Abstract
The solubility and digestion efficiency are two crucial factors that might affect the identification of integral membrane proteins (IMPs). In this work, 1% (v/v) ionic liquid (IL), 1-butyl-3-methyl imidazolium tetrafluoroborate (BMIM BF4), added in NH4HCO3 buffer (pH 8.3), was applied as a sample preparation buffer for IMPs analysis. Compared to the commonly used sodium dodecyl sulfate and methanol methods, the number of identified IMPs from rat brain by microcolumn reversed phase liquid chromatography (μRPLC)-electrospray ionization tandem mass spectrometry (ESI-MS/MS) was improved by over three times, which might be due to the fact that BMIM BF4 offered high solubilizing ability for IMPs and good compatibility for tryptic digestion. Furthermore, compared to Rapigest and urea methods, with BMIM BF4 method, the number of identified IMPs from rat brain could be improved 25% and 80%, respectively, which might be contributed to the good solubilizing ability and high thermal stability of such IL. With the sample treated by BMIM BF4 method, by 2D-nanoSCX-RPLC-ESI-MS/MS, 1,450 non-redundant proteins and 7,978 unique peptides were identified from rat brain, and 418 proteins contained at least one predicted transmembrane domain, with false discovery rates of less than 1% for peptide identification, and at least two identified unique peptides per protein. All these results demonstrate that the BMIM BF4 method is of high potential for the large-scale identification of IMPs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Integral membrane proteins (IMPs) lie at the critical junctions between intracellular compartments, cells, and their environments, which play unique roles in mediating various cellular processes, including intercellular communication, vesicle trafficking, ion transport, protein translocation/integration, and the propagation of signaling cascades [1–3]. However, the identification of IMPs meets great challenges due to their hydrophobicity [4–7].
Recently, shotgun approach has emerged as a powerful technique for membrane proteome identification and quantification [8–14], by which proteins should be degraded into peptides by either enzymatic or chemical digestion, followed by the separation and identification by high-performance liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS). Therefore, for IMPs analysis, improvement on the solubility of such hydrophobic proteins in suitable solvent systems that are of good compatibility with enzymatic digestion is of great significance.
In the past few years, several kinds of solvent systems have been applied in IMPs analyses [15], including chaotropes [16–18], detergents [7, 10, 18–21], organic solvents [7, 18, 22–26], and organic acids [8, 27]. Among them, urea is one of the most commonly used chaotropes with strong denaturation ability. Although it can be easily removed before MS analysis, its capacity to extract most IMPs, particularly polytopic proteins, is limited. Furthermore, urea can also cause problematic carbamylation of N-termini and lysine residues, when heated above 37 °C. Although sodium dodecyl sulfate (SDS), the strongest denaturation reagent, is also often used in membrane protein analysis, it is rather difficult to remove before MS analysis. In addition, MS compatible surfactants, such as Rapigest, are among the most promising additives to enhance IMPs solubilization without interfering with LC-MS. Yates et al. [10] made the global analysis of mammalian brain proteome with MS compatible surfactants, and 5,000 proteins were identified from 1.8 mg of rat brain homogenate, among which 1,897 proteins were annotated by Gene Ontology as membrane proteins, and 1,225 proteins were predicted to contain at least one transmembrane domain (TMD). However, Rapigest is expensive, and the cleavage step at low pH could result in hydrophobic peptides loss due to co-precipitation with the hydrophobic part of RapiGest [28]. Organic solvents, especially 60% (v/v) methanol, are also effective to destroy membrane structure but could attenuate enzyme activity. Organic acids with high concentration are of strongly solubilizing ability for membrane proteins, and CNBr is usually used for membrane proteins digestion. However, due to the high toxicity of CNBr, great caution should be made during sample preparation, and the high concentration of acid might lead to unexpected modifications [29].
Ionic liquids (ILs) are liquids composed completely of ions, and thus have unique physicochemical properties, including broad liquid range, low to negligible vapor pressures, high thermal stability, and high solubility for different classes of analytes [30–42]. Therefore, ILs environment is quite different from that of all polar or non-polar organic solvents [32], which is capable of forming virtually all possible types of interactions with solutes [33, 34].
Due to the above-mentioned unique properties, ILs are regarded as a new class of green solvents [30, 35] and have been successfully applied in sample extraction [31, 36, 37], chromatography separation [38, 39], and MS identification [40–42]. Anderson et al. [31] applied ILs containing tris(pentafluoroethyl)trifluorophosphate anion for the extraction of polycyclic aromatic hydrocarbons. Compared to other commonly used ILs, such ILs are substantially more hydrophobic and hydrolytically stable, making them ideal candidates for the direct immersion extraction from aqueous matrices. Chang et al. [39] obtained excellent chiral recognition ability in capillary electrophoresis, HPLC, and capillary gas chromatography by applying (R)-N,N,N-trimethyl-2-aminobutanol-bis (trifluoromethanesulfon)imidate. Armstrong et al. [40] tried different ILs as matrices for the ultraviolet matrix-assisted laser desorption ionization MS analysis of peptides, proteins, and poly(ethylene glycol) (PEG-2000), which could produce high signal intensity, resulting in lower limits of detection than comparable solid matrices.
However, to the best of our knowledge, the application of ILs in IMPs preparation for shotgun membrane proteomics analysis has not been reported. The ideal ILs for membrane protein analysis should have the properties such as high thermal stability, good solubility and solubilizing ability, good compatibility for enzymatic digestion, and easy removal before MS analysis. Since 1-butyl-3-methyl imidazolium tetrafluoroborate (BMIM BF4) can meet such requirements, in our recent work, it was applied for the analysis of IMPs extracted from rat brain. Compared to commonly used SDS, methanol, Rapigest, and urea methods, more IMPs were identified, which demonstrates that BMIM BF4 method is of high potential for the analysis of IMPs.
Materials and methods
Materials
BMIM BF4 was obtained from Shanghai Cheng Jie Chemical Co. Ltd. (Shanghai, China). Methanol and protease Inhibitor Cocktail Set I were ordered from Merck (Darmstadt, Germany). SDS was purchased from Biomol GmbH (Hamburg, Germany). Rapigest was bought from Waters Corp. (Milford, MA, USA). Urea was obtained from Shenyang Chemical Reagents (Shenyang, China). Bacteriorhodopsin from Halobacterium salinarium and trypsin TPCK-treated (bovine pancreas) were ordered from Sigma-Aldrich (St. Louis, MO, USA). Bovine serum albumin (BSA) was bought from Sino-American Biotech (Luoyang, China). Dithiothreitol (DTT) and iodoacetamide (IAA) were from Acros (Morris plains, NJ, USA). Deionized water purified by a Milli-Q system (Millipore, Milford, MA, USA) was used in all experiments. SCX beads (UNI MC5SP-500, 5 μm, 500 Å pore) were purchased from Shenzhen Nano-micro Technology Inc. (Shenzhen, China). C18 silica particles (5 μm, 200 Å pore) were bought from Bona (Tianjin, China).
Sample preparation
Rat brain (∼0.3 g) was homogenized by Tissue Tearor from Biospec Products (Bartlesville, OK, USA) in 2 mL of lysis buffer, composed of 2 M NaCl, PBS, and 1% (v/v) Protease Inhibitor Cocktail Set I, at approximately 10,000 rpm for 1 min. The suspension was centrifuged at 12,000×g for 20 min at 4 °C. With supernatant discarded, the pellet was re-extracted by vortexing in 3 mL of high pH buffer (0.1 M Na2CO3, 1% (v/v) protease inhibitor, pH 11.3). After 30 min incubation on ice, the soluble part was removed under the same centrifugation conditions. The pellet was treated again by high pH buffer, following by ultrasonication (Cole-Parmer, Vernon Hills, IL, USA) in 2 mL of 1% (v/v) protease inhibitor for 2 min on ice at power 100% pulse duration. After protein concentration measurement by Bradford method, the cell lysates were precipitated with methanol/chloroform to remove lipids. After centrifugation, the pellet was lyophilized in a SpeedVac (Thermo Fisher, San Jose, CA, USA), and stored at −80 °C before use.
Solubility measurement for bacteriorhodopsin
Equal aliquots of lyophilized bacteriorhodopsin (50 μg each) were solubilized in 80 μL of five solvent systems, including (a) 50 mM NH4HCO3 buffer, (b) 50 mM NH4HCO3 buffer containing 60% (v/v) methanol, (c) 50 mM NH4HCO3 buffer containing 2 M urea, (d) 50 mM NH4HCO3 buffer containing 0.1% (w/v) Rapigest, and (e) 50 mM NH4HCO3 buffer containing 1% (v/v) BMIM BF4. The five samples were firstly sonicated for 0.5 h in a water bath at room temperature, and then put in the water bath at 37 °C for 10 min. After vortex for 1 min, the samples were centrifuged to remove insoluble materials, and the supernatant was respectively collected, followed by quantification by Bradford method at 595 nm with BSA as the standard protein.
In-solution digestion of BSA
BSA (6 mg/mL, dissolved in 1 mL 1 M urea, 100 mM NH4HCO3, pH 8.3) was denatured at 90 °C for 10 min and reduced and alkylated with standard protocol, followed by respective dilution with different solutions, including 100 mM NH4HCO3, 60% (v/v) methanol, 0.1% (w/v) SDS, 1% (v/v) BMIM BF4, 2 M urea, and 0.1% (w/v) Rapigest, to the final protein concentration of 1 mg/mL. The above-mentioned six samples were digested by trypsin with a trypsin/protein ratio (w/w) of 1:40 at 37 °C for 0.5 h, respectively. The obtained digested products (10 μg for each sample) were run on a 12% acrylamide gel, and stained with Coomassie staining.
Protein solubilization and digestion
Lyophilized protein pellets (200 μg) were resuspended in five different buffers for trypsin digestion, respectively. BMIM BF4-assisted solubilization and proteolysis of IMPs, briefly named as BMIM BF4 method, were performed as follows: 300 μL of 1% (v/v) BMIM BF4 dissolved in 100 mM NH4HCO3 (pH 8.3) was used to re-suspend 200 μg of membrane protein pellets via vortex. The suspension was sonicated for 1 h in a water bath at room temperature, and then kept at 90 °C for 20 min for thermal denaturation. Subsequently, the sample was cooled, followed by reduction in 13 mM DTT at 56 °C for 2 h. After cysteines were alkylated in darkness in 32 mM IAA for 1 h at room temperature, tryptic digestion was performed with a trypsin/protein ratio (w/w) of 1:25 at 37 °C for 24 h. After digestion, the solution was acidified with 90% (v/v) formic acid (1% final) to stop proteolysis. Followed by centrifugation, the supernatants were desalted with a C18 solid-phase cartridge, during which ILs could be removed simultaneously. Finally, the digests were lyophilized, and re-dissolved in 80 μL of deionized water containing 0.1% (v/v) formic acid, prior to microcolumn reversed phase liquid chromatography (μRPLC)-electrospray ionization (ESI)-MS/MS analysis.
SDS and organic solvent-assisted solubilization and proteolysis of IMPs, briefly named as SDS method and methanol method, were performed according to the published protocol [7]. Rapigest-assisted solubilization and proteolysis of IMPs, briefly named as Rapigest method, were performed according to the published protocol [10] with slight modifications. Urea-assisted solubilization and proteolysis of IMPs, briefly named as urea method, were performed according to the published protocol [18]. The final digests were re-dissolved in 80 μL of deionized water containing 0.1% (v/v) formic acid, prior to μRPLC-ESI-MS/MS analysis. The details of SDS, methanol, Rapigest, and urea methods are shown in Electronic Supplementary Material “Supporting material I.”
μRPLC-ESI-MS/MS analysis
Digests obtained by the above-mentioned five methods were respectively analyzed by duplicate μRPLC-ESI-MS/MS experiments according to our previous protocols [43]. μRPLC was performed on a paradigm GM4 μHPLC system (Michrom Bioresources, Auburn, CA, USA), and ESI-MS/MS was performed on LCQDUO (Thermal-Fisher, San Jose, CA, USA). A homemade C18 column (300 μm i.d. ×35 cm) was used for peptides separation, with the flow rate of 5 μL/min. Two percent (v/v) acetonitrile with 0.1% (v/v) formic acid (buffer A) and 98% (v/v) acetonitrile with 0.1% (v/v) formic acid (buffer B) were used to generate a 420-min gradient, set as follows: 0% B for 10 min, to 5% B in 10 min, to 40% B in 380 min, to 80% B in 10 min, and kept at 80% B for 10 min. The sample injection volume was 20 μL for each μRPLC-ESI-MS/MS analysis.
LCQ mass spectrometer was operated at positive ion mode. The spray voltage was 2.5 kV, and the heated capillary temperature was 150 °C. Total ion current chromatograms and mass spectra covering the mass range from m/z 400 to 2,000 were recorded with Xcalibur software (version 1.4). MS/MS spectra were acquired by data-dependent acquisition mode with three precursor ions selected from one MS full scan. Precursor selection was based on parent ions intensity, and the normalized collision energy for MS/MS scanning was 35%. μRPLC solvent gradient and mass spectrometer scan were controlled by the Xcalibur data system (version 1.4).
2D-nanoSCX-RPLC-ESI-MS/MS analysis
The membrane protein pellets extracted from rat brain were solubilized and digested according to 1% (v/v) BMIM BF4 method, as mentioned in “Protein solubilization and digestion” section, and about 160 μg digested products were obtained. After lyophilized, the digests were re-dissolved in 100 μL of 2% (v/v) acetonitrile with 0.1% (v/v) formic acid, prior to 2D-nanoSCX-RPLC-ESI-MS/MS analysis.
LTQ coupled with a quaternary Surveyor pump (Thermo-Fisher, San Jose, CA, USA) were used for 2D-nanoSCX-RPLC-ESI-MS/MS, similar with previous report [44]. Water solution with 0.1% (v/v) formic acid (buffer A′), acetonitrile with 0.1% (v/v) formic acid (buffer B′), and 2,000 mM NH4Ac (pH 3) were applied. The optimized flow rate was ∼200 nL/min.
The tryptic digest was loaded automatically onto an SCX trap column (2 cm, 150 μm i.d./375 μm o.d.) packed with polystyrene SCX particles (5 μm, 500 Å). Then, a series of stepwise elution with 20, 60, 80, 100, 140, 180, 300, 500, and 1,000 mM NH4Ac was used to elute peptides onto a home-packed reversed phase capillary column (14 cm, 75 μm i.d./375 μm o.d.) packed with C18 silica particles (5 μm, 300 Å), and each salt step lasted 15 min. After the nanoRPLC system was re-equilibrated for 5 min with buffer A′, a binary gradient was applied, from 0% to 10% B′ for 10 min, to 40% B′ for 75 min, and to 80% B′ for 5 min. After each run, the column was flushed by 80% B′ for 15 min, and further equilibrated with A′ for 15 min. The 2D-nanoSCX-RPLC-ESI-MS/MS analysis was run in triplicates, and the injection volume was 20 μL (32 μg) for each run.
LTQ was operated at positive ion mode. The temperature of the ion transfer capillary was set at 180 °C. The spray voltage was set at 2.2 kV, and the normalized collision energy was set at 35.0%. One microscan was set for each MS and MS/MS scan. All MS and MS/MS spectra were acquired in the data dependent mode. The mass spectrometer was set that one full MS scan was followed by nine MS/MS scans on the nine most intense ions. The dynamic exclusion function was set as follows: repeat count 1, repeat duration 30 s, and exclusion duration 180 s. System control and data collection were done by Xcalibur software (version 2.0).
Database search and interpretation of MS/MS data sets
For μRPLC-ESI-MS/MS analysis, protein identification was performed using BioWorks Software 3.1 for LCQ with SEQUEST search program. The rat database was ipi.RAT.v3.26. fasta (41494 protein entries), and revered sequences were appended to the database for the evaluation of false discovery rate (FDR). Cysteine residues were searched as static modification of 57.0215 Da. Peptides were searched using fully tryptic cleavage constraints, and up to two internal cleavages sites were allowed for tryptic digestion. The mass tolerances were 2 Da for parent masses and 1 Da for fragment masses. Peptides were considered as positive identification if Xcorr was higher than 1.9 for singly charged peptides, 2.2 for doubly charged peptides, and 3.75 for triply charged peptides. ∆Cn cutoff value was set to control the FDR of peptide identification less than 3%, determined by the calculation based on the reversed database. FDRs were calculated by using the following equation: FDR = n(rev)/n(forw), where n(forw) and n(rev) are the number of peptides identified in proteins with forward (normal) and reversed sequence, respectively.
For 2D-nanoSCX-RPLC-ESI-MS/MS analysis, protein identification was performed using BioWorks Software 3.3.1 for LTQ with SEQUEST search program. Cysteine residues were searched as static modification of +57.0215 Da, and methionine residues were searched as variable modifications of +15.9949 Da. Other parameters for database searching were same as those for μRPLC-ESI-MS/MS analysis. After database searching, to control the FDR and reduce the apparent redundancy in protein identification, our results were aligned by a software tool (BuildSummary) using the following criterion: ∆Cn value was set at 0.1, and Xcorr values were adjusted to ensure the FDR of peptide identification less than 1%. When the same peptides were assigned to multiple proteins, the assigned proteins were clustered into a “protein group” [45]. For bioinformatics analysis, only the protein with the highest sequence coverage in each “protein group” was analyzed.
Bioinformatics analysis
For results obtained by μRPLC-ESI-MS/MS analysis, the TMDs of the identified proteins were predicted according to the algorithm at http://psort.ims.u-tokyo.ac.jp/form.html. For results obtained by 2D-nanoSCX-RPLC-ESI-MS/MS analysis, the TMHMM (www.cbs.dtu.dk/services/TMHMM/) algorithm was used to predict TMDs from identified proteins. Proteins with at least one predicted TMD are regarded as IMPs. The cellular components and molecular functions based on the Gene Ontology (GO) consortium were assigned with GoMiner. Grand average of hydropathicity (GRAVY) values and isoelectric points (pIs) of the identified peptides were calculated with the ExPASy Server tool: ProtParam (http://expasy.org/tools/protparam.html).
Results and discussion
ILs are of great capacity to interact with different solutes via all possible types of interaction [33, 34] and might be helpful to solubilize IMPs. Besides good solubility, the choice of ILs for membrane protein analysis should follow additional two criterions: compatibility with tryptic digestion and facility to remove before MS/MS analysis.
Effect of additives on solubilizing ability
In order to compare the solubilizing ability of different additives for IMPs, bacteriorhodopsin was chosen as the sample, and 50 mM NH4HCO3 buffers, without and respectively with 60% (v/v) methanol, 2 M urea, 0.1% (w/v) Rapigest, and 1% (v/v) BMIM BF4 added, were chosen as solvent systems. Firstly, to eliminate the effect of the four additives (60% (v/v) methanol, 2 M urea, 0.1% (w/v) Rapigest, and 1% (v/v) BMIM BF4) on the protein concentration measurement by Bradford method at 595 nm, the absorbance of 50 mM NH4HCO3 buffer containing the four kinds of additives was measured, which was further compared with that of 50 mM NH4HCO3 buffer. The results showed that the additives were of similar absorbance to 50 mM NH4HCO3, which indicated that these additives did not affect the protein concentration measurement by Bradford method. Then, by Bradford method, the measured soluble protein concentrations were respectively as 0.03, 0.087, 0.093, 0.086, and 0.159 mg/mL. With the solubilizing ability of 50 mM NH4HCO3 buffer set as 1, that of the rest solvent systems was 2.9, 3.1, 2.9, and 5.3, respectively, which indicated that the solubilizing ability of 1% (v/v) BMIM BF4 is the best among such five solvent systems.
The good solubilizing ability of BMIM BF4 for IMPs is due to its special features. ILs are capable of most types of interactions (e.g., dispersive, π–π, n–π, hydrogen bonding, dipolar, and ionic/charge-charge) [34]. BMIM BF4 is composed of 1-butyl-3-methyl imidazolium (cations) and BF4 (anions). In water solution, because of hydrogen-bonding structure between imidazolium cations and BF4 anions and π–π interaction of imidazole rings, hydrogen-bonded nanostructures might present, which contain hydrophobic domains (alkyl chains) and hydrophilic domains (ions) [46]. Therefore, IMPs might be solubilized by the hydrogen-bonded nanostructures through hydrophobic, ionic, and π–π interactions. Such interaction might exist between alkyl chains, ions, imidazole rings, and IMPs, respectively. In addition, the hydrogen bonding in the nanostructures might be helpful to stabilize IMPs.
Effect of additives on trypsin activity
To study the effects of additives, including methanol, urea, BMIM BF4, Rapigest, and SDS, on trypsin activity, BSA was chosen as the sample. For comparison, 100 mM NH4HCO3 (pH 8.3), commonly used buffer for trypsin digestion, was also used as an additive for contrast. Through the digests analysis by SDS-polyacrylamide gel electrophoresis (shown in Fig. S1 in Electronic Supplementary Material “Supporting material I”), it was found that with 1% (v/v) BMIM BF4 and 2 M urea as additives, there was no evident BSA band, which indicated that BSA was completely digested into peptides, similar to that obtained with 100 mM NH4HCO3 as additive. With 0.1% (w/v) Rapigest as additive, there was no evident BSA band, but there was only one band between 20,100 and 14,400 Da. However, with 60% (v/v) methanol and 0.1% (w/v) SDS as additives, the digestion of BSA was not sufficient, and many bands between 66,200 and 14,400 Da were observed. These results demonstrate that the compatibility of 1% (v/v) BMIM BF4, 2 M urea, and 0.1% (w/v) Rapigest with trypsin is better than that of 60% (v/v) methanol and 0.1% (w/v) SDS. Furthermore, because of very weak retention on C18 particles, BMIM BF4 could be removed easily with a C18 trap column by 2% (v/v) acetonitrile washing, making it a good candidate for IMPs analysis.
Comparison of identified IMPs with different sample preparation methods
With the experimental setup shown in Fig. 1, insoluble proteins (200 μg) extracted from rat brain were respectively solubilized and digested with methanol, SDS, BMIM BF4, urea, and Rapigest methods, followed by the further analysis by μRPLC-ESI-MS/MS in duplicate runs. The base peak spectrums are shown as Fig. S2 in Electronic Supplementary Material “Supporting material I,” and the identified proteins and peptides are shown in Electronic Supplementary Material “Supporting material II.”
As shown in Table 1, although by different methods, the ratios of identified IMPs versus total proteins were similar, ranging from 26% to 36%, which might be determined by the IMPs extraction method. In addition, compared with SDS and methanol methods, over three times improvement on the identified IMPs number was obtained with BMIM BF4 method, which might be due to its high solubilizing ability, good compatibility with trypsin, and easy removal before MS/MS identification.
Compared to Rapigest and urea methods, with BMIM BF4 method, not only more peptides and proteins were identified but also the number of identified IMPs was improved by 25% and 80%, respectively. The results might be due to three reasons. Firstly, from the results of solubilizing ability for bacteriorhodopsin, it could be concluded that 1% (v/v) BMIM BF4 is of better solubilizing ability than those of 2 M urea and 0.1% (w/v) Rapigest. Secondly, when heated above 37 °C, urea could cause problematic carbamylation of N-termini and lysine residues. However, BMIM BF4 is of high thermal stability, avoiding problematic modifications. Thirdly, for the removal of Rapigest before MS analysis, the cleavage step at low pH could result in hydrophobic peptides loss due to co-precipitation with the hydrophobic part of RapiGest [28]. However, BMIM BF4 could be removed easily with a C18 trap column before MS analysis, avoiding loss of hydrophobic peptides.
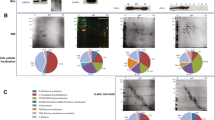
The overlapping of identified IMPs by BMIM BF4, SDS, and methanol methods is shown in Fig. 2a. A total of 99 IMPs are identified by such three methods, and IMPs identified by methanol and SDS methods are, respectively, 74% and 85% covered by those identified by BMIM BF4 method. In addition, the overlapping of identified IMPs by BMIM BF4, Rapigest, and urea methods is shown in Fig. 2b. A total of 122 IMPs are identified by such three methods, and IMPs identified by Rapigest and urea methods are, respectively, 71% and 74% covered by those identified by BMIM BF4 method. All these results show that BMIM BF4 method is better than other four commonly used methods for the analysis of IMPs extracted from rat brain.
In addition, the identified IMPs in the insoluble fraction of rat brain by μRPLC-ESI-MS/MS with different methods were compared with respect to the number of TMDs, predicted based on the algorithm at http://psort.ims.u-tokyo.ac.jp/form.html. As illustrated in Fig. 3, the identified TMDs could be divided into four sections: [1, 3), [3, 5), [5, 8), and [8, ∞), and IMPs identified by BMIM BF4 method are distributed in all sections, with number higher than those identified by other methods.
Besides comparisons of identified IMPs, the peptides identified with the five methods were also compared with respect to the GRAVY values, as shown in Fig. 4. Interestingly, by these five methods, similar peptide distribution profilings were obtained. However, with BMIM BF4 method, more hydrophilic peptides (peptides with negative GRAVY values) and hydrophobic peptides (peptides with positive GRAVY values) were identified, when compared with those obtained by other methods. Especially, two peptides with GRAVY values higher than 1.5 were identified with BMIM BF4 method. These results demonstrated that BMIM BF4 method might be beneficial to the identification of peptides with high hydrophobicity.
Although by μRPLC-ESI-MS/MS with an LCQ MS the identified peptides and IMPs number are not very high, the above-mentioned results demonstrate that compared to other presently applied methods, sample preparation with BMIM BF4 method is of superiority to study IMPs from rat brain.
Global analysis of IMPs in rat brain analyzed by 2D-nano-SCX-RPLC-ESI-MS/MS
To achieve the global IMPs analysis in rat brain, 32 μg of tryptic digests of membrane protein pellet from rat brain was automatically injected onto a 2-cm-long SCX trap column, eluted by nine salt steps, and analyzed by nano-RPLC-ESI-MS/MS, with the total analysis time of 21 h. The base peak spectrums are shown as Fig. S3 in Electronic Supplementary Material “Supporting material I.” After database searching, in triplicate runs, in total, 1,450 non-redundant proteins and 7,978 unique peptides were identified from rat brain, and 418 proteins contained at least one predicted TMD, with FDR less than 1% for peptide identification and at least two identified unique peptides per protein. The details of proteins and peptides identification are shown in Electronic Supplementary Material “Supporting material III.”
For the identified non-redundant IMPs, the distribution of identified IMPs with respect to the number of TMDs is shown in Fig. 5 and compared with those from IPI RAT database version 3.26. With at least two identified unique peptides per protein (gray bars), 418 IMPs were identified, and about 46% (194) of total IMPs are of one predicted TMD, and about 54% (224) are of two to 19 TMDs. For the IPI RAT database version 3.26 (white bars), 10,332 proteins contain at least one TMD. With respect to the number of TMDs, the percentages of identified IMPs with one to four TMDs were higher compared with IPI RAT database version 3.26, and the percentage of identified IMPs with six or seven TMDs was lower.
The distributions of cellular components and molecular functions of identified proteins were further analyzed, according to Gene Ontology (GO) information obtained with GoMiner, and the comparison with the IPI RAT database version 3.26 was also made. As shown in Fig. S4 in Electronic Supplementary Material “Supporting material I,” for the identified proteins with at least two unique peptides per protein (gray bars), 67.14% of identified annotated proteins were mapped on membrane, among which the ratios of macromolecular complex, Golgi apparatus, cytoplasmic membrane-bounded vesicle, endoplasmic reticulum, and mitochondrion were higher than those obtained with IPI RAT database version 3.26 (white bars). In addition, as illustrated in Fig. S5 in Electronic Supplementary Material “Supporting material I,” for the proteins identified with at least two unique peptides per protein (gray bars), 87.95% of the identified annotated proteins are of binding function, followed by catalytic activity (55.94%), transporter activity (20.81%), molecular transducer activity (11.29%), and structural molecule activity (9.94%), among which the percentages of proteins with binding function, catalytic activity, and transporter activity were higher compared with IPI RAT database version 3.26 (white bars).
As shown in Table 2, in total, four voltage-gated calcium ion channels, seven voltage-gated potassium ion channels, and two voltage-gated sodium ion channels were identified from rat brain. All these results show that ILs method, with BMIM BF4 added in sample preparation buffer, is powerful to discover ionic channels on membranes.
The GRAVY values and pIs of the identified unique peptides were also analyzed. As shown in Fig. 6, the peptides are mainly distributed in three sections: (−1.0, −0.5], (−0.5, 0], and (0, 0.5]. The GRAVY values of the identified unique peptides range from −2.958 to 2.65, which indicates that with BMIM BF4 method, peptides with high hydrophobicity and hydrophilicity could be identified efficiently. As shown in Fig. 7, most of identified unique peptides are acidic peptides (pI < 7) and are mainly distributed in (4, 5] and (5, 6] sections. The pIs of identified unique peptides range from 3.33 to 12.3, which shows that with BMIM BF4 method, acidic and basic peptides could be identified efficiently.
Furthermore, with BMIM BF4 method, 47 peptides containing partial or whole TMD were identified and recognized as transmembrane peptides, as shown in Table S1 in Electronic Supplementary Material “Supporting material I.” The TMDs number of IMPs identified with transmembrane peptides ranges from one to 12, and eight and three IMPs are of one and 12 TMDs, respectively. These results show that with BMIM BF4 method, transmembrane peptides could be identified from IMPs with low and high TMD, which is important and challenging for IMPs analysis.
Conclusions
IL, BMIM BF4, added in NH4HCO3 buffer (pH 8.3) was successfully applied for analysis of the IMPs extracted from rat brain. The experimental results indicate that compared to other commonly used membrane proteins treatment methods, with BMIM BF4 method, more IMPs could be identified, not only due to high thermal stability, good solubilizing ability, and compatibility for trypsin digestion but also benefited from easy removal during desalting so that the interference on MS/MS detection is ignorable. By such a method, the large-scale and comprehensive analysis of IMPs could be easily achieved. Furthermore, the systematic study on the applicability of other kinds of ILs for IMPs analysis is undergoing, which might be helpful to discover the in-depth mechanisms about the interaction between ILs and IMPs and find other suitable ILs for membrane proteomics study.
References
Ott CM, Lingappa VR (2002) J Cell Sci 115:2003–2009
Torres J, Stevens TJ, Samsó M (2003) Trends Biochem Sci 28:137–144
Braun RJ, Kinkl N, Beer M, Ueffing M (2007) Anal Bioanal Chem 389:1033–1045
Wu CC, Yates JR 3rd (2003) Nat Biotechnol 21:262–267
Rabilloud T (2003) Nat Biotechnol 21:508–510
Shaw AR, Li L (2003) Curr Opin Mol Therap 5:294–301
Zhang N, Chen R, Young N, Wishart D, Winter P, Weiner JH, Li L (2007) Proteomics 7:484–493
Washburn MP, Wolters D, Yates JR 3rd (2001) Nat Biotechnol 19:242–247
Wu CC, MacCoss MJ, Howell KE, Yates JR 3rd (2003) Nat Biotechnol 21:532–538
Chen EI, McClatchy D, Park SK, Yates JR 3rd (2008) Anal Chem 80:8694–8701
Wang N, MacKenzie L, De Souza AG, Zhong H, Goss G, Li L (2007) J Proteome Res 6:263–272
Nielsen PAa, Olsen JV, Podtelejnikov AV, Andersen JR, Mann M, Wiśniewski JR (2005) Mol Cell Proteomics 4:402–408
Zhou J, Lin Y, Deng XC, Shen JY, He QY, Chen P, Wang XC, Liang SP (2008) J Proteome Res 7:1778–1783
Chick JM, Haynes PA, Bjellqvist B, Baker MS (2008) J Proteome Res 7:4974–4981
Speers AE, Wu CC (2007) Chem Rev 107:3687–3714
Wei J, Sun J, Yu W, Jones A, Oeller P, Keller M, Woodnutt G, Short JM (2005) J Proteome Res 4:801–808
Zhang H, Lin Q, Ponnusamy S, Kothandaraman N, Lim TK, Zhao C, Kit HS, Arijit B, Rauff M, Hew CL, Chung MCM, Joshi SB, Choolani M (2007) Proteomics 7:1654–1663
Ruth MC, Old WM, Emrick MA, Meyer-Arendt K, Aveline-Wolf LD, Pierce KG, Mendoza AM, Sevinsky JR, Hamady M, Knight RD, Resing KA, Ahn NG (2006) J Proteome Res 5:709–719
Lu A, Wiśniewski JR, Mann M (2009) J Proteome Res 8:2418–2425
Blackler AR, MacCoss MJ, Wu CC (2007) In 55th ASMS Conference on Mass Spectrometry, 3–7 June, Indianapolis, IN
Speers AE, Blackler AR, Wu CC (2007) Anal Chem 79:4613–4620
Blonder J, Terunuma A, Conrads TP, Chan KC, Yee C, Lucas DA, Schaefer CF, Yu LR, Issaq HJ, Veenstra TD, Vogel JC (2004) J Invest Dermatol 123:691–699
Chick JM, Haynes PA, Molloy MP, Bjellqvist B, Baker MS, Len ACL (2008) J Proteome Res 7:1036–1045
Blonder J, Goshe MB, Moore RJ, Pasa-Tolic L, Masselon CD, Lipton MS, Smith RD (2002) J Proteome Res 1:351–360
Blonder J, Conrads TP, Yu L-R, Terunuma A, Janini GM, Issaq HJ, Vogel JC, Veenstra TD (2004) Proteomics 4:31–45
Ye XY, Johann DJ Jr, Hakami RM, Xiao Z, Meng ZJ, Ulrich RG, Issaq HJ, Veenstra TD, Blonder J (2009) J Proteomics 73:112–122
Blackler AR, Speers AE, Ladinsky MS, Wu CC (2008) J Proteome Res 7:3028–3034
Masuda T, Tomita M, Ishihama Y (2008) J Proteome Res 7:731–740
Yu Y-Q, Gilar M, Gebler JC (2004) Rapid Commun Mass Spectrom 18:711–715
Berthod A, Ruiz-Ángel MJ, Carda-Broch S (2008) J Chromatogr A 1184:6–18
Yao C, Pitner WR, Anderson JL (2009) Anal Chem 81:5054–5063
Fitzwater G, Geissler W, Moulton R, Plechkowa NV, Robertson A, Seddon KR, Swindall J, Joo KW (2005) Ionic liquids: sources of innovation, Report Q002. QUILL, Belfast
Poole CF (2004) J Chromatogr A 1037:49–82
Anderson JL, Ding J, Welton T, Armstrong DW (2002) J Am Chem Soc 124:14247–14254
Plaquevent J-C, Levillain J, Guillen F, Malhiac C, Gaumon A-C (2008) Chem Rev 108:5035–5060
Wang J, Pei Y, Zhao Y, Hu Z (2005) Green Chem 7:196–202
Wang J-H, Cheng D-H, Chen X-W, Du Z, Fang Z-L (2007) Anal Chem 79:620–625
Polyakova Y, Jin YZ, Zheng JZ, Row KH (2006) J Liquid Chromatogr Relat Technol 29:1687–1701
Yuan LM, Han Y, Zhou Y, Meng X, Li Y, Zi M, Chang YX (2006) Anal Lett 39:1439–1449
Armstrong D, Zhang L-K, He L, Gross M (2001) Anal Chem 73:3679–3686
Tholey A, Zabet-Moghaddam M, Heinzle E (2006) Anal Chem 78:291–297
Giménez E, Benavente F, Barbosa J, Sanz-Nebot V (2010) Anal Bioanal Chem 398:357–365
Tao D, Zhu G, Sun L, Ma J, Liang Z, Zhang W, Zhang L, Zhang Y (2009) Proteomics 9:2029–2036
Wang F, Dong J, Jiang X, Ye M, Zou H (2007) Anal Chem 79:6599–6606
Zhou H, Dai J, Sheng QH, Li RX, Shieh CH, Guttman A, Zeng R (2007) Electrophoresis 28:4311–4319
Dupont J (2004) J Braz Chem Soc 15:341–350
Acknowledgements
The authors are grateful for the financial support from National Basic Research Program of China (2007CB914100), National Natural Science Foundation (20935004), and Knowledge Innovation Program of Chinese Academy of Sciences (KJCX2YW.H09).
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the special issue Analytical and Bioanalytical Science in China with Guest Editors Lihua Zhang, Qiankun Zhuang, and Yukui Zhang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 3.70 MB)
Rights and permissions
About this article
Cite this article
Sun, L., Tao, D., Han, B. et al. Ionic liquid 1-butyl-3-methyl imidazolium tetrafluoroborate for shotgun membrane proteomics. Anal Bioanal Chem 399, 3387–3397 (2011). https://doi.org/10.1007/s00216-010-4381-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4381-5