1