Abstract
A very sensitive and highly specific heterogeneous immunoassay system, based on surface-enhanced Raman scattering (SERS) and gold nanoparticles, was developed for the detection of bacteria and other pathogens. Two different types of gold nanoparticles (citrate-stabilized gold nanosphere and hexadecyltrimethylammonium bromide (CTAB)-stabilized gold nanorod particles) were examined and this immunoassay was applied for the detection of Escherichia coli. Raman labels were constructed by using these spherical and rod-shaped gold nanoparticles which were first coated with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) and subsequently with a molecular recognizer. The working curve was obtained by plotting the intensity of the SERS signal of the symmetric NO2 stretching of DTNB at 1,333 cm−1 versus the concentration of the E. coli. The analytical performance of gold particles was evaluated via a sandwich immunoassay, and linear calibration graphs were obtained in the E. coli concentration range of 101–105 cfu/mL with a 60-s accumulation time. The sensitivity of the Raman label fabricated with gold nanorods was more than three times higher than spherical gold nanoparticles. The selectivity of the developed sensor was examined with Enterobacter aerogenes and Enterobacter dissolvens, which did not produce any significant response. The usefulness of the developed immunoassay to detect E. coli in real water samples was also demonstrated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The identification and quantification of microorganisms have become a key point in biodefense, food safety, diagnostics, and drug discovery research. Identification of a microorganism is usually realized by traditional approaches such as culture-based and colony-counting methods which, in most cases, require several handling steps. The traditional methods are time consuming and inconvenient [1, 2]. Hence, rapid identification and detection of pathogens are needed. Rapid detection of bacteria is particularly important to allow timely intervention in managing health problems. Several rapid methods have been reported such as polymerase chain reaction, flow cytometry, and immunomagnetic separation (IMS) assays. IMS was coupled with various detection methods based on fluorophore, enzymes, and quantum dot (QD)-labeled antibodies [2–10]; and optical spectroscopies, among which vibrational and fluorescence spectroscopies are the most preferred methods in immunoanalytical measurements [11].
Surface-enhanced Raman scattering (SERS) has been used to obtain molecular information through sharp and easily distinguishable vibrational bands. This information is commonly used for detection of (bio)molecules [12]. The SERS technique may provide up to 1014-fold enhancement in Raman signal intensity, and this is sufficient to detect pico- to femtomolar amounts of biomolecules [12, 13]. Since the discovery of SERS, there has been growing interest in its use as a quantitative tool in the analytical sciences [14–16]. In particular, SERS has been used for biological sensing [17–19], and the usage of water as a solvent in Raman spectroscopy provides a facility for working directly with biological samples [20]. In recent microbiology studies, much attention has focused on SERS due to its attractive capabilities for single molecule detection, obtaining fingerprint signatures of the organism and rapid response time by using gold and silver nanoparticles [21, 22]. Nevertheless, improved instrument technology makes Raman instruments simple and useful, and some experiments have also shown that Raman scattering is more enhanced with some special substrates [11]. For this aim, special substrates are prepared by using gold and silver nanoparticles. SERS substrates constructed from gold and silver nanoparticles have been used for microorganism [23–25], protein [26, 27], and nucleotide [28] detection. Nanoparticle-based biosensors for detection of pathogens have also been developed by using magnetic nanoparticles [29, 30] and silver nanoparticles [31–34]. Nanoparticle cluster arrays provided SERS signals and enabled spectral discrimination of bacteria species [35–38]. In particular, silver and gold colloidal nanoparticle-based SERS can be used in routine and rapid screening of bacteria [36]. Most studies involve adding an external supply of gold or silver nanoparticles to the constituents of microorganisms and the SERS signals are collected from the outer cell membrane of bacteria; however, it is difficult to introduce a nanometer-sized colloidal metal particle into the cell membrane [37].
UV resonance Raman spectroscopy was also used to reproducibly acquire information-rich Raman fingerprints from bacteria [39]. An aggregated silver colloid substrate was used to analyze a collection of bacteria Escherichia coli [40]. However, one of the main drawbacks of the technique for microbiological samples with SERS has been the inability to acquire meaningful information from a region of the sample matrix where both the SERS substrate and biomass are present [41]. The potential of SERS for detection and identification of bacterial pathogens with species and strain specificity on gold-particle-covered glassy substrates was demonstrated [42]. The detection and identification of bacterial samples by SERS have been conducted by mixing suspensions of bacteria with a suspension of colloidal silver particles and its limit of detection (LOD) was reported to be 103 cfu/mL [43–48]. SERS has also been used to detect bacteria captured by polyclonal antibodies sorbed onto protein-A-modified silver nanoparticles. The selectivity and discrimination of the proposed technique were achieved by using an appropriate antibody [49].
Gold nanoparticles are one of the most important nanomaterials which are used as Raman labels to enhance SERS signal intensity. They have been successfully applied to labeling technology because of their controllable size distribution, long-term stability, and biocompatibility with antibodies, antigen proteins, DNA, etc. [50]. Gold nanorods have been shown to have distinct optical properties that depend on their shape. In particular, they have two plasmon absorption bands, one the transverse plasmon (TP) band, and the other the longitudinal plasmon (LP) band. The high-energy band corresponds to the oscillation of the electron perpendicular to the rod axis (TP) and the other absorption band, which is red-shifted, is caused by the oscillation of electrons along the rod axis (LP). The position and intensity of these bands can be affected by changes in the dielectric constant around the vicinity of nanoparticles. The elongated nanoparticles also have higher sensitivity to the local dielectric environment and observed SERS signal intensity is substantially increased [51].
In this paper, gold nanorods are introduced to the research of the SERS-based sandwich immunoassay for rapid detection and enumeration of E. coli. SERS signal intensities regarding two different types of gold nanoparticles, spherical citrate-stabilized gold nanoparticles and hexadecyltrimethylammonium bromide (CTAB)-stabilized gold nanorods, were compared. Gold nanoparticles and capture surfaces were characterized with transmission electron microscopy (TEM), UV–Vis spectroscopy, and atomic force microscopy (AFM) measurements. The analytical performance of the SERS-based sandwich immunoassay system with respect to linear range, detection limit, response time, and selectivity is presented and discussed. The ability of the immunoassay to detect E. coli in real water samples was investigated and the results are compared with the experimental results from plate-counting methods.
Experimental
Chemicals
Biotin-conjugated rabbit anti-E. coli polyclonal antibodies were obtained from Abcam plc. (Cambridge, UK). Hydrogen tetrachloroaurate (HAuCl4), 3-mercaptopropionic acid (3-MPA), hexadecyltrimethylammonium bromide (CTAB), l-ascorbic acid (AA) 98% ethanolamine, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), trisodium citrate dehydrate, absolute ethanol, and Tween 20 were obtained from Sigma-Aldrich (Taufkirchen, Germany). Silver nitrate (AgNO3) and sodium borohydride (NaBH4) were obtained from Merck (Darmstadt, Germany). 5,5′-Dithiobis(2-nitrobenzoic acid) (DTNB) was obtained from Acros (Morris Plains, NJ). Immunopure avidin was obtained from Pierce Biotechnology (Rockford, IL). N-Hydroxysulfosuccinimide sodium salt (NHS) was obtained from Pierce Biotechnology (Bonn, Germany). NaCl, Na2HPO4, and KH2PO4 were from J.T. Baker (Deventer, the Netherlands), used as phosphate-buffered saline (PBS). All solutions were prepared with ultrapure water (18 MΩ cm) to reach the desired concentrations.
Microorganism
Escherichia coli K12 strain, Enterobacter aerogenes, and Enterobacter dissolvens were obtained from Refik Saydam National Type Culture Collections, Ankara, Turkey. The cultures for assay were grown on tryptic soy broth (TSB; Merck KgaA, Germany) at 37 °C for 18 h. Bacteria samples were prepared by diluting the culture in 0.1 M PBS at pH of 7.5. Also, for E. coli detection, VRB agar was obtained from Merck (Darmstadt, Germany) in order to enumerate the number of colony forming units (cfu). For this aim, diluting and plating methods were used and incubation was implemented at 37 °C for 18 h.
Instrumentation
Absorption spectra of spherical gold and rod-shaped gold nanoparticle solutions were recorded with an Agilent 8453 UV–Vis spectrophotometer (Agilent Technologies, Inc., Santa Clara, CA) with a photodiode array detector. TEM measurements were performed on a JEOL 2100 HRTEM instrument (JEOL Ltd., Tokyo, Japan). TEM samples were prepared by pipetting 10 µL of nanoparticle solution onto TEM grids that were then allowed to stand for 10 min. DeltaNu Examiner Raman microscope (Deltanu Inc., Laramie, WY) with a 785-nm laser source, a motorized microscope stage sample holder, and a CCD detector was used to detect E. coli. Instrument parameters were as follows: ×20 objective, 30-µm laser spot size, 100-mW laser power, and 60-s acquisition time. Baseline correction was performed for all measurements. Sandwich immunoassays on the gold-coated slides were also visualized step by step with atomic force microscopy (AFM, XE-100E; Park Systems Corp., Suwon, Korea). The measurements were performed in the non-contact mode by using 910 M-NSC14/Cr–Au-type cantilevers with 0.3 to 0.5-Hz scanning speed. Before AFM measurements, the samples were washed with ultrapure water (18.3 mΩ cm) and allowed to dry in air. In order to minimize the deformation of E. coli, which is caused by dehydration, the bacteria-containing samples were measured right after drying in air.
Synthesis of gold nanoparticles
The spherical gold nanoparticles were synthesized according to Sutherland’s procedure [52] in a 500-mL round-bottom flask equipped with a condenser. 500 mL of 0.01% HAuCl4 solution was prepared and boiled with stirring. Then 7.5 mL of 1% sodium citrate solution was poured into the boiling solution. Approximately 60 s later the color of the solution turned to the color of red wine. Boiling was continued for 15 min, after which the solution was cooled to make it ready for use.
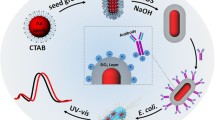
Gold nanorods were prepared by using a seed-mediated growth technique with slight modification [53]. Seed solution was prepared by mixing 7.5 mL of 0.1 M CTAB and 250 µL of 0.01 M HAuCl4 solution. Once mixed, 600 µL of 0.01 M ice-cold NaBH4 was added rapidly to the resulting solution, and allowed to stand for 5 min to form seed solution.
To prepare rod-shaped gold nanoparticles, 4.75 mL of 0.1 M CTAB, 1 mL of 0.01 M HAuCl4, and 60 µL of 4 × 10−3 M AgNO3 were mixed, respectively; the resulting color was dark orange. When the dark orange color was observed, 250 µL of 0.1 M AA was added dropwise to the resulting solution to afford the stock solution. The solution turned colorless after adding 250 µL AA. Then, 5 µL seed solution was added to the stock solution. The final mixture was stirred for a few seconds and allowed to stand for 3 h at room temperature to afford nanorods.
Preparation of Raman labels
Equal quantities (1 mg) of spherical gold nanoparticles and gold nanorods were used as Raman labels by assembling DTNB on nanoparticles. The self-assembled monolayer (SAM) was prepared by using 50 mM DTNB and it was formed on rod-shaped and spherical gold nanoparticles in absolute ethanol overnight to track the SERS signal. After creating the self-assembled monolayer, modification of the carboxylate groups of DTNB was performed.
Sandwich immunoassay protocol
The assay was prepared in two steps. First, the surface of gold-coated slides was modified with 100 mM 3-MPA in ethanol overnight. For surface activation of carboxylate groups, slides were immersed for 30 min in 0.05 M NHS and 0.2 M EDC solution prepared in 0.1 M MES buffer at pH 6.5. After surface activation, gold-coated slides were immersed in MES buffer containing 0.5 mg/mL avidin for 30 min. Then, 0.1 mg/mL biotinylated antibody was bound to gold-coated slides via biotin–avidin affinity. To avoid nonspecific interaction 10% (v/v) ethanolamine was used to block active groups on the surface for 1 h. Then, surfaces were immersed individually in each bacterium concentration (101–105 cfu/mL) for 30-min incubation. At the end of the incubation period, slides were washed with PBST solution (mixture of 0.1 M PBS buffer at pH 7.4 and 0.05% Tween). In the second step, 50 mM DTNB was formed as a self-assembled monolayer on spherical and rod-shaped gold nanoparticles in absolute ethanol overnight. Then, the same biotinylated antibody binding procedure was followed for nanoparticles. The two components of the assay, i.e., bacteria on the slides and nanoparticles which had been stamped with DTNB, were then merged by immersing the gold-coated slides into the gold nanoparticle solution for 30 min.
When the assay was completed, a washing procedure was begun by using PBST for slides surfaces. After washing with PBST, the slides were put in an ultrasound bath containing ultrapure water for 10 s. In the last step of the washing procedure, slides were washed with deionized water in preparation for SERS measurement and AFM imaging. The cross specificity of the developed immunoassay was determined by following the same assay procedure using E. aerogenes and E. dissolvens, which are also in the group of coliform bacteria. The SERS intensities obtained for each bacteria were compared with the results obtained for E. coli. The developed immunoassay was tested with the actual water samples collected from a lake, tap water, and bacteria-inoculated tap water. For this purpose, E. coli was enumerated by using the developed immunoassay in lake water and tap water. The results were compared with those obtained from the plate-counting method. For the plate-counting method, the serial dilutions of water samples were prepared in PBS, and a 100-µL sample from the serial dilutions was plated on VRB agar and incubated at 37 °C for 24 h. The purple colonies were counted to determine the amount of E. coli in the water samples.
Results and discussion
Particle characterization
Figure 1a and 1b shows representative TEM images of spherical gold nanoparticles and gold nanorods, respectively. As shown in Fig. 1a, spherical gold nanoparticles have an average size of 20 nm. The shape of nanorods is determined by the particles’ aspect ratio which is defined as the length of a particle divided by its width. Aspect ratio, length, and width of the synthesized gold nanorods were determined as approximately 3, 45 nm, and 15 nm, respectively (Fig. 1b). Figure 2 shows the extinction spectrum for each colloid, which is used to examine the shape of gold nanoparticles. The first plasmon band of the gold nanorods belongs to the transverse plasmon band at 524 nm, and the second plasmon band belongs to the longitudinal plasmon band at 665 nm. On the other hand, spherical gold nanoparticles have a plasmon band at 521 nm as shown in Fig. 2.
Assay design
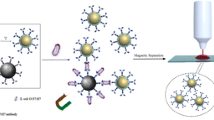
Scheme 1 illustrates the overall strategy for using the SERS-based sandwich immunoassay for E. coli detection. This schema, originally developed by Narayanan and co-workers, was adapted to our system [54]. Briefly, a sensing surface is constructed by attaching biotinylated polyclonal antibodies on avidin-coated gold surface. Raman label preparation involves first the covalent linkage of SERS reporter molecule (DNTB) to the gold nanoparticles. In this study, commercially available DTNB was chosen as the SERS reporter molecule owing to its ability to generate strong Raman signals. Antibody binding to the nanoparticle surfaces was accomplished by typical avidin–biotin interaction. Once E. coli was captured by the antibody-coated gold slide after a 30-min incubation period, the reporter-labeled secondary antibody was attached to the bacteria surface. SERS measurement was performed after removing the nonspecifically bound reporter-labeled antibody by applying the washing procedure.
Characterization of surface
Atomic force microscopy (AFM) measurements were performed in addition to SERS experiments in order to demonstrate the appropriate surface modification of gold-coated slides, the immobilization of E. coli, and the existence of gold nanoparticles on the bacteria. The roughness values for different surfaces, which are obtained in a step by step manner, are given in Table 1. It can be seen from Fig. 3a that the bare gold-coated slide surface is clean enough and the arithmetic average (R a) and root mean square roughness (R q) values are 0.250 and 0.384 nm, respectively, for this surface. After the self-assembly of the 3-MPA monolayer on the surface, as presented in Fig. 3b, the R a and R q values slightly increased to 0.281 and 0.387 nm, respectively. In Fig. 3c, avidin clusters on the surface can be seen. This image is consistent with images given in the literature. The approximate size of the avidin knobs is measured as 200 nm, and since the sizes of the avidin knobs are higher than a single avidin molecule we concluded that avidin exists as clusters on the surface [55]. Coating of the surface with avidin and then addition of antibody to the sequence both result in considerable increases in R a and R q values as expected, and these values are measured as 0.981 and 1.326 nm for avidin and 2.422 and 3.297 nm for the antibody being the top layer on the surface. So, it can be concluded from the topographic evaluations of the samples via AFM as seen in Fig. 3a–d as well as the roughness values that the surface modification of the gold-coated slides is achieved in a step by step manner.
In Fig. 4a–c, the error and the 3D topography images signify that the bacteria are immobilized on the modified surface and the surfaces of the bacteria are coated with gold nanorods and spherical gold nanoparticles, respectively. In Fig. 4c the 3D topography image of the spherical gold nanoparticle surface on E. coli gives more insight into the changes observed at the surface of the bacteria upon addition of spherical gold nanoparticles. From the AFM image presented in Fig. 4a, it can be seen that the bacteria have a size of 2.2 μm, and the size and shape of the E. coli are very similar to those in images presented in the literature [56, 57]. In Fig. 4b and c, the gold nanoparticles and nanorods on the bacteria can be seen. The increase in the roughness values can be considered as additional evidence that the bacteria are coated with nanoparticles. The R a and R q values for the single bacterium are 20.716 and 24.116 nm, respectively, as shown in Fig. 4a. After coating with gold nanoparticles the R a and R q values become 50 and 91 nm for spherical gold nanoparticles and 74 and 98 nm for gold nanorods.
Heterogeneous detection of E. coli using SERS
The SERS spectra for E. coli assays conducted by using Raman label constructed from spherical gold nanonparticles and gold nanorods are presented in Figs. 5 and 6. The spectra contain features which are attributable to Raman label (reporter molecule) and are dominated by bands representative of the DSNB-based adlayer (e.g., the symmetric nitro stretch (ν s (NO2)) at 1,336 cm−1 and an aromatic ring stretching mode at 1,588 cm−1) [58]. Figure 7 displays a typical SERS response of the immunoassay system after addition of various concentrations of E. coli. In each case, the intensity of NO2 band, which is the strongest of the observed features, is used to construct the dose–response plots as shown in Fig. 7. The error bars in the calibration curve represent the standard deviations in the signals obtained from five measurements. The SERS signal tracks with E. coli concentration and become distinguishable when 101 bacteria exist in a sample matrix. The SERS signal exhibits a linear dependence within the concentration range of 4.9 × 101–4.9 × 105 cfu/mL for spherical gold nanoparticles and 7.5 × 101–7.5 × 105 cfu/mL for gold nanorods. The dose–response curve was analyzed and good linear fits were acquired for both type of nanoparticles with high correlation coefficients (R 2), 0.999 and 0.989 for spherical and rod-shaped nanoparticles, respectively. An assessment of the slopes of the two dose–response plots indicates that the response from the gold nanorods is approximately more than 3.2 times stronger than that for the spherical gold nanoparticles. The LODs for E. coli with the gold nanorods and the spherical gold nanoparticles were found to be 4 and 5 cfu/mL, respectively, defined as the equivalent of the average signal of the blank sample plus three times its standard deviation.
The selectivity of the developed immunoassay was investigated by using other coliform bacteria, which can be found in water as a result of fecal contamination. For this purpose, E. aerogenes (3.1 × 105 cfu/mL) and E. dissolvens (4.3 × 105 cfu/mL) were subjected to the same procedure as that of E. coli solution, and the SERS intensities were measured. The results are given in Fig. 8. The intensities for E. aerogenes and E. dissolvens in high concentrations were lower than the detection limit of the developed assay, which shows the high specificity of the immunoassay.
To check the accuracy of the applied method, real sample analysis has been performed for E. coli detection and obtained results were compared with the classical counting methods. Samples that were taken from lake and tap water were used with the developed immunoassay method and SERS signal was followed in order to detect bacteria concentration via the calibration curve. Raman signal was not observed in tap water samples. On the other hand, SERS signal was obtained for the lake water sample. Bacteria concentration was determined to be 2.5 × 102 cfu/mL according to the calibration curve for spherical gold nanoparticles; a similar result was also obtained with gold nanorods, 2.9 × 102 cfu/mL. In comparison, E. coli concentration was also determined with conventional plate-counting method. No E. coli was detected in tap water, and there was 2.0 × 102 cfu/mL E. coli in the lake sample based on the counting method. The results obtained with SERS measurements were close to results of classical counting methods. To confirm the accuracy of the assay, recovery experiments were also carried out with tap water. Known concentrations of E. coli ranging from 3.5 × 103 to 3.5 × 105 cfu/mL were added to tap water from stock bacteria solution (3.5 × 108 cfu/mL), and enumeration was implemented by using both spherical gold nanoparticles and gold nanorods. Results were obtained from the calibration curves for the spherical gold nanoparticles and gold nanorods as 1.0 × 103, 3.0 × 104, 3.9 × 105 cfu/mL and 1.6 × 103, 2.0 × 104, 3.2 × 105 cfu/mL, respectively.
The total analysis time, which includes capturing the bacteria (30 min) on the gold slide, SERS-labeled antibody interaction (30 min), SERS measurement and other activities (approximately 10 min), was less than 70 min. The time for the developed assay seems quite short when compared with other novel immunoassays. For example, ELISA for the detection of E. coli took approximately 1 day [59]. The time of analysis for real-time PCR was more than 5 h, and it was 170 min by quartz crystal microbalance biosensor and 2 h by immunomagnetic separation with QD-labeled secondary antibodies [2, 59]. Analysis time of the developed methods was comparable to that of a surface plasmon resonance (SPR) biosensor (30 min) for E. coli detection with a high number of parallel measurements that is limited in SPR biosensors [60].
Conclusion
Rapid detection and enumeration of E. coli were achieved by using a SERS-based sandwich immunoassay. Gold nanoparticles exhibit unique electronic and optical properties that are critically related to their size and shape. It is possible to control the aspect ratio of gold nanorods by appropriately adjusting the synthesis conditions. Therefore, the optical properties of gold nanorods are varied and they could be used in designing the SERS-based immunoassay system for E. coli detection. SERS results showed that gold nanorods substantially enhance the sensitivity of immunoassay method as compared with that of spherical gold nanoparticles. The intensity enhancement was found to be at least threefold when compared with the spherical gold nanoparticles. Real sample analysis has been performed for E. coli detection and obtained results are in good agreement with the classical counting methods. The selectivity and discrimination of the proposed method was secured by using an appropriate antibody corresponding to the E. coli.
Most water quality standards were set at 1,000 coliforms per 100 mL and the health goal for total coliforms at zero for drinking water [61]. The LOD value of the developed immunoassay is lower then the requirement of the water quality standards. On the other hand the sensitivity of the assay described here is not high enough to detect such low concentrations required for drinking water. But it can be improved with a preconcentration or preincubation step. In addition, SERS spectra of the Raman labels have characteristic sharp bands. This approach opens the possibility to design multiplex and simultaneous detection of bacteria; employing them in food and water samples requires further investigation.
References
Dudak FC, Boyacı İH (2009) Biotechnol J 4:1003–1011
Dudak FC, Boyacı İH, Jurkevica A, Hossain M, Aguilar ZP, Halsall HB, Seliskar CJ, Heineman WR (2009) Anal Bioanal Chem 393:949–956
Fu Z, Rogelj S, Kieft LT (2005) Int J Food Microbiol 99:47–57
Kaclikova E, Pangallo D, Oravcova K, Drahovska H, Kuchta T (2005) Lett Appl Microbiol 41:132–135
D’Souza JM, Wang L, Reeves P (2002) Gene 297:123–127
Yamaguchi N, Sasada M, Yamanaka M, Nasu M (2003) Cytometry 54:27–35
Sachidanandham R, Yew-Hoong GK, Laa PC (2005) Biotechnol Bioeng 89:24–31
Liu Y, Li Y (2002) J Microbiol Methods 51:369–377
Gehring GA, Irwin LP, Reed SA, Tu IS, Andreotti EP, Akhavan-Tafti H, Handley SR (2004) J Immunol Methods 293:97–106
Hara-Kudo Y, Konuma H, Nakagawa H, Kumagai S (2000) Lett Appl Microbiol 30:151–154
Brown LO, Doorn SK (2008) Langmuir 24:2277–2280
Woo MA, Lee SM, Kim G, Baek JH, Noh MS, Kim JE, Park SJ, Minai-Tehrani A, Park SC, Seo YT, Kim YK, Lee YS, Jeong DH, Cho MH (2009) Anal Chem 81:1008–1015
Lin CC, Yang YM, Chen YF, Yang TS, Chang HC (2008) Biosens Bioelectron 24:178–183
Moskovits M (2006) Top Appl Phys 103:1–17
Haynes CL, Yonzon CR, Zhang X, Van Duyne RP (2005) J Raman Spectrosc 36:471–484
Vo-Dinh T (1995) Sens Actuators B29:183–189
Vo-Dinh T (1998) TrAC-Trends Anal Chem 17:557–582
Vo-Dinh T, Yan F, Stokes DI (2005) Methods Mol Biol 300:255–283
El-Kovedi M, Keating CD (2004) Concepts, applications and perspectives. In: Niemeyer C, Mirkin CA (eds) Nanobiotechnology. Wiley-VCH, Weinheim, pp 429–443
Smith E, Dent G (2005) Introduction, basic theory and principles. Modern Raman spectroscopy: a practical approach. Wiley, Chichester, England, pp 1–20
He L, Liu Y, Lin M, Mustapha A, Wang Y (2008) Sens & Instrumen Food Qual 2:247–253
Efrima S, Zeiri L (2008) J Raman Spectrosc 40:277–288
Dutta RK, Sharma PK, Pandey AC (2009) Dig J Nanomater Biostruct 4:83–87
Liu Y, Chao K, Nou X, Chen Y (2008) Sens & Instrumen Food Qual 3:100–107
Montaya JR, Armstrong RL, Smith GB (2003) Proc SPIE-Int Soc Opt Eng 5085:144–152
Fabris L, Dante M, Nguyen TQ, Tok JBH, Bazan GC (2008) Adv Funct Mater 18:1–8
Drachev VP, Thoreson MD, Nashine V, Khaliullin EN, Amotz DB, Davisson VJ, Shalaen VM (2005) J Raman Spectrosc 36:648–656
Bell SEJ, Sirimuthu NMS (2006) J Am Chem Soc 128:15580–15581
Lin YS, Tsai PJ, Wang MF, Chen YC (2004) Anal Chem 76:7162–7168
El-Boubbou K, Gruden C, Huang XF (2007) J Am Chem Soc 129:13392–13393
Varshney M, Li YB (2007) Biosens Bioelectron 22:2408–2414
Naja G, Bouvrette P, Hrapovic S, Luong JT (2007) Analyst 132:679–686
Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, Ziegler LD (2005) J Phys Chem B 109:312–320
Kalele SA, Kundu AA, Gosavi SW, Deobagkar DN, Deobagkar DD, Kulkarni SK (2006) Small 2:335–338
Yan B, Thubagere A, Premasiri WR, Ziegler LD, Dal Negrus L, Reinhard BM (2009) ACS Nano 3:1190–1202
Culha M, Adıguzel A, Yazıcı MM, Kahraman M, Sahin F, Gulluce M (2008) Appl Spectrosc 62:1226–1232
Jarvis RM, Law N, Shadi IT, O’Brien P, Lloyd JR, Goodacre R (2008) Anal Chem 80:6741–6746
Goodacre R, Timmins EM, Burton R, Kaderbhai N, Woodward AM, Kell DB, Rooney P (1998) J Microbiol 144:1157–1170
Lopez-Diez EC, Goodacre R (2004) Anal Chem 76:585–591
Jarvis RM, Goodacre R (2004) Anal Chem 76:40–47
Jarvis RM, Brooker A, Goodacre R (2004) Anal Chem 76:5198–5202
Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, Ziegler LD (2005) J Phys Chem B 109:312–320
Sengupta A, Mujacic M, Davis EJ (2006) Anal Bioanal Chem 386:1379–1386
Jarvis R, Clarke S, Goodacre R (2006) Top Appl Phys 103:397–408
Zeiri L, Bronk BV, Shabtai Y, Czeye J, Efrima S (2002) Colloids Surf A Physicochem Eng Asp 208:357–362
Zeiri L, Bronk BV, Shabtabi Y, Eichler J, Efrima S (2004) Appl Spectrosc 58:33–40
Efrima S, Bronk BV (1998) J Phys Chem B 102:5947–5950
Kahraman M, Yazici MM, Sahin F, Culha M (2007) J Biomed Opt 12(5):054015/1–054015/6
Liu Y, Chen YR, Nou X, Chao K (2007) Appl Spectrosc 61:824–831
Xu W, Xu S, Ji X, Song B, Yuan H, Ma L, Bai Y (2005) Colloids Surf B Biointerfaces 40:169–172
Orendorff CJ, Gole A, Sau TK, Murphy CJ (2005) Anal Chem 77:3261–3266
Sutherland WS, Winefordner JD (1992) J Colloid Interface Sci 48:129–141
Nikoobakht B, El-Sayed MA (2003) Chem Mater 15:1957–1962
Radha Narayanan R, Lipert RJ, Porter MD (2008) Anal Chem 80:2265–2271
Tero R, Misawa N, Watanabe H, Yamamura S, Nambu S, Nonogaki Y, Urisu T (2005) e-J Surf Sci Nanotechn 3:237–243
Udomrat S, Praparn S, Puntheeranurak T (2009) J Microscopy Soc Thail 23:38–41
Jing W, Shiying H, Lina X, Ning G (2007) Chin Sci Bull 52:2919–2924
Grubisha DS, Lipert RS, Park HY, Driskell J, Porter MD (2003) Anal Chem 75:5936–5943
Lazcka O, Del Campo FJ, Munoz FX (2007) Biosens Bioelectron 22:1205–1217
Dudak FC, Boyacı IH (2007) Food Res Int 40:803–807
US Environmental Protection Agency (2008) Ground water & drinking water. http://www.epa.gov/safewater. Accessed 21 Mar 2010
Acknowledgment
The authors are grateful for the financial support provided by The Scientific and Technological Research Council of Turkey; Project Number: 107 T682-COST MP0701.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Temur, E., Boyacı, İ.H., Tamer, U. et al. A highly sensitive detection platform based on surface-enhanced Raman scattering for Escherichia coli enumeration. Anal Bioanal Chem 397, 1595–1604 (2010). https://doi.org/10.1007/s00216-010-3676-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-3676-x