Abstract
It has been shown that the coelenterazine analog, coelenterazine-v, is an efficient substrate for a reaction catalyzed by Renilla luciferase. The resulting bioluminescence emission maximum is shifted to a longer wavelength up to 40 nm, which allows the use of some “yellow” Renilla luciferase mutants for in vivo imaging. However, the utility of coelenterazine-v in small-animal imaging has been hampered by its instability in solution and in biological tissues. To overcome this drawback, we ligated coelenterazine-v to Ca2+-triggered coelenterazine-binding protein from Renilla muelleri, which apparently functions in the organism for stabilizing and protecting coelenterazine from oxidation. The coelenterazine-v bound within coelenterazine-binding protein has revealed a greater long-term stability at both 4 and 37 °C. In addition, the coelenterazine-binding protein ligated by coelenterazine-v yields twice the total light over free coelenterazine-v as a substrate for the red-shifted R. muelleri luciferase. These findings suggest the possibility for effective application of coelenterazine-v in various in vitro assays.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Renilla luciferase is an enzyme that is responsible for the bioluminescence of the soft coral Renilla [1]. The enzyme catalyzes the oxidative decarboxylation of coelenterazine in the presence of O2, resulting in the formation of coelenteramide in its excited state and CO2 as the reaction products [2]. The excited coelenteramide relaxes to its ground state with the production of blue light with a maximum at 480 nm. The bioluminescence in vivo results from the radiationless energy transfer from excited coelenteramide to green-fluorescent protein, which then emits green light with an emission maximum of 509 nm [3]. In addition, the Renilla bioluminescence in vivo also involves a third protein called Ca2+-triggered coelenterazine-binding protein (CBP). The CBP is a stable complex of coelenterazine and a single-chain polypeptide with molecular mass about 20 kDa [4]. The primary structure of CBP from Renilla reniformis, as determined by direct amino acid sequencing [5], reveals three canonic Ca2+-binding sites characteristic for the family of EF-hand Ca2+-binding proteins [6, 7]. On addition of calcium, the coelenterazine bound with CBP becomes available for bioluminescence reaction with luciferase, thereby linking the bioluminescence with the nerve net of coral [8]. Recently, a synthetic gene encoding R. reniformis CBP was synthesized from oligonucleotides [9] based on the published amino acid sequence of R. reniformis CBP [5], and the full-size cDNA encoding the CBP from R. muelleri (RM-Luc) has been cloned as well [10]. It was shown that Renilla luciferase bioluminescence is more efficient using the recombinant CBP with bound coelenterazine as the substrate than with coelenterazine alone [10]. The determination of crystal structures of CBP with coelenterazine [11] and apo-CBP bound with calcium ions [12] has allowed the reasonable mechanistic suggestion as to why oxygen addition to coelenterazine is inhibited within the CBP cavity, whereas it occurs readily in Ca2+-regulated photoproteins [13, 14], and as to how CBP might interact with luciferase to provide a higher bioluminescent yield of reaction.
Following the cloning of luciferase from the firefly Photinus pyralis [15], luciferase genes have become valuable components of biological research. They are used ubiquitously as reporter genes in cell culture experiments, and their use as reporters has been extended into the context of small-animal imaging [16]. The firefly type luciferases, however, are not optimal for employment in imaging. These luciferases are not particularly small (~62 kDa) and are dependent on firefly luciferin, ATP, molecular oxygen, and magnesium for activity. The dependence on ATP especially would hinder the application of these luciferases in vivo, as serum ATP concentrations are generally below 10 nM [17]. Luciferases that use coelenterazine as their substrate (e.g., from Renilla [18], Metridia [19], and Gaussia [20]) are more appropriate for application as bioluminescent reporters, as these enzymes are not ATP dependent and in general require only molecular oxygen in addition to coelenterazine for luminescence. Among them, the Renilla luciferase is the most usable [21].
The major limitation for the application of Renilla and other coelenterazine-dependent luciferases for bioluminescence imaging in small animals is that they emit blue light (~480 nm), which is preferentially absorbed by biological tissues [22]. In the case of Renilla luciferase, which has its spectral maximum at 480 nm, only ~3% of the emitted photons are of wavelength above 600 nm where tissue attenuation is minimized. Consequently, the coelenterazine-dependent luciferases have diminished sensitivity at non-superficial locations.
A few years ago, mutants of Renilla luciferase with long-wavelength shifts of bioluminescence spectrum up to 66 nm (peak at 547 nm) have been described [23]. These mutants revealed greater stability and higher light emission than native luciferase. In addition, it is notable that when the coelenterazine-v, an analog of coelenterazine, was used as a substrate, an orange-peaked emission resulted (λ max = 588 nm) [23]. Coelenterazine-v, which is a significantly more efficient substrate of Renilla luciferase than native coelenterazine, only shifts the bioluminescence of native luciferase from blue to green [24]. However, despite very promising results with coelenterazine-v, its application in vivo remains limited. This coelenterazine analog appears to be extremely unstable in solutions and exhibits an order of magnitude increase in a background autochemiluminescence in experiments in vivo. Therefore, the development of alternative coelenterazine analogs, or overcoming this coelenterazine-v instability, might be very important for developing small-animal imaging techniques using red-shifted Renilla luciferase mutants.
In the present study, we demonstrate that coelenterazine-v analog stability in vitro can be greatly increased on binding within the Ca2+-triggered coelenterazine-binding protein from R. muelleri, and that CBP with this coelenterazine analog is a more efficient substrate of a long-wavelength shifted mutant of R. muelleri luciferase than coelenterazine-v alone.
Materials and methods
Materials
Native coelenterazine was purchased from Prolume Ltd. (USA). All chemicals were used as received without further purification.
Molecular biology
To introduce the 5′-terminal TEV-specific protease site and attB1/attB2 flanking recombination sites into RM-Luc coding sequence for subsequent cloning with the Gateway system (Invitrogen), the coding sequence for native R. muelleri luciferase (NCBI accession number is EF535511) from pET-19b plasmid [10] was amplified by two-step PCR using “AccuPrime Pfx supermix” (Invitrogen). The first PCR with the gene-specific forward S-RM-TEV (5′-GAAAACCTGTATTTTCAGGGCATCATGACGTCAAAAGTTTACGATCC-3′) and reverse A-RM-attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTGTTCATT TTTAAGAACACGC-3′) primers was performed for 27 cycles (denaturation, 25 s at 95 °C; annealing, 60 s at 56 °C; extension, 60 s at 68 °C). For the second PCR, the first PCR product was used as a template and the full-size gene was amplified under a similar condition with the second forward S-attB1-TEV (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAAAACCTGTATTTTCAGGGCATC-3′) and the same reverse A-RM-attB2 primers. To generate an entry clone, pDONR221 (Invitrogen) was used as an entry vector in BP reaction of recombination.
The resulting pDONR221-RM plasmid harboring the native R. muelleri luciferase sequence was used as a template for site-directed mutagenesis in order to construct the red-shifted R. muelleri luciferase (RM-Y) mutant. The following amino acid substitutions A55T, C124A, C130A, A143M, M253L, S287L, A123T, D154M, E155G, D162E, I163L, and V185L were sequentially introduced into the RM-Luc coding sequence using designed oligonucleotide primers and Quick-Change site-directed mutagenesis kit (Stratagene). These mutations were selected based on the substitutions leading to the red-shifted bioluminescence of R. reniformis luciferase (mutant RLuc8.6-535) [23]. The plasmids harboring mutations were verified by DNA sequencing.
To generate the expression plasmid, pDEST-566 vector harboring the N-terminal His6-tag, a carrier protein–maltose-binding protein (MBP), and the attR1/attR2 sites [25] was used in LR reaction of recombination, which was performed according to the protocol supplied with the kit (Gateway Cloning, Invitrogen).
Synthesis of coelenterazine-v
Coelenterazine-v was synthesized using the described procedures [26, 27]. To produce pure coelenterazine-v, the product was passed over a Silica Gel 60 column (EMD Chemicals) and eluted with a linear gradient of methylene chloride to methanol. The pure coelenterazine-v was characterized by 1H NMR (300 MHz) in CD3OD made acidic with the addition of one drop of DCl. The chemical shifts [δ: 8.62 (d, J = 9.4, 1 H), 8.60 (d, J = 9.1, 1 H), 7.56 (d, J = 9.4, 1 H), 7.53–7.47 (m, 2 H), 7.38–7.11 (m, 6 H), 7.08 (d, J = 2.4, 1 H), 6.81–6.73 (m, 2 H), 4.55 (s, 2 H), 4.27 (s, 2 H), (phenolic OH is not observed)] of synthesized coelenterazine-v do not exactly correspond to reported values, but the acidic conditions used here likely account for the differences [28]. The calculated mass amounted to 447.48; the determined mass was 448.2 (M+) and 446.4 (M−). The ε 238 nm in acidic methanol equaled to 23,110 M−1 cm−1.
Purification of RM-Y mutant luciferase and CBP
For protein production, Escherichia coli BL21-CodonPlus (DE3)-RIPL strain (Stratagene) was transformed with plasmid isolated from LR-RM-Y clones. Transformed E. coli cells were cultivated with vigorous shaking at 37 °C in LB medium containing 200 μg/mL of ampicillin and induced with 0.5 mM IPTG when the culture reached an A 600 of 0.8–1.0. After addition of IPTG, the cultivation was continued for 8 h at 20 °C.
For purification of luciferase, the cell paste was resuspended in Ni-binding buffer (0.2 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 0.5 mM PMSF, 50 mM Na2HPO4/KH2 PO4 buffer, pH 7.1) and disrupted with ultrasound (1 min × 6) at 0 °C. The mixtures were passed over a 5-mL HisTrap Fast Flow column (Amersham Biosciences) and eluted with an imidazole gradient (0–0.5 M) in 0.3 M NaCl, 50 mM Na2HPO4/KH2PO4 buffer pH 7.1. The luciferase peak was collected and dialyzed against TEV protease cleavage buffer (1 mM DTT, 1 mM EDTA, 20 mM Tris–HCl, pH 8.0). MBP-tag was cleaved by incubation of the fusion protein with TEV protease at ratio 50:1 (w/w) overnight at room temperature; then the samples were loaded on a Ni-NTA resin column and the flow-through was collected. The collected sample of RM-Y was dialyzed against 1 mM EDTA, 20 mM Tris–HCl pH 8.0 and loaded on an anion exchange HiTrap Q column (Amersham Biosciences). The active fractions were collected and concentrated on Amicon centrifugal filters (Millipore) in 25 mM NaCl, 0.5 mM EDTA, 20 mM Tris–HCl pH 7.0.
Plasmid pET22b-CBP3 containing the gene encoding CBP from R. muelleri (NCBI accession number EF535511) [10] without any purification tags, was used to transform E. coli cells, strain BL21-CodonPlus (DE3)-RIPL (Stratagene). For protein production, transformed cells were cultivated with vigorous shaking at 37 °C in LB media containing 200 μg/mL of ampicillin and induced with 1 mM IPTG when the culture reached an A 600 of 0.8–1.0. After addition of IPTG, the cultivation was continued for 3 h at 37 °C.
Most apo-CBP produced was accumulated inside E. coli cells in inclusion bodies that can be easily isolated by centrifugation. The purification was performed as previously reported for recombinant obelin [29] with modification as described in Ref. [10]. The apo-CBP sample in 6 M urea obtained after chromatography on DEAE Sepharose Fast Flow was refolded by desalting on 26/10 HiPrep Desalting column (Amersham Biosciences) in 5 mM EDTA, 20 mM Tris-HCI pH 7.0 buffer, and then mixed with coelenterazine-v (or native coelenterazine) in an equimolar concentration to protein. The mixture was incubated for 3 h at 4 °C, followed by ion-exchange chromatography on UnoQ-10 column (Bio-Rad) to separate CBP from apo-protein and from unbound substrate. The fraction of CBP was collected and concentrated on Amicon centrifugal filters (Millipore) in 25 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl pH 7.0. Protein concentration was measured with DC Bio-Rad protein assay kit. According to SDS-PAGE analysis, the RM-Y luciferase mutant and CBP were of high purity.
Bioluminescence assay
The bioluminescence was measured at room temperature with a Mithras LB940 plate luminometer (Berthold Technologies) by injecting 10 μL of RM-Y (17.8 ng/mL) into the wells containing 100-μL solution of different concentrations of (a) coelenterazine-v in 25 mM NaCl, 0.5 mM EDTA, 50 mM Tris–HCl pH 7.0; or (b) CBP–coelenterazine-v in 25 mM NaCl, 50 mM CaCl2, 50 mM Tris–HCl pH 7.0. Luminescence data were corrected for a methanol inhibition effect (5–25%) on RM-Y activity. Coelenterazine-v in methanol was prepared by dilution from stock solution of coelenterazine-v in methanol. The concentration was determined using the extinction coefficient for coelenterazine-v (ε 238 nm = 23,110 M−1 cm−1) in methanol. The specific bioluminescent activity was determined as integral signal (integration time 20 s).
Thermostability assays
Thermostability was assayed using the purified CBP–coelenterazine-v (0.52 mg/mL in 25 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl pH 7.0) by incubating samples at 37 or 4 °C. Aliquots of 10 μL were removed over periods of time and assayed with RM-Y luciferase. For measurement, the CBP–coelenterazine-v aliquot was mixed in luminometer cuvette with 500 μL of RM-Y luciferase in 25 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl pH 7.0, and then the bioluminescence reaction was initiated by rapid injecting 200 μL of 40 mM CaCl2 in 50 mM Tris–HCl pH 7.0. The coelenterazine-v stability in methanol was tested by storing stock solution (0.24 mM) at −20 °C. Aliquots of 5 μL were removed over periods of time and tested with RM-Y luciferase by rapid injection of 5-μL aliquot into cuvette with 700 μL of RM-Y luciferase in the same buffer as above. In both cases, the RM-Y luciferase concentration in cuvette was 9 ng/mL.
Spectral measurements
Bioluminescence spectra were measured with an AMINCO spectrofluorimeter (Thermo Spectronic, USA) using a slit width of 4 nm and a scan rate of 16 nm/s. All spectra were corrected for intensity change over the scan and for the spectral sensitivity of the instrument with the program supplied by AMINCO. The absorption spectra were obtained with a UVIKON 943 Double Beam UV/VIS spectrophotometer (Kontron Instruments, Italy). The measurements were carried out at room temperature.
Calcium concentration–effect curve
The measurements were carried out as previously described [10, 30] with EDTA-free solutions of CBP and luciferase. EDTA was removed from the CBP and luciferase by gel filtration on a DSalt plastic column (Pierce). The column was equilibrated and eluted with 150 mM KCl, 5 mM PIPES, pH 7.0, which had been previously passed two times through freshly washed beds of Chelex-100 chelating resin. The fractions containing either CBP or luciferase were identified by the bioluminescence assay. To avoid possible contamination with EDTA, only the first few protein fractions to come off the column were used for the determination of Ca2+ concentration–effect curves. Ca–EGTA buffers (total [EGTA] = 2 mM) were used to establish Ca2+ concentrations below 10−5 M, and simple dilutions of CaCl2 (in a Chelex-scrubbed solution of 150 mM KCl, 5 mM PIPES, pH 7.0) for the range of Ca2+ concentrations from 10−6 to 10−2 M. The Ca2+ buffers were prepared by the two stock-solution method described elsewhere [31]. The peak light intensity (L) was measured after 10 μl of CBP solution (2.32 × 10−8 M) was injected into 1 mL of the test solution that contained a constant amount of luciferase (1.16 × 10−9 M), so that CBP and luciferase in the cuvette were in the ratio of 20:1. All measurements were done at 20 °C maintained with the temperature-stabilized cuvette block of the luminometer.
Results and discussion
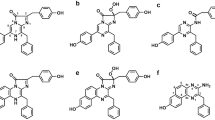
The chemical structure of coelenterazine-v differs from that of the native coelenterazine by an additional –CH═CH– group bridging C5 and C20 atoms (Scheme 1). The introduction of an additional conjugated double bond leads to a planar conformation of the 6-(p-hydroxy) phenyl group relative to pyrazine ring and results in an increase of rigidity of the overall structure of the molecule. This steric difference of coelenterazine-v when compared with the native coelenterazine drastically decreases the bioluminescence light output of Ca2+-regulated photoproteins and Oplophorus luciferase, but significantly increases the efficiency of the reaction catalyzed by Renilla luciferase [24]. There was no detectable effect of structure changes on binding coelenterazine-v to apo-CBP. The apo-CBP binds coelenterazine-v very rapidly from Ca2+ free solution, similar to that as the native coelenterazine, thus implying there that is sufficient space to accommodate this coelenterazine analog within the CBP cavity despite the presence of the additional –CH═CH– group (Fig. 1). Probably for the same reason, the analog coelenterazine-e, which has –CH2–CH2– linker in place of the –CH═CH–, also effectively ligates to apo-CBP [9]. Consequently, the procedure for preparation of CBP bound with coelenterazine-v required only small modifications from that previously reported for CBP binding with native coelenterazine [10]. In the first step, the purified apo-CBP was refolded and then incubated with coelenterazine-v. To yield a homogeneous holoprotein free of apo-CBP and unbound coelenterazine-v, it was additionally purified on a UnoQ anion exchange column. It should be noted that DTT also was not required for the conversion of apo-CBP to CBP bound with coelenterazine-v. The absorption spectrum of the homogeneous CBP loaded with coelenterazine-v displays two main maxima at 286/462 and two minor maxima at 350/380 nm (Fig. 2a). In addition, there are two apparent shoulders at 310 and 490 nm. The 286 nm/462 nm absorption ratio amounts to 1.6. The absorbance maximum of coelenterazine-v bound with protein is red-shifted from that in methanol (λ max = 455 nm). This effect might be caused by a more hydrophobic environment of coelenterazine-v within the protein. Extinction coefficients of CBP with coelenterazine-v were determined as ε 285 nm = 16.2 mM−1 cm−1 and ε 462 nm = 10.6 mM−1 cm−1. Addition of calcium to CBP leads to conformational changes in the protein making coelenterazine-v solvent exposed and available for reaction with Renilla luciferase. In the presence of Ca2+, the visible maximum is shifted to 450 nm and the absorbance decreases slowly to zero after less than 3 h of incubation at 4 °C. This effect is attributed to coelenterazine-v dissociation and autooxidation (Fig. 2b).
a Absorption spectra of purified CBP–coelenterazine-v in EDTA-buffer and freshly prepared coelenterazine-v in methanol (gray lines). b Changes of absorption spectrum of CBP–coelenterazine-v on calcium addition. Arrows show increase (at 375 nm) and decrease (at 460 nm) of coelenterazine-v absorbance bands; 0–3 h, time after calcium addition. All spectra are normalized at the short wavelength maximum
The luciferases emitting a large percentage of photons in red to near-infrared wavelengths (600–900 nm) have advantages in bioluminescent imaging in intact animals. Hence, the examination of CBP bound with coelenterazine-v as a bioluminescent substrate was carried out using the pure R. muelleri luciferase mutant (RM-Y), which has a red-shifted bioluminescence spectrum. To express Renilla luciferase mutant as a soluble protein, RM-Y was fused with maltose-binding protein, which greatly enhances solubility of the proteins upon their expression in E. coli cells [25]. With this approach, the yield of the pure RM-Y mutant amounts to ~50–60 mg/L of cell culture.
Figure 3 shows the spectra of Ca2+-triggered bioluminescence of RM-Y luciferase mutant with CBP, loaded with either coelenterazine-v or native coelenterazine, as a substrate. As expected, the use of CBP bound with coelenterazine-v produces a longer-wavelength bioluminescence (λ max = 574 nm) as compared with CBP loaded with native coelenterazine (λ max = 535 nm). Visually, the bioluminescence has the yellow-orange and green-yellowish color, respectively (Fig. 3b). It needs to be noted that the reaction of the RM-Y luciferase mutant with the free coelenterazine-v or native coelenterazine produces identical bioluminescence spectra to those when the substrates are CBP bound (data not shown). It is interesting that the bioluminescence spectrum with coelenterazine-v shows a diminished contribution of a shoulder at 420 nm, which corresponds to the excited electronic state of coelenteramide in neutral form (Fig. 3a). It should also be noted that the RM-Y mutant of R. muelleri luciferase has a 25% greater activity and improved thermostability at 37 °C, similar to the RLuc8.6-535 mutant of R. reniformis luciferase [23].
a Normalized bioluminescence spectra of Ca2+-triggered RM-Y luciferase mutant with CBP bound with native coelenterazine (greenish) and CBP bound with coelenterazine-v (orange). b Photo of calcium-triggered (40 mM) luminescence of RM-Y luciferase mutant (0.4 μg/mL) mixed with CBP bound with either native coelenterazine (left) or coelenterazine-v (right)
Figure 4 shows the concentration dependence of the bioluminescence integrated intensity when RM-Y mutant is assayed with either CBP bound with coelenterazine-v or free coelenterazine-v. The total emitted light with CBP is about 2.2 times higher than that with free coelenterazine-v over most of the range of substrate concentrations, i.e., this difference is practically the same as obtained for CBP loaded by native coelenterazine and wild-type R. muelleri luciferase [10].
Log–log plots of RM-Y luciferase bioluminescence assay with CBP–coelenterazine-v (empty circles) or free coelenterazine-v (filled circles). RLU is integral bioluminescence intensity in relative light units. The luciferase concentration is 4.9 nM. The data on the plots are average of three measurements
Generally, the implementation of imaging experiments in vivo requires reporter enzymes and compounds which are sufficiently stable at 37 °C. As mentioned above, one of the limitations of the use of coelenterazine-v is its instability in solutions. Hence, we also tested whether the binding of coelenterazine-v with CBP stabilizes this coelenterazine analog in solution. For that, CBP bound with coelenterazine-v was incubated both at 4 and 37 °C. The coelenterazine-v diluted in methanol and stored in the dark at −20 °C was used as a control. The quantity of the residually active coelenterazine-v was determined in a bioluminescence assay with RM-Y mutant luciferase. A methanol solution of coelenterazine-v possesses poor stability even when kept at low temperature; there was only ~1% of residual activity detected after 9 days of storage at −20 °C (Fig. 5a). In contrast, the coelenterazine-v bound within CBP (0.52 mg/mL) is stable in 1 mM EDTA, 25 mM NaCl, 50 mM Tris–HCl pH 7.0 over half a year at 4 °C and preserves about 80% activity after 32 h of incubation even at 37 °C (Fig. 5b). It should be noted that there was no effect of room light on the stability of the coelenterazine-v bound in CBP.
It is well-known that free coelenterazine and its analogs are unstable in neutral aqueous solution, undergoing a slow autooxidation over several hours [14]. In an aprotic solvent, coelenterazine is much less stable, reacting with oxygen to generate chemiluminescence. As the “aprotic reaction” is triggered by base, the reaction mechanism is proposed to be by oxygen attack at C2 on the coelenterazine-N7 anion [32, 33]. It has been proposed that in the CBP coelenterazine-binding pocket, hydrogen bonding should result in a net dipole across the ring system, as the C3-oxygen is an acceptor resulting in its obtaining a partial positive charge and at the other end of the molecule, the 6-(p-hydroxy) phenyl group and the N7, are H-bond donors, leaving them with partial negative charge [11]. Thus, the partial negative charge on the coelenterazine-N7 bound in CBP should render this position much less acidic than in free solution. Apparently, the same mechanism of stabilization has been implemented in CBP bound with coelenterazine-v. Interactions within the binding site shift the pK and thus reduce the probability of the coelenterazine-v anion formation, the target of oxygen attack.
Figure 6 shows two representations of Ca2+ concentration–effect curve for the mixture of CBP–coelenterazine-v and RM-Y mutant luciferase. The curves are log–log plots with light intensities expressed in terms of the ratio either L/L max or L/L int. The utility of representation of Ca2+ concentration–effect curves in terms of the ratio L/L int has already been discussed elsewhere [34]. The peak light intensity (L) is measured from a sample when mixed with Ca2+-containing solution under a particular set of circumstances. L max and L int are the peak light intensity and total light, respectively, recorded from an identical sample when mixed with saturating [Ca2+] under the same conditions. The curves are sigmoid. The mixture of CBP–coelenterazine-v with RM-Y mutant luciferase begins to respond to changes of [Ca2+] starting from 10−8 M and saturates at approximately 10−5.5 M. The mixture of CBP bound with native coelenterazine and wild-type Renilla luciferase responds to [Ca2+] changes from 10−7 M to approximately 10−5 M under the same conditions [10]. Thus, CBP–coelenterazine-v with RM-Y mutant luciferase reveals a slightly higher affinity to calcium than CBP bound with native coelenterazine and wild-type R. muelleri luciferase. The curve spans have a vertical range approximately of 2.5 log units. This is higher by approximately one order as compared with the span of the Ca2+ concentration–effect curve for the mixture of CBP–coelenterazine and wild-type R. muelleri luciferase. It should be noted, however, that the CBP–coelenterazine-v and RM-Y mutant luciferase mixture exhibits a high level of Ca2+-independent luminescence, which is approximately the same as for CBP loaded by native coelenterazine and wild-type R. muelleri luciferase [10], but exceeding the level of Ca2+ free luminescence of Ca2+-regulated photoproteins by almost two orders of magnitude [34]. The slope of the Ca2+ concentration–effect curve amounts to 1.2, indicating that binding of only two Ca2+ to CBP–coelenterazine-v is enough to trigger the RM-Y bioluminescence, whereas for Ca2+-regulated photoproteins, three calcium ions are necessary [13, 34].
Conclusion
A shift of the bioluminescence emission from Renilla luciferase is very desirable for in vivo imaging and development of in vitro dual-color bioluminescence assays [35, 36], and this is achieved by using a coelenterazine analog, coelenterazine-v, as a substrate. In the present study, we show a potential utility of CBP from R. muelleri as a coelenterazine-v protector against oxidation and as a carrier protein, which allows ready turnover of substrate with Renilla luciferase reaction upon Ca2+ binding. An important finding is that the reaction of the R. muelleri luciferase mutant RM-Y with CBP–coelenterazine-v produces more than twice the total light over that with the free substrate. Thus, at low concentrations of the reaction components, the coelenterazine-v bound within CBP would generate a brighter bioluminescence signal than would free coelenterazine, thereby increasing the assay sensitivity. In addition, this approach for coelenterazine-v protection might be applied in vivo by expressing apo-CBP within mammalian cells by using the same technology developed for the intracellular use of Ca2+-regulated photoproteins [37–39].
Abbreviations
- CBP:
-
Ca2+-triggered coelenterazine-binding protein
- RM-Luc:
-
Native Renilla muelleri luciferase
- RM-Y:
-
Long-wavelength shifted Renilla muelleri luciferase mutant
References
Cormier MJ, Lee J, Wampler JE (1975) Bioluminescence. Recent advances. Annu Rev Biochem 44:255–272
Hori K, Wampler JE, Matthews JC, Cormier MJ (1973) Identification of the product excited states during the chemiluminescent and bioluminescent oxidation of Renilla (sea pansy) luciferin and certain of its analogs. Biochemistry 12:4463–4468
Ward WW, Cormier MJ (1979) An energy transfer protein in coelenterate bioluminescence. Characterization of the Renilla green-fluorescent protein. J Biol Chem 254:781–788
Charbonneau H, Cormier MJ (1979) Ca2+-induced bioluminescence in Renilla reniformis. Purification and characterization of a calcium-triggered luciferin-binding protein. J Biol Chem 254:769–780
Kumar S, Harrylock M, Walsh KA, Cormier MJ, Charbonneau H (1990) Amino acid sequence of the Ca2+-triggered luciferin binding protein of Renilla reniformis. FEBS Lett 268:287–290
Moncrief ND, Kretsinger RH, Goodman M (1990) Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol 30:522–562
Kawasaki H, Nakayama S, Kretsinger RH (1998) Classification and evolution of EF-hand proteins. Biometals 11:277–295
Cormier MJ (1978) Applications of Renilla bioluminescence: an introduction. Methods Enzymol 57:237–244
Inoyue S (2007) Expression, purification and characterization of calcium-triggered luciferin-binding protein of Renilla reniformis. Protein Expr Purif 52:66–73
Titushin MS, Markova SV, Frank LA, Malikova NP, Stepanyuk GA, Lee J, Vysotski ES (2008) Coelenterazine-binding protein of Renilla muelleri: cDNA cloning, overexpression, and characterization as a substrate of luciferase. Photochem Photobiol Sci 7:189–196
Stepanyuk GA, Liu ZJ, Markova SV, Frank LA, Lee J, Vysotski ES, Wang BC (2008) Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.7 Å: why it is not a calcium-regulated photoprotein. Photochem Photobiol Sci 7:442–447
Stepanyuk GA, Liu ZJ, Vysotski ES, Lee J, Rose JP, Wang BC (2009) Structure based mechanism of the Ca2+-induced release of coelenterazine from the Renilla binding protein. Proteins 74:583–593
Vysotski ES, Lee J (2004) Ca2+-regulated photoproteins: structural insight into the bioluminescence mechanism. Acc Chem Res 37:405–415
Shimomura O (2006) Bioluminescence: chemical principles and methods. World Scientific, Singapore
de Wet JR, Wood KV, Helinski DR, DeLuca M (1985) Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci USA 82:7870–7873
Contag CH, Spilman SD, Contag PR, Oshiro M, Eames B, Dennery P, Stevenson DK, Benaron DA (1997) Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol 66:523–531
Yegutkin GG, Samburski SS, Jalkanen S (2003) Soluble purine-converting enzymes circulate in human blood and regulate extracellular ATP level via counteracting pyrophosphatase and phosphotransfer reactions. FASEB J 17:1328–1330
Lorenz WW, McCann RO, Longiaru M, Cormier MJ (1991) Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc Natl Acad Sci USA 88:4438–4442
Markova SV, Golz S, Frank LA, Kalthof B, Vysotski ES (2004) Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa. A novel secreted bioluminescent reporter enzyme. J Biol Chem 279:3212–3217
Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO (2005) Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11:435–443
Greer LF 3rd, Szalay AA (2002) Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence 17:43–74
Weissleder R (2001) A clearer vision for in vivo imaging. Nat Biotechnol 19:316–317
Loening AM, Wu AM, Gambhir SS (2007) Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Meth 4:641–643
Inouye S, Shimomura O (1997) The use of Renilla luciferase, Oplophorus luciferase, and apoaequorin as bioluminescent reporter protein in the presence of coelenterazine analogues as substrate. Biochem Biophys Res Commun 233:349–353
Kataeva I, Chang J, Xu H, Luan CH, Zhou J, Uversky VN, Lin D, Horanyi P, Liu ZJ, Ljungdahl LG, Rose J, Luo M, Wang BC (2005) Improving solubility of Shewanella oneidensis MR-1 and Clostridium thermocellum JW-20 proteins expressed into Esherichia coli. J Proteome Res 4:1942–1951
Kishi Y, Tanino H, Goto T (1972) The structure confirmation of the light-emitting moiety of bioluminescent jellyfish. Tetrahedron Lett 13:2747
Kakoi H, Inoue S (1980) A new synthesis of Watasenia prelucifein by cyclization of 2-amino-3-benzyl-5-(p-hydroxyphenyl)pyrazine with p-hydroxyphenylpyruvic acid. Chem Lett 9:299–300
Shimomura O, Musicki B, Kishi Y (1988) Semi-synthetic aequorin. An improved tool for the measurement of calcium ion concentration. Biochem J 251:405–410
Vysotski ES, Liu ZJ, Rose J, Wang BC, Lee J (2001) Preparation and X-ray crystallographic analysis of recombinant obelin crystals diffracting to beyond 1.1 Å. Acta Crystallogr D Biol Crystallogr 57:1919–1921
Illarionov BA, Frank LA, Illarionova VA, Bondar VS, Vysotski ES, Blinks JR (2000) Recombinant obelin: cloning and expression of cDNA, purification and characterization as calcium indicator. Methods Enzymol 305:223–249
Klabusay M, Blinks JR (1996) Some commonly overlooked properties of calcium buffer systems: a simple method for detecting and correcting stoichiometric imbalance in CaEGTA stock solutions. Cell Calcium 20:227–234
McCapra F, Chang YC (1967) The chemiluminescence of a Cypridina luciferin analogue. Chem Commun 19:1011–1012
Goto T (1968) Chemistry of bioluminescence. Pure Appl Chem 17:421–441
Markova SV, Vysotski ES, Blinks JR, Burakova LP, Wang BC, Lee J (2002) Obelin from the bioluminescent marine hydroid Obelia geniculata: cloning, expression, and comparison of some properties with those of other Ca2+-regulated photoproteins. Biochemistry 41:2227–2236
Frank LA, Borisova VV, Markova SV, Malikova NP, Stepanyuk GA, Vysotski ES (2008) Violet and greenish photoprotein obelin mutants for reporter applications in dual-color assay. Anal Bioanal Chem 391:2891–2896
Rowe L, Dikici E, Daunert S (2009) Engineering bioluminescent proteins: expanding their analytical potential. Anal Chem 81:8662–8668
Chiesa A, Rapizzi E, Tosello V, Pinton P, de Virgilio M, Fogarty KE, Rizzuto R (2001) Recombinant aequorin and green fluorescent protein as valuable tools in the study of cell signalling. Biochem J 355:1–12
Pozzan T, Mongillo M, Rudolf R (2003) The Theodore Bücher lecture. Investigating signal transduction with genetically encoded fluorescent probes. Eur J Biochem 270:2343–2352
Michelini E, Cevenini L, Mezzanotte L, Coppa A, Roda A (2010) Cell-based assays: fuelling drug discovery. Anal Bioanal Chem. doi:10.1007/s00216-010-3933-z
Acknowledgments
This work was supported by grant 09-04-12022 of the Russian Foundation for Basic Research, “Molecular and Cell Biology” program of Russian Academy of Sciences, by the SB RAS grant No.2, and by the SB RAS Lavrentiev grant for Young Scientists.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stepanyuk, G.A., Unch, J., Malikova, N.P. et al. Coelenterazine-v ligated to Ca2+-triggered coelenterazine-binding protein is a stable and efficient substrate of the red-shifted mutant of Renilla muelleri luciferase. Anal Bioanal Chem 398, 1809–1817 (2010). https://doi.org/10.1007/s00216-010-4106-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4106-9