Abstract
In this study, gas chromatography mass spectrometry (GC-MS) and two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC-TOFMS) were employed for the metabolic footprinting of a pair of immortalized human uroepithelial cells namely HUC-1 (nontumorigenic) and HUC T-2 (tumorigenic). Both HUC-1 and HUC T-2 cell lines were cultivated in 1 mL of Ham’s F-12 media. Subsequent to 48 h of incubation, 200 μL of cell culture supernatant was protein-precipitated using 1.7 mL of methanol and an aliquot of 1.5 mL of the mixture was separated, dried, trimethylsilyl-derivatized, and analyzed using GC-MS and GC×GC-TOFMS. Metabolic profiles were analyzed using multivariate data analysis techniques to evaluate the changes of the metabolomes. Both GC-MS and GC×GC-TOFMS analyses showed distinct differences in metabolic phenotypes of the normal and tumorigenic human bladder cells (partial least squares-discriminant analysis (PLS-DA) of GC×GC-TOFMS data; two latent variables, R 2 X = 0.418, R 2 Y = 0.977 and Q 2 (cumulative) = 0.852). Twenty metabolites were identified as being statistically different between the two cell types. These metabolites revealed that several key metabolic pathways were perturbed in tumorigenic urothelial cells as compared to the normal cells. Application of GC×GC-TOFMS offered several advantages compared to classical one-dimensional GC-MS which include enhanced chromatographic resolution (without increase in analytical run time), increase in sensitivity, improved identification of metabolites, and also separation of reagent artifacts from the metabolite peaks. Our results reinforced the advantages of GC×GC-TOFMS and the role of metabolomics in characterizing bladder cancer biology using in vitro cell culture models.

Metabolic footprinting of tumorigenic and nontumorigenic uroepithelial cells using GCxGCTOFMS
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bladder cancer (BC) is one of the common cancers diagnosed in clinics with an estimated 70,980 new cases in 2009 in the USA [1]. Although many urinary assays have been utilized, most of the diagnostic assays do not achieve the specificity and sensitivity of cystoscopy [2]. Therefore, alternative noninvasive diagnostic methods such as the use of biomarkers in detecting BC become important. While clinical cancer metabonomics appear to be a direct approach to discover biomarkers related to cancer biology, many confounding factors such as age, gender, diet, gut microbiome, pathology, and pharmacological interventions may complicate the analysis and mask the important cancer-derived metabolic fluxes. Recently, profiling of extracellular metabolites has been recognized as a complementary tool for the physiological characterization of microorganisms and cells [3–5]. The strategy of analyzing the phenotypes of cells or tissues by profiling the metabolites that they excrete or fail to assimilate from their extracellular environment is defined as metabolic footprinting or exometabolomics [3]. On the other hand, analyzing intracellular metabolites is commonly referred to metabolic fingerprinting [6]. Metabolic footprinting presents several advantages compared to metabolic fingerprinting. As the intracellular metabolism is dynamic and the turnover rate of most metabolites is extremely fast, ranging from milliseconds to minutes, metabolic fingerprinting demands a rapid quenching of cellular metabolism followed by an effective separation of intracellular and extracellular metabolites [7]. These experimental requirements impose technical difficulties and may result in misinterpretation of the metabolic fluxes. Secondly, there are several biochemical processes that are specifically related to the extracellular media, such as the degradation of complex substrates, and these can be assessed only by measuring the degradation products in the extracellular medium [8]. Hence, metabolic footprinting may provide complementary information on the perturbations of metabolites that can be used to interpret the in vivo pathophysiological status. The analytical platforms commonly exploited in metabolic footprinting studies are nuclear magnetic resonance spectroscopy [9, 10], liquid chromatography mass spectrometry (LC-MS) [11], and gas chromatography mass spectroscopy (GC-MS) [8, 12]. Among the various analytical platforms investigated, GC-MS proved to be a potentially useful method based on its high sensitivity, peak resolution, and reproducibility [13]. Availability of GC-MS electron impact (EI) spectral library further facilitates the identification of diagnostic biomarkers and aids the subsequent mechanistic elucidation of the biological or pathological variations [14]. Although one-dimensional GC-MS offers significant chromatographic resolution, peak overlapping is inevitable due to the large number of extracellular metabolites. Poor chromatographic separation may result in incorrect biomarker identification, inaccurate estimation of metabolite concentrations, and poor predictive chemometric models. Comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC-TOFMS) is a maturing technique noted for its ability to analyze complex mixtures and has been successfully applied in metabolomic investigations [15–19]. GC×GC-TOFMS is superior in terms of chromatographic peak resolution when compared to conventional gas chromatographic techniques [13, 16, 18]. In a GC×GC system, analytes eluting from the first column, over a defined time window, are focused and released (via a process termed modulation) onto a second column where further separation is achieved. Typically, a nonpolar column is selected as the first column and relatively polar column as the second column [16, 18]. This configuration achieves orthogonality of retention on the two columns, allowing much more complete separation of metabolites in complex matrices. Additionally, the fast spectral acquisition rate (up to 500 Hz) of TOFMS enables deconvolution of the mass spectra of closely eluting peaks if the spectra are sufficiently distinct. The ability to deconvolute the mass spectra robustly and accurately is paramount in the analysis of moderately complex samples in metabolic footprinting. The principles and instrumentation of comprehensive two-dimensional gas chromatography have been reviewed in detail in several papers previously [20, 21].
The overarching aim of this study was to apply GC×GC-TOFMS-based metabolomics for the metabolic footprinting of a pair of immortalized human uroepithelial cells, nontumorigenic HUC-1 and tumorigenic HUC T-2. As these cells share the same genome, their differential metabolic phenotypes are related to the transformation of the normal urothelial cells to the tumorigenic cells via exposure to polycyclic hydrocarbon chemical carcinogen, 3-methylcholanthrene (MCA). Since the exposure to polycyclic hydrocarbon from tobacco smoke has been linked to the development of several cancers including BC; the in vitro biomarkers arising from the metabolic footprinting of HUC-1 and HUC T-2 may facilitate the mechanistic understanding of the carcinogenesis of BC. Although application of GC×GC-TOFMS has been proven to be important in metabolomics analysis of complex samples such as urine [16] or tissues [22], its significance in analyzing relatively less complex samples such as metabolic footprinting samples has not been explored so far. Nevertheless, it is possible that one-dimensional GC-MS may be adequate for the analysis of metabolic footprinting samples. Therefore, the secondary objective of our study was to compare GC-MS and GC×GC-TOFMS to note the specific advantages or drawbacks of each technique in analysis of moderately complex samples such as cell culture media. In addition, a subset of samples was analyzed in GC-TOFMS mode to verify the necessity of two-dimensional separations in conjunction to deconvolution of mass spectra of metabolite peaks.
Materials and methods
Chemicals
N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) and methoxyamine HCl in pyridine (MOX® reagent) were purchased from Pierce (Rockford, IL,USA). Alkane standard mixture (C10 to C40) and sodium sulfate (anhydrous) were obtained from Sigma-Aldrich (St. Louis MO, USA). SV-HUC-1 (CRL-9520) and MC-SV-HUC T-2 (CRL-9519) cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). Ham’s F-12 nutrient mixture powder, 2 mM l-glutamine, and 50 units penicillin/50 μg streptomycin per milliliter were purchased from Invitrogen (Invitrogen Corporation, CA, USA). All other chemicals were of analytical grade.
Cell culture in complete media
Both HUC-1 and HUC T-2 cells were cultured on 24-well plates at 1 × 105 cell density per well in 1 mL of Ham’s F-12 media (10% FBS, 2 mM l-glutamine, 50 units penicillin/50 μg streptomycin per milliliter) and incubated at 37 °C in the presence of 5% CO2. After 24 h, the cell medium in each plate was removed and replaced with 1 mL of fresh medium. Each sample was incubated further for 24 h. The media obtained from HUC-1 and HUC T-2 cells were subsequently harvested and snapped frozen using liquid nitrogen. Three replicates of Ham’s F-12 medium without cells which were used as quality controls (QC) were treated in the same way as HUC-1 and HUC T-2 cell culture medium samples. All the samples were stored at −80 °C until further analysis.
Monitoring cell proliferation
Plated separately in quadruplicates in a 96-well plate and incubated at 37 °C with 5% CO2 for 24 and 48 h were 1 × 104 each of the HUC-1 and HUC T-2 cells. CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, USA) was used to measure the viability of HUC-1 and HUC T-2 after 24 and 48 h. Briefly, 20 μL of the CellTiter 96® AQueous One Solution Reagent was added to each well containing 100 μL of culture medium and incubated in the dark at 37 °C for 2 h. The absorbance of each well was measured at 490 nm using GENios Pro™ microplate reader (Tecan, Switzerland). To confirm the cell proliferation rate, 1 × 105 cells per well were plated into a six-well plate, and the cell number per well was enumerated after 24 and 48 h using a hemocytometer.
Sample preparation for GC-MS and GC×GC-TOFMS analysis
All the samples were thawed at room temperature (23 ± 3 °C). Subsequently, 1.7 mL of methanol was added to 200 μL of each medium sample. The mixture was vortex-mixed at high speed for 5 min, centrifuged subsequently for 10 min at 10,000×g at 4 °C. A 1.5 mL of each supernatant was then carefully separated and evaporated to dryness at 40 °C under a gentle stream of nitrogen gas using TurboVap LV (Caliper Life Science, Hopkinton, MA, USA). One hundred microliters of toluene (dried over anhydrous sodium sulfate) was added to each dry residue, mixed for 1 min, and dried again at 50 °C under nitrogen gas. The dried metabolic extract was derivatized first with 50 μL of methoxyamine (20 mg/mL) for 2 h at 60 °C. Subsequently, 100 μL of MSTFA with 1% TMCS was added to the mixture and heated for 1 h at 60 °C to form trimethylsilyl (TMS) derivatives. TMS derivatives were cooled, and 100 μL of supernatant was transferred into GC vial and subjected to GC-MS and GC×GC-TOFMS analysis independently, where the HUC-1, HUC T-2, and control samples were randomized to avoid analytical bias. In addition, a subset of samples used for GC×GC-TOFMS analysis was also subjected to one-dimensional GC-TOFMS analysis.
GC-MS analysis
Analyses were performed on a Shimadzu QP2010 GC-MS system (Shimadzu, Kyoto, Japan) equipped with HP-5MS (Agilent J&W Scientific, Folsom, CA) 30 m × 250 μm (i.d.) fused silica capillary column. Helium was used as the carrier gas at 1.2 mL/min, and the injector split ratio was set to 1:5. The injector and source temperatures were 250 and 200 °C, respectively. Oven temperature was kept at 60 °C for 2 min, increased at 8 °C/min to 310 °C where it was held for 2 min. The MS was operated in EI mode (70 eV). Data acquisition was performed in the full scan mode from m/z 40 to 600 with a scan time of 0.5 s.
GC×GC-TOFMS analysis
A Pegasus GC×GC-TOFMS (LecoCorp., St. Joseph, MI, USA) was utilized for the analysis. The instrument was equipped with an Agilent 7890 GC and Pegasus IV TOFMS (LecoCorp., St. Joseph, MI, USA) featuring a dual-stage, quad-jet thermal consumable-free modulator (CFM) and an independently temperature-controlled secondary oven. CFM is a recently introduced modulator where the nitrogen gas is chilled by passing it through a coil immersed in a dewar of silicone oil. The silicone oil is chilled by the cold probe from a closed-loop immersion cooler (FTS, Stone Ridge, NY, USA) which was set at the temperature of −80 °C. Helium was used as the carrier gas at 1.5 mL/min in corrected constant flow mode, and the injector split ratio was set to 1:20. A 30 m × 250 μm (i.d.) × 0.25 μm DB-1 (Agilent J&W Scientific, Folsom, CA) and a 1.5 m × 100 μm (i.d.) × 0.100 μm Rxi®-17 (Restek Corp., Bellefonte, PA, USA) fused silica capillary columns were used as the primary and secondary columns, respectively. Primary oven temperature was programmed at 70 °C for 0.2 min and increased at 10 °C/min to 270 °C where it was held for 5 min. Secondary oven temperature was always maintained at 10 °C higher than the primary oven temperature. The thermal modulator was set to 45 °C higher relative to the primary oven. A modulation time of 2 s with hot pulse of 0.6 s was used. The injector, transfer line, and ion source temperatures were maintained at 220, 200, and 250 °C, respectively, throughout each analysis. The MS was operated in EI mode (70 eV) at the detector voltage of 1,650 V. Data acquisition was performed in the full scan mode from m/z 40 to 600 with an acquisition rate of 100 Hz. One-dimensional GC-TOFMS analysis method parameters were similar to GC×GC-TOFMS except that split ratio was set four times lower at 1:5, and acquisition rate was set to 20 Hz.
GC-MS data preprocessing
Data preprocessing was performed using the Shimadzu GCMSsolution (version 2.5). A reference compound table was built by registering peaks from multiple chromatograms. Subsequently, the compound table was used to extract and calculate peak area values from all the chromatograms. To obtain accurate peak areas, single mass was specified for the quantification of each compound. Additionally, two reference ions were used along with the quantifying mass to cross-validate the peak identity of each compound. Identity of GC-MS detected peaks was established by comparing the mass spectra and the retention indices of the peaks with those available in the NIST mass spectral library (Wiley registry) and internally compiled spectral libraries. Peaks with similarity index more than 70% were assigned compound names while other peaks were listed as unknown (UN) compounds. The total ion intensity for each detected peak was normalized against the sum of the peak intensities within that sample chromatogram using custom scripts developed within Matlab (Mathworks, Natick, MA, USA).
GC×GC-TOFMS data preprocessing
Each chromatogram obtained from GC×GC-TOFMS analysis was processed for baseline correction, smoothing, noise reduction, deconvolution, library matching, and area calculation using the ChromaTOF software (version 4.21, LecoCorp.). Area of each peak was calculated using the unique mass of each derivatized metabolite. Only peaks with signal to noise ratio (S/N) greater than 100 were utilized for further analysis. All GC×GC-TOFMS detected peaks were identified by comparing both the MS spectra and the retention indices with those available in the NIST mass spectral library (Wiley registry) and our in-house spectral library. Subsequently, the Statistical Compare feature of ChromaTOF software was utilized to generate data table in which all the peak information from different chromatograms were aligned. Statistical Compare utilized a mass spectral match criterion of 70% when aligning the multidimensional peak data comprising sample names, metabolites, retention time (RT), mass, and integrated peak area. Quantification mass for each peak in the data table is selected from the unique mass that is most common to all matching peaks within the retention window (two times of peak width). Subsequently, this unique mass is used to calculate the peak areas from each chromatogram. Similarly, the best quality peak from matching peaks was then selected whose name was used as analyte name in the data table. The resulting data table comprised of observations (samples and controls) with each of them in turn described by variables (peak intensities) aligned according to their RT and unique mass pairs as identifiers. The data table was exported as a .csv file. The .csv file was then processed using our in-house developed Matlab scripts to retain only observations (sample identities) and variables (area values) in the data table and remove other details which were not necessary for the immediate chemometric analysis. In addition, peaks which were not present in at least 50% of the samples were removed from the data table. Total area normalization was performed based on total ion chromatograms (TIC) to correct minor variations occurring due to sample preparation and analysis.
Chemometric data analysis
Normalized data obtained from GC-MS or GC×GC-TOFMS analysis were exported to SIMCA-P (version 12.0, Umetrics, Umeå, Sweden) to perform principal component analysis (PCA) where grouping trends and outliers in the data were observed. Prior to PCA analysis, normalized data were mean-centered and unit variance scaled. DModX plot was calculated to check for any outliers. After initial overview of the GC-MS and GC×GC-TOFMS data using PCA analysis, the data were subjected to separate PLS-DA where each model was built and utilized to identify marker metabolites that accounted for the differentiation of HUC-1 and HUC T-2 cells. Chemical identification of marker metabolites peaks was further confirmed by cross-referencing with the Golm metabolite library [23] and the Human Metabolome Database (HMDB) [24]. Further, metabolic pathway interpretation of marker metabolites was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [25].
Results and discussion
Cancer and normal cell derived from different sources may have differences in their metabolic phenotypes unrelated to the tumorigenicity of the cells but rather reflective of their inherent genetic differences. Therefore, to compare the metabolic profiles of normal and tumor cells, it is important to use cells derived from the same origin. The HUC-1 and HUC T-2 cells obtained from ATCC were originally produced by immortalizing uroepithelial cells with the SV-40 virus to give HUC-1 cells which were further treated with a chemical carcinogen, MCA, to produce HUC T-2 cells. HUC-1 is nontumorigenic and has a balanced chromosome composition, while HUC T-2 is tumorigenic and aneuploid. As the rate at which cells proliferate will affect the quantity of metabolic products observed, the growth rates of both cell lines were determined using the MTS assay. As shown in Fig. 1a, the proliferation rates of both cells lines after 24 and 48 h were similar (p > 0.05). Therefore, any differences observed in the metabolic footprints were possibly related to metabolic perturbations. To further confirm our observation, we enumerated the cells at 24 and 48 h of culture. As shown in Fig. 1b, the cell numbers were not statistically significant (p > 0.3) at both the time points. These data confirmed that both cell lines were proliferating at similar rates.
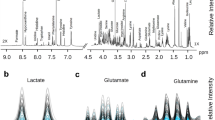
The sample preparation utilized for GC-MS analysis was evaluated and validated by us and other researchers previously [26, 27]. Parameters related to first-dimension chromatographic separation such as gradient temperature program of primary oven, injector, and transfer line temperatures were optimized independently prior to optimization of parameters related to second-dimension separation in GC×GC-TOFMS method. The GC-TOFMS parameters were tested for analytical reproducibility and applied for metabolic profiling of large number of urine samples [28]. For the GC×GC-TOFMS analysis, the method parameters were optimized with respect to following criteria: (1) detection of maximum number of peaks, (2) optimum sensitivity of detection without column overloading or detector saturation, (3) separation of artifact peaks from metabolites of interest, (4) shorter analytical runtime, (5) increased chromatographic resolution of peaks, and (6) modulation of majority of peaks into three to five segments of modulation cycles to retain the first-dimension separation. It is worth noting that, since two columns are connected in series, carrier gas flow rates cannot be selected independently. Therefore, a compromise is generally chosen that provides acceptable separation in both dimensions [29]. The higher carrier gas flow rate (1.5 mL/min) selected in this study could have compromised the second-dimension resolution of selected metabolites. However, the loss of resolution was found to be less significant possibly due to the flat slope of the Golay curves for narrow-bore column [30]. Recently, it was also shown that carrier gas flow rates have minimal influence on retention time, order of peak elution, and peak width in second dimension of GC×GC-TOFMS analysis [31]. Moreover, lower carrier gas flow rates not only increase analytical run time but also increase subsequent data processing time due to the high data acquisition rate used in GC×GC-TOFMS analysis. Typical GC-MS and GC×GC-TOFMS chromatograms obtained from the analysis of HUC-1 cell culture supernatant are shown in Fig. 2. The representative GC×GC-TOFMS TIC is depicted as a surface plot (Fig. 2b), and it underscores the increased peak capacity, improved analyte detectability, and enhanced resolution gained by the two-dimensional separation technique. Analytical run time of each sample was 35 and 25 min for GC-MS and GC×GC-TOFMS, respectively. Visual inspection of GC×GC-TOFMS chromatograms to identify major difference between the two sample groups was performed using several strategies proposed by Shellie et al. [19]. In GC×GC-TOFMS analysis, the derivatized metabolites were not only separated chromatographically but also via software deconvolution of the mass spectra. The MS spectra of most of the TMS derivatives were dominated by 73 and 147 mass ions while the characteristic ions of each metabolite were low in abundance. This phenomenon might result in inaccuracies in peak deconvolution and area integration of coeluting metabolites [32]. Therefore, the background TMS ions (m/z 73 and 147) were specified in the data processing method of the ChromaTOF software for two purposes. Firstly, the software performed two levels of peak identification when comparing the peak MS spectra with library spectra, one with the specified masses and one without the masses. Both matches must pass the threshold for assigning the compound name. Secondly, while selecting a mass for peak area integration, specified TMS derivative masses were given lower preference by using higher S/N threshold. When derivative masses were specified in the data processing method, we observed that analyte names were more accurately assigned due to enhanced library matching.
Data preprocessing
In GC-MS analysis, a total of 65 derivatized peaks were detected across all the samples after removal of reagent artifact and column bleed peaks. On the other hand, an average of over 450 derivatized compounds was detected in each chromatogram via GC×GC-TOFMS analysis post-deconvolution. Although, a large number of analytes were detected in GC×GC-TOFMS analysis, many of them belonged to derivatizing reagent artifacts or column bleed. One key advantage of GC×GC-TOFMS was that the artifacts were chromatographically separated from the metabolites due to the second-dimension separation (Electronic Supplementary Material, Fig. S1). To consistently remove these artifacts from all the chromatograms, the classification feature of ChromaTOF software was utilized where chromatographic regions belonging to artifacts were demarcated. Such artifacts or chromatographic noise, if not removed, would significantly disturb the subsequent chemometric data analysis [18]. Subsequent to the removal of artifacts, peak information belonging to different chromatograms was aligned in a data table. Due to the varied number of peaks found and inconsistencies in assigning compound name for the same peak in each chromatogram, aligning peaks in suitable matrix format was particularly challenging. Few strategies had been reported to address this challenge [16, 19, 33]. However, these strategies typically required manual data processing in conjunction with existing instrument software [19] or development of new software [33]. This challenge was suitably addressed in our study by using the Statistical Compare feature of ChromaTOF software where it accurately aligned a large percentage of data. However, in a few instances, we observed that some of the analytes were represented multiple times in the data table based on the similar analyte names and quantitation masses assigned to the common peaks. Multiple-represented peaks were counter-checked in the raw data and suitably corrected, and finally, only a single analyte name was retained in the data table for each metabolite. Subsequent to alignment of peak information, the final data table comprised of 286 derivatized peaks which were collated from the GC×GC-TOFMS analysis of both HUC-1 and HUC T-2 cells. The number of peaks was more than fourfold higher compared to that observed using one-dimensional GC-MS.
Chemometric data analysis
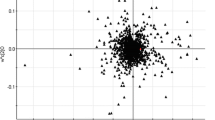
Chemometric data analysis was necessary to generate interpretable models for the complex intercorrelation of data [34]. In PCA (two components, R 2 X = 0.563 and Q 2 (cum) = 0.384) of GC-MS data, all samples were found within Hotelling's T2 (0.95) ellipse, and the DModX values were below critical values, suggesting no outliers in the data. On the other hand, when GC×GC-TOFMS data were subjected to PCA, score plot revealed moderate outliers belonging to the respective HUC-1 and HUC T-2 groups as these observations were lying outside Hotelling's T2 (0.95) ellipse. While DModX values of these two observations were below the critical value, they were still excluded from further chemometric data analysis. Performance statistics of PCA model showed R 2 X = 0.325 and Q 2 (cumulative) = 0.118. Clear grouping trends of HUC-1 and HUC T-2 were observed in the PCA score plot with some minor overlap in both GC-MS and GC×GC-TOFMS analysis. While PCA provided a simplified representation of the information contained in the spectra, it did not consider additional information such as class information. Therefore, PCA was followed by a supervised analysis technique such as PLS-DA that aided our identification of a list of potential biomarkers which are statistically significant and which separate one class from another [35, 36]. PLS-DA score plot (Fig. 3a) obtained from GC-MS data analysis revealed clear separation of HUC-1 and HUC T-2 samples (three latent variables, R 2 X = 0.624, R 2 Y = 0.903, and Q 2 (cum) = 0.690). Similarly, for GC×GC-TOFMS data, the PLS-DA model calculated based on 22 observations contained three latent variables, showing the performance statistics of R 2 X = 0.418, R 2 Y = 0.977, and Q 2 (cumulative) = 0.852 (Fig. 3b). Loadings, regression coefficients, and variable importance plots (VIPs) revealed variables (metabolites) that separated the two classes. In total, 17 and 81 metabolites showed a VIP >1 in GC-MS and GC × GC-TOFMS analyses, respectively, suggesting that the overall metabolic profiles of the two cell lines were distinctly different. Using a VIP cutoff of 1.5 and p value (Welch t test) less than 0.05, five and 20 marker metabolites were generated using the GC-MS and GC × GC-TOFMS platforms, respectively. These results confirmed that the GC × GC-TOFMS platform was capable of profiling more statistically significant marker metabolites when compared to the one-dimensional separation technique. The marker metabolites are shown in Table 1. Validity of the PLS-DA model obtained from GC × GC-TOFMS analysis was verified by prediction of cell media derived from QC samples. From the prediction score plot (Electronic Supplementary Material, Fig. S2), it can be seen that QC samples were closely clustered, suggesting analytical variation was minimal. In addition, variations in the levels of marker metabolites were further verified by calculating percentage coefficient of variation (% CV) of similar marker metabolites in QC samples. These quantified marker metabolites showed an average and median % CV values of 14.1% and 16.4%, respectively. Such low analytical variation in QC samples validated that the observed differences in metabolite concentrations were related to biological differences between HUC-1 and HUC T-2 cells rather than analytical variation.
Metabolic pathway evaluation
Both KEGG and HMDB databases were explored to connect the marker metabolites to metabolic pathways. Metabolic perturbations were mainly detected in the area of propionate metabolism and other pathways related to energy metabolism. A number of interesting observations could be inferred from the metabolic fluxes. Firstly, glycine was found to be at higher concentrations while 3-phosphoglyceric acid was at lower concentrations in cancer cell media compared to that of normal cells. Glycine is formed from the glycolytic intermediate 3-phosphoglycerate and is an important source of one-carbon units for the synthesis of nucleotides, through serine. Increased glycine coupled to decreased 3-phosphoglycerate suggested possible upregulation of hypoxia-inducible factor-1 (HIF-1) signaling pathway. Higher expression of HIF-1 correlates to the upregulation of HIF-1 signaling pathway observed clinically in BC patients [37, 38]. HIF-1 is also upregulated in several cancer cell types, resulting in the increased expression of proteins involved in a range of metabolic pathways and increased rate of glycolysis [39]. Increased glycolysis was evident in our study based on the higher amount (approximately fourfold, p < 0.0005) of glucose consumption observed in cancer HUC T-2 cells as compared to normal HUC-1 cells. However, increased glucose consumption can also be due to increased expression of glucose transporters and type II hexokinase which are commonly observed in cancer cells [40, 41].
Advantages of GC×GC-TOFMS
Several advantages of GC×GC-TOFMS in metabolomic analysis of biofluids such as urine were noted previously when compared to GC-MS [22]. Compared to urine samples, cell culture media samples are relatively less complex. Therefore, it is possible that GC×GC-TOFMS may not offer significant advantages in metabolic footprinting analyses of cell culture media. Hence, comparison between the two analytical technologies is warranted to justify the use of GC×GC-TOFMS for cellular metabolic footprinting. Several advantages of GC×GC-TOFMS compared to GC-MS were noted in our study. Firstly, GC×GC-TOFMS showed fourfold increase (286 versus 65) in the number of metabolites detected compared to one-dimensional GC-MS. Secondly, for GC×GC-TOFMS, artifact peaks arising from column bleed or derivatizing agent could be chromatographically resolved from the metabolite peaks (Electronic Supplementary Material, Fig S1) and automatically removed from the data tables using classification-based rules. Artifact peaks appeared in several GC×GC modulation cycles which were distinct from metabolite peaks. The number of modulation cycles in which a particular peak appears could be visualized using the subpeak display (Fig S1 of Electronic Supplementary Material). In contrast, the identification of artifact peaks in GC-MS demanded experience, knowledge, and manual intervention. Thirdly, a fivefold increase in sensitivity was observed when GC×GC-TOFMS was used. The increased sensitivity was achieved at fourfold higher split ratio for GC×GC-TOFMS analysis as compared to that of GC-MS. Lastly, due to second-dimension separation and software peak deconvolution function, the total analytical run time was reduced for GC×GC-TOFMS analysis from 35 to 25 min. One could argue that the increase in detected peaks could be solely attributed to the software peak deconvolution function and one-dimensional GC-TOFMS would suffice for the metabolomic analysis instead of two-dimensional GC×GC-TOFMS. To address this pertinent question, a number of samples were analyzed in GC-TOFMS mode to examine the necessity of two-dimensional separation. One representative cell medium analyzed illustrates the comparison of various outcome parameters of one- and two-dimensional GC-TOFMS-based analyses (Table S1, Electronic Supplementary Material). Compared to GC-TOFMS, a higher number of peaks were detected using two-dimensional separation. More importantly, the deconvolution purity of peaks was also significantly enhanced. Majority of peaks (64%) detected using GC×GC-TOFMS had good deconvolution purity (less than one). In contrast, only 35% of peaks showed good deconvolution purity in GC-TOFMS analysis. It should be emphasized that artifact peaks were further resolved from the metabolite peaks when GC×GC-TOFMS was used.
Conclusions
In this study, GC×GC-TOFMS was demonstrated as a powerful tool for analyzing metabolic footprinting cell culture media. Application of GC×GC offered several advantages including enhanced chromatographic resolution (without increase in analytical run time), improved identification of metabolites, and also separation of reagent artifacts from the metabolite peaks. Results from our GC×GC-TOFMS metabolic footprinting study indicated that it was possible to discriminate in vitro metabolic profiles in relation to tumorigenic and nontumorigenic uroepithelial cell types. A number of metabolic pathways related to energy metabolism were found to be perturbed in cancer cells. The unique in vitro metabolic profiles generated by GC×GC-TOFMS revealed metabolic perturbations in BC that should be further cross-validated with in vivo biomarker profiling of the malignancy.
References
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) CA Cancer J Clin 59:225–249
Lotan Y, Roehrborn CG (2002) J Urol 167:75–79
Kell DB, Brown M, Davey HM, Dunn WB, Spasic I, Oliver SG (2005) Nat Rev Microbiol 3:557–565
Mapelli V, Olsson L, Nielsen J (2008) Trends Biotechnol 26:490–497
Kaderbhai NN, Broadhurst DI, Ellis DI, Goodacre R, Kell DB (2003) Comp Funct Genomics 4:376–391
Fiehn O (2002) Plant Mol Biol 48:155–171
Buchholz A, Hurlebaus J, Wandrey C, Takors R (2002) Biomol Eng 19:5–15
Villas-Boas SG, Noel S, Lane GA, Attwood G, Cookson A (2006) Anal Biochem 349:297–305
Abel CB, Lindon JC, Noble D, Rudd BA, Sidebottom PJ, Nicholson JK (1999) Anal Biochem 270:220–230
Miccheli AT, Miccheli A, Di Clemente R, Valerio M, Coluccia P, Bizzarri M, Conti F (2006) Biochim Biophys Acta 1760:1723–1731
Dunn WB, Brown M, Worton SA, Crocker IP, Broadhurst D, Horgan R, Kenny LC, Baker PN, Kell DB, Heazell AE (2009) Placenta 30:974–980
Pope GA, MacKenzie DA, Defernez M, Aroso MA, Fuller LJ, Mellon FA, Dunn WB, Brown M, Goodacre R, Kell DB, Marvin ME, Louis EJ, Roberts IN (2007) Yeast 24:667–679
Pasikanti KK, Ho PC, Chan EC (2008) J Chromatogr B Analyt Technol Biomed Life Sci 871:202–211
Want EJ, Nordstrom A, Morita H, Siuzdak G (2007) J Proteome Res 6:459–468
Almstetter MF, Appel IJ, Gruber MA, Lottaz C, Timischl B, Spang R, Dettmer K, Oefner PJ (2009) Anal Chem 81:5731–5739
Li X, Xu Z, Lu X, Yang X, Yin P, Kong H, Yu Y, Xu G (2009) Anal Chim Acta 633:257–262
Ralston-Hooper K, Hopf A, Oh C, Zhang X, Adamec J, Sepulveda MS (2008) Aquat Toxicol 88:48–52
Mohler RE, Dombek KM, Hoggard JC, Pierce KM, Young ET, Synovec RE (2007) Analyst 132:756–767
Shellie RA, Welthagen W, Zrostlikova J, Spranger J, Ristow M, Fiehn O, Zimmermann R (2005) J Chromatogr A 1086:83–90
Cortes HJ, Winniford B, Luong J, Pursch M (2009) J Sep Sci 32:883–904
Ong RC, Marriott PJ (2002) J Chromatogr Sci 40:276–291
Welthagen W, Shellie RA, Spranger J, Ristow M, Zimmermann R, Fiehn O (2005) Metabolomics 1:65–73
Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D (2005) Bioinformatics 21:1635–1638
Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L (2007) Nucleic Acids Res 35:D521–D526
Mendes P (2002) Brief Bioinform 3:134–145
Pasikanti KK, Ho PC, Chan EC (2008) Rapid Commun Mass Spectrom 22:2984–2992
Koek MM, Muilwijk B, van Stee LL, Hankemeier T (2008) J Chromatogr A 1186:420–429
Pasikanti KK, Esuvaranathan K, Ho PC, Mahendran R, Kamaraj R, Wu QH, Chiong E, Chan EC (2010) J Proteome Res 9:2988–2995
Dalluge J, Beens J, Brinkman UA (2003) J Chromatogr A 1000:69–108
Donato P, Tranchida PQ, Dugo P, Dugo G, Mondello L (2007) J Sep Sci 30:508–526
Ong R, Marriott P, Morrison P, Haglund P (2002) J Chromatogr A 962:135–152
Begley P, Francis-McIntyre S, Dunn WB, Broadhurst DI, Halsall A, Tseng A, Knowles J, Goodacre R, Kell DB (2009) Anal Chem 81:7038–7046
Oh C, Huang X, Regnier FE, Buck C, Zhang X (2008) J Chromatogr A 1179:205–215
Holmes E, Antti H (2002) Analyst 127:1549–1557
Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J (2008) Anal Chem 80:115–122
Trygg J, Holmes E, Lundstedt T (2007) J Proteome Res 6:469–479
Theodoropoulos VE, Lazaris A, Sofras F, Gerzelis I, Tsoukala V, Ghikonti I, Manikas K, Kastriotis I (2004) Eur Urol 46:200–208
Ioachim E, Michael M, Salmas M, Michael MM, Stavropoulos NE, Malamou-Mitsi V (2006) Urol Int 77:255–263
Griffin JL, Shockcor JP (2004) Nat Rev Cancer 4:551–561
Dang CV, Semenza GL (1999) Trends Biochem Sci 24:68–72
Pedersen PL, Mathupala S, Rempel A, Geschwind JF, Ko YH (2002) Biochim Biophys Acta 1555:14–20
Acknowledgments of research support
This study was supported by the National University of Singapore (NUS) grant R-148-000-100-112 provided to E.C.Y.C and National Medical Research Council grant R-176-000-119-213 provided to K.E., P.C.H., R.M., and E.C.Y.C. GC × GC-TOFMS was kindly sponsored by the NUS grant R-279-000-249-646. K.K.P is supported by NUS President's Graduate Fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 675 kb)
Rights and permissions
About this article
Cite this article
Pasikanti, K.K., Norasmara, J., Cai, S. et al. Metabolic footprinting of tumorigenic and nontumorigenic uroepithelial cells using two-dimensional gas chromatography time-of-flight mass spectrometry. Anal Bioanal Chem 398, 1285–1293 (2010). https://doi.org/10.1007/s00216-010-4055-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4055-3