Abstract
Competent laboratories monitor genetically modified organisms (GMOs) and products derived thereof in the food and feed chain in the framework of labeling and traceability legislation. In addition, screening is performed to detect the unauthorized presence of GMOs including asynchronously authorized GMOs or GMOs that are not officially registered for commercialization (unknown GMOs). Currently, unauthorized or unknown events are detected by screening blind samples for commonly used transgenic elements, such as p35S or t-nos. If (1) positive detection of such screening elements shows the presence of transgenic material and (2) all known GMOs are tested by event-specific methods but are not detected, then the presence of an unknown GMO is inferred. However, such evidence is indirect because it is based on negative observations and inconclusive because the procedure does not identify the causative event per se. In addition, detection of unknown events is hampered in products that also contain known authorized events. Here, we outline alternative approaches for analytical detection and GMO identification and develop new methods to complement the existing routine screening procedure. We developed a fluorescent anchor-polymerase chain reaction (PCR) method for the identification of the sequences flanking the p35S and t-nos screening elements. Thus, anchor-PCR fingerprinting allows the detection of unique discriminative signals per event. In addition, we established a collection of in silico calculated fingerprints of known events to support interpretation of experimentally generated anchor-PCR GM fingerprints of blind samples. Here, we first describe the molecular characterization of a novel GMO, which expresses recombinant human intrinsic factor in Arabidopsis thaliana. Next, we purposefully treated the novel GMO as a blind sample to simulate how the new methods lead to the molecular identification of a novel unknown event without prior knowledge of its transgene sequence. The results demonstrate that the new methods complement routine screening procedures by providing direct conclusive evidence and may also be useful to resolve masking of unknown events by known events.

Molecular toolbox for the identification of genetically modified organisms
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past decade, genetically modified (GM) crops have become widely adopted. Since 1996, a total of 670 approvals in 55 countries have been granted for 144 events in 24 crops. The number of GM crops, events, and countries where they are cultivated is steadily increasing [1]. Currently, 33 GM crops are commercialized worldwide, and more than 90 novel events are in advanced stages of the development, authorization, or commercialization pipeline and may enter the market in the near future [2, 3]. Competent authorities worldwide therefore face the increasingly complex task to monitor the presence of existing and novel genetically modified organisms (GMO) in the food/feed chain and in the environment. To facilitate GMO labeling and traceability, authorization for commercialization requires the development and validation of an event-specific detection method for each GM event, together with appropriate reference material. Currently, 37 officially validated event-specific methods are available to competent laboratories in the European Union (EU), including detection methods for GMOs approved in the EU and some asynchronously authorized GMOs [4].
Occasionally, unauthorized GM events unintentionally enter the food/feed chain, such as Starlink, Bt10, LLRice601, and Bt63 [5–9]. The socioeconomic risks associated with the release of unapproved events [2, 10, 11] raise the need for early discovery of such incidents. Two classes of unauthorized events are discriminated: (1) GM events that are officially registered for commercialization and approved in some, but not all jurisdictions, e.g., due to differences in the authorization procedure or timeline (asynchronous authorization). For these, DNA sequence information from the GM event or an event-specific detection method may be available from the jurisdiction that has approved the event; (2) GM events that are not (yet) officially registered in any jurisdiction. In this report, we refer to these GMOs as “unknown events.” The difficulty in detecting unknown events is that lack of officially available information at the DNA sequence level precludes the design of an analytical detection method. In addition, such events have usually not been subject to official safety assessment. Here, we focus on the development of analytical detection strategies to detect and identify unknown events. We outline the possible scenarios that may be followed (Fig. 1, steps 1–13) and demonstrate newly developed methods to complement existing routine analytical procedures.
In principle, strategies for the detection of GMOs, including unknown events, can be targeted or nontargeted, both at the level of product sampling and at the level of analytical detection. First, at the level of product sampling, if information on the GMO composition of products and their distribution on the market is available, a targeted sampling of products can be performed (step 1). Otherwise, the discovery of unknown events depends on nontargeted random sampling of products (step 2). Second, at the level of analytical detection, a targeted strategy requires transgene sequence information for the development of a corresponding analytical test per event. Systematically performing such tests leads to direct evidence for the presence or absence of all known events in a given sample (step 3). Likewise, if information can be collected on the release of a novel event, including the transgene sequence information, a dedicated test may be designed for targeted analytical confirmation. Elsewhere, we have described how knowledge technologies may be used to collect documented information on the release of novel GMOs and how this may complement product sampling schemes and analytical testing [11]. Recently, an advertisement indicated the commercial availability of a novel GMO-derived product [11, 12]. The product contains recombinant human intrinsic factor (rhIF) collected from transgenic Arabidopsis thaliana leaves and vitamin B12. It is intended for patients suffering from vitamin B12 deficiency. Here, as a first objective of this report, we first report on the molecular analytical confirmation of the identity of this novel GMO by sequencing of the insert (step 3). Thus, we demonstrate the feasibility of the targeted approach.
This procedure was clearly different from the current routine analysis. In daily practice, competent laboratories receive blind samples for analysis and have to establish the GMO composition by identification and/or quantification of GM events. In the vast majority of the cases, information on the GMO content and/or DNA sequence level is not available, and a screening analysis has to be performed (steps 4–8). The routine screening analysis is usually divided into two parts to increase the efficiency and reduce the cost (steps 4–8). First, screening tests are performed to establish whether or not a particular product contains any GMOs. These tests detect commonly used transgenic elements (so-called screening elements) such as the CaMV 35S promoter (p35S), nopaline synthase terminator (t-nos), or the bar/pat, nptII, cry, and epsps genes [13–25] (step 4). Such screening tests may identify negative samples with only a limited number of tests (step 5). If positive samples are identified, subsequent analytical evidence must be obtained to identify the causative events (steps 6–13). A short list of candidate known events is deduced from the combinatorial presence of one or more positively detected screening elements. This method is called the “matrix approach” because a table (mathematical matrix) listing all screening elements that may be detected per event is used to identify the candidates. Finally, these candidate events are tested using qualitative or quantitative event-specific real-time polymerase chain reaction (RT-PCR) assays to establish the GMO composition of the sample (steps 6 and 7).
In addition, the screening approach may indicate the presence of unknown events, provided that the unknown event contains at least one of the screening elements. However, the matrix approach is inherently limited in three ways. First, the presence of unknown events is only inferred when positively detected screening elements cannot be explained by the presence of known events (step 8). Therefore, the evidence for unknown events is indirect because it is based on a set of negative observations: the failure to detect any known event. Second, the evidence for the presence of an unknown event is inconclusive because the screening method by itself only detects a common transgenic sequence but does not identify the causative event per se. Third, in the screening procedure, an unknown event may remain unnoticed if the presence of known events does explain the detection of screening elements. Increasingly, more products are expected to contain a low-level presence of one or more known events, which, in turn, may “mask” the presence of novel unknown events. This is because the number of commercialized events and the scale of commercial cultivation are continuously increasing [1, 2]. For instance, in the main GM-producing countries (USA, Argentina, Brazil, Canada, India, China), GM crop adoption rates for maize, soybean, cotton, and canola have reached 80–90% of the crop cultivation area [26]. To resolve these issues, several methods have been suggested to detect unknown events, including differential quantitative PCR [27], GM fingerprinting by genome walking [28, 29], the use of high-density GM microarray chips [30], or next-generation high-throughput sequencing. Differential quantitative PCR aims to test whether a given sample contains an unknown (or unapproved) GMO by comparing the number of detected molecules of a common sequence (such as a screening element) to the number of detected molecules identifying the known (approved) GMOs (determined by event-specific PCR). A significant difference infers the presence of an unknown GMO. This approach is useful in detecting masked unknown events, but it does not identify the causative event. While proof of concept has been demonstrated for some, these methods await further development before implementation in routine analysis is possible. In addition, the level of expertise required and the high cost associated with alternative methods, such as high-density GM microarray analysis or next-generation high-throughput sequencing, reduce the applicability of these methods to a limited number of stakeholders and/or a limited number of samples.
As a second objective of this report, we set out to test whether it would have been possible to identify the novel GMO if it had been provided to a laboratory as a routine blind sample. For this purpose, we had to develop new methods to identify unknown events because these are lacking from the routine screening procedure (see above). Identification of unknown events can be based on amplification and characterization of screening element flanking sequences. So, in the second part of this report, we demonstrate the extended procedure by purposefully treating the novel GMO-derived product mentioned above as a blind sample, i.e., without making use of prior transgene sequence information. In this way, we simulate how unknown events (exemplified by this novel GMO), in general, can be identified using the new procedure. Proof of concept is based on the two most frequently detected screening elements during routine analysis (p35S and t-nos), which were also present in the novel GMO material. So, the procedure can be directly applied to any other unknown event that contains p35S and/or t-nos elements. To provide a complete description of the procedure, we first performed the existing routine analyses (steps 4–8). Then, we used two methods for amplification of screening element flanking sequences. (1) We used conventional PCR amplification to amplify intervening sequences between multiple screening elements (step 9). This step simply uses the same primers from the screening tests but now in all-against-all combinations. (2) We developed a novel fluorescent anchor-PCR GM fingerprinting method (step 10) to amplify sequences flanking the p35S and/or t-nos elements, optionally followed by sequencing of amplicons (step 11) to identify unknown events. In parallel, we established a collection of in silico calculated fingerprints of known events to support interpretation of anchor-PCR fingerprints of blind samples (step 12). We used a test set of 13 known events with sufficient sequence information. Such a collection of fingerprints may be useful in quickly determining whether the amplicons in a blind sample are derived from known events, unknown events, or a mixture thereof. So, the anchor-PCR fingerprint may lead to direct evidence of the presence of an unknown event in a sample (step 13). Thus, it complements the indirect and/or inconclusive evidence obtained in the routine screening approach used today.
Experimental methodology
DNA extraction, PCR, Q-PCR analysis, and sequencing
A novel GMO-derived product was obtained as described elsewhere [11]. The product contains rhIF collected from dried powdered transgenic A. thaliana leaves [12]. Genomic DNA (gDNA) extraction was performed using the DNeasy Plant Mini Kit extraction procedure (Qiagen). High purity and integrity of the DNA extract were confirmed by agarose gel electrophoresis and spectrophotometry (Nanodrop ND1000). Screening tests for p35S and t-nos elements [13] were performed using a commercial kit for detection (Diagenode) according to the manufacturer’s instructions, on an ABI7000 real-time PCR system (Applied Biosystems). The real-time PCR-based ready-to-use multitarget analytical system developed by the EC-JRC was used for the event-specific simultaneous detection of 39 events (all EU-approved and unapproved events for which a method was submitted to the Community Reference Laboratory for GM Food and Feed) and of the corresponding plant species [31]. The system consists of 96-well prespotted plates containing lyophilized primers and probes for the individual detection of all GM events and of reference genes for rice, maize, soybean, cotton, potato, sugar beet, and oilseed rape. A positive and a negative control sample were provided together with the system. The assay was performed with the TaqMan Universal PCR Master Mix, on an ABI7000 real-time PCR system (Applied Biosystems).
Various primer pair combinations were used to amplify overlapping fragments of the T-DNA insert by conventional PCR. Primers were designed using Primer3 software on the left border region, the right border region, the antibiotic resistance selection marker, and the expressed trait present in the novel GM event. The p35S and t-nos primers were the same as those used in the screening tests (Supplemental Table 1). Each reaction contained 1.5 mM MgCl2, 400 nM of each primer, 2.5 U Amplitaq DNA polymerase (Applied Biosystems), 200 µM dNTPs, and 25 ng of genomic DNA in a total volume of 50 µl. Amplification was performed with the following program: 95 °C 10:00; 35 cycles at 95 °C 0:30, 58 °C 0:30, and 72 °C 5:00, final extension at 72 °C 5:00. PCR products were analyzed by 1.2% agarose gel electrophoresis, purified using MSB Spin PCRapache purification kit (Invitek), and were used directly for sequencing using the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems).
Fluorescent anchor-PCR
Existing protocols [32–34] were adapted leading to the following setup: adapters were designed for use with the restriction enzymes BfaI, MboI, BamHI, EcoRI, NcoI, XbaI, and XmaI. Adapters were designed such that the restriction site is lost after adapter ligation. Enzymatic digestion of genomic DNA and adapter ligation are performed simultaneously to increase the efficiency of the adapter ligation. Restriction–ligation reactions are performed overnight at 25 °C, in a total volume of 10 µl. Each reaction contains 100 ng genomic DNA, 50 pmol of the respective adapter, 24 U T4 ligase (NEB), 5 U (BfaI, MboI), 10 U (XmaI, NcoI), or 20 U (EcoRI, BamHI, XbaI) restriction enzyme (NEB), and 1× T4 ligase buffer (containing 1 mM ATP, NEB) supplemented with an extra 600 µM ATP (Roche Diagnostics). Subsequently, a touchdown PCR is performed for each combination of the respective adapter primer and one of the four anchor primers (p35S-F, p35S-R, tNOS-F, tNOS-R), with the following PCR program: 95 °C 10:00; eight cycles at 95 °C 0:15 (decreasing from 66 to 59 °C at 1 °C per cycle over eight cycles) for 0:30, 72 °C 2:00; 25 cycles at 95 °C 0:15, 58 °C 0:30, 72 °C 2:00; final extension at 72 °C 5:00. Each reaction contained 1.5 mM MgCl2, 500 nM of each primer, 0.25 U of Jumpstart DNA polymerase (Sigma), 200 µM dNTPs, and 1 µl of the undiluted ligation mixture in a total volume of 10 µl. Primary anchor-PCR products are 100-fold diluted prior to the second PCR. For each primary anchor-PCR product, three separate PCRs are performed in parallel, using three anchor primers that are labeled with HEX, FAM, or NED. These anchor primers are nested with respect to the first anchor primer. Amplification was performed with the same reaction composition as the first anchor-PCR but with the following program: 95 °C 10:00; ten cycles at 95 °C 0:15, 63 °C 0:30, 72 °C 2:00; 20 cycles at 95 °C 0:15, 60 °C 0:30, 72 °C 2:00; final extension at 72 °C 5:00. All primers are listed in Supplemental Table 1. These fluorescently labeled anchor-PCR amplicons are pooled, together with a ROX-labeled Genescan-500 or Genescan-1000 length marker (Applied Biosystems), and analyzed by capillary electrophoresis on a ABI3130 Genetic Analyzer, using POP-7 polymer in a 50-cm capillary array (Applied Biosystems). Fragment analysis was performed using GeneMapper v4.0 software (Applied Biosystems) using the Microsatellite Default analysis method for peak calling.
In silico calculation of anchor-PCR fingerprints
DNA sequences of 13 known events were retrieved from the GMO Detection Method Database [35]. These events were selected because at least several hundred base pairs of sequence upstream or downstream of the p35S or t-nos screening elements were available, allowing us to in silico calculate the GM fingerprints. All anchor-PCR primer binding sites and restriction enzyme cleavage sites were determined using VectorNTI (Invitrogen). The amplicon length is defined as the distance between the start of the anchor primer binding site to the next restriction site plus the length of the adapter primer. Amplicon lengths were systematically calculated in an Excel worksheet for all adapter/anchor primer combinations per event, up to a maximal length of about 1,500 bp. Multiple amplicons per adapter were retained (maximal of ten) if multiple restriction sites occurred within 1,500 bp from the respective anchor primer.
Results and discussion
Targeted approach for the analytical confirmation of the identity of a novel GMO event
A search for documented information on novel GM events revealed an advertisement indicating the commercial availability of a novel GMO-derived product [11]. Here, we first report on the analytical confirmation of the molecular identity of the novel event. Using documented information on the putative transgene sequence, 16 primers were designed to amplify 18 partially overlapping fragments of the insert for sequencing (Fig. 2). A contiguous sequence of about 5 kb, spanning the T-DNA insert from the left border region to the right border region was obtained. This sequence was consistent with available published data of a construct carrying such trait [36, 37]. Together, these data revealed the molecular identity of the product and confirmed the documented evidence that the product contained a novel GMO event [11].
Targeted confirmation of event identity by amplification and sequencing of fragments of a novel event. Shaded vertical gray bars indicate p35S and t-nos screening elements, and red arrows indicate screening primers. Red lines indicate the fragments that bridge between the screening elements, which were amplified using p35S-F/t-nos-R or t-nos-F/p35S-R primer combinations, respectively
Extension of the screening approach for the identification of an unknown event
Next, we developed an extension to the routine screening procedure to facilitate the detection and identification of unknown events in blind samples. Here, we demonstrate it by purposefully treating the novel GMO as a blind sample, i.e., without making use of prior transgene sequence information. First, routine screening tests for p35S and t-nos [13] (see “Experimental methodology”) revealed the presence of the p35S and t-nos elements (Fig. 3). In a blind sample, this is the first indication that the sample contains GM material. To identify the origin of the p35S and t-nos elements, the sample was subsequently tested for the presence of 39 EU-approved or known unapproved events using validated event-specific quantitative real-time PCR methods. For this purpose, a real-time PCR-based ready-to-use multitarget analytical system was developed by the Molecular Biology and Genomics Unit of the JRC Institute for Health and Consumer Protection [31]. Analysis of the novel GMO material with this system revealed that none of the tested events could be detected (Fig. 3), suggesting that the p35S and t-nos elements originate from another and therefore “unknown” event. Also, none of the reference genes for the seven plant species that may contain approved events were detected. So, even without prior information on sample composition, this indicates that the sample contains a GMO of a plant species other than rice, maize, soybean, cotton, potato, sugar beet, and oilseed rape, supporting the evidence that it is an unknown event. So, performing currently available methods on blind samples is sufficient to indicate the presence of an unknown event, provided that the event contains screening element(s) included in the matrix approach.
GMO analysis using a combination of screening methods and event-specific methods performed on a novel GMO-derived product. Left panels: positive observation of p35S (above) and t-nos (below) RT-PCR detection methods reveals the presence of GMO material. Right panels: negative observation of multitarget RT-PCR assay verifies the absence of 39 EU-approved and some unapproved events and of the corresponding plant species. Together, these data indicate the presence of an unknown event in a blind sample
Amplification of intervening sequences: bridging between multiple screening elements
The most direct way to subsequently identify an unknown event is to obtain a unique event-specific sequence flanking a positively detected screening element. Because two different screening elements were detected, a conventional PCR was used to amplify the intervening sequences, assuming that they originated from the same locus. Since the screening tests alone are sufficient to identify which primers are functional in a given blind sample, no other information on the event sequence is required for this approach. Simply, all-against-all respective combinations of the four p35S and t-nos screening primers were tested, and the relative orientation and distance between pairs of primers were inferred from amplified fragments. Amplification and sequencing of two fragments (p35S-F/tNOS-R, 1,761 bp; tNOS-F/p35S-R, 1,189 bp; Fig. 2) identified the expressed trait (rhIF) and a fragment of the transformation vector, respectively. Together, these straightforward analyses revealed the GMO identity and confirmed, by direct evidence, that the detected screening elements originated from a novel event. Although it was successful in our case study, this approach may fail if: (1) the screening elements do not originate from the same event; (2) two linked elements are spaced too far apart for efficient PCR amplification; or (3) severe DNA degradation (e.g., in highly processed food or feed samples) hampers amplification of long fragments.
Establishment of a fluorescent anchor-PCR method
If a single screening element is detected or if amplification of intervening sequences fails, the flanking sequences of screening elements are amplified by anchor-PCR. A novel fluorescent anchor-PCR GM fingerprinting method was developed (see “Experimental methodology”) using anchor primers targeting the p35S and t-nos elements to provide a direct link to the screening approach (Fig. 4). After enzymatic digestion of gDNA and ligation of adapters, a first touchdown PCR amplifies fragments using an adapter primer and an anchor primer in a screening element. For each primary PCR product, three second PCRs are performed in parallel, using three nested fluorescently labeled anchor primers. These three fluorescently labeled PCR products are pooled to yield a triplet of anchor-PCR amplicons that can be characterized by automated capillary electrophoresis (CE). A typical simple anchor-PCR profile consists of a triplet of three amplicons with the correct fluorescent label order. The length difference between the individual amplicons corresponds to the distance between the primers in the screening element. Any amplicon that does not occur as part of a triplet structure is considered a false positive and is excluded from further analysis. A CE profile containing multiple triplets is referred to as a complex profile (see below). A complete collection of profiles (all combinations of adapters/anchor primers (Ad/AP)) is referred to as a GM fingerprint for a given sample. For clarity, we further refer to “amplicons” instead of “triplets of amplicons” throughout the text. Optimal amplicon length for PCR amplification and CE separation is between 150 and 1,000 bp, although amplicons of about 1,270 bp have also been detected.
A collection of in silico calculated anchor-PCR fingerprints of known events
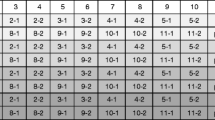
In parallel, we established a collection of in silico calculated anchor-PCR fingerprints of known events. Comparison of experimentally determined fingerprints to this collection identifies amplicons in a test sample that are derived from an unknown event. In the AgBios database, about 120 different events worldwide are listed [38]. However, the transgene DNA sequence information of the integrated locus is publicly available for only a subset of these events. For 13 known events, sufficient sequence information was available in the GMO Detection Method Database [35], and these were selected as a test set to in silico calculate a GM fingerprint (see “Experimental methodology”). Multiple amplicons per adapter were retained (maximal 10) if multiple restriction sites occurred within 1,500 bp from the anchor primer. This is to account for the possibility that partial digestion could occur in a given test sample and that all resulting amplicons should be present in the collection for comparison. In practice, if enzymatic digestion and adapter ligation reactions are complete, then all restriction sites are transformed into adapter-ligated ends. In this case, only the shortest fragment (reaching to the first restriction site) flanking the screening element will be amplified and detected. In reaction conditions with partial digestion, at least the shortest fragment will be amplified but also some of the longer amplicons resulting from adjacent restriction sites (because the first restriction site was not cut in a small percentage of the molecules). It is important to note that amplification of the shortest fragment is the most efficient and that partial digestion may be beneficial because it allows us to amplify additional and longer amplicons simultaneously, without losing the shortest amplicons. As such, partial digestion may result in an increase in the resolution of the fingerprint. In conclusion, in the collection of in silico calculated fingerprints, a number of amplicons are present that will not be detected under optimal restriction digestion/adapter ligation conditions. These are merely present for comparison in case partial digestion does occur in a given test sample. Amplicons were grouped per Ad/AP combination and were ordered by increasing length (Supplemental Table 2). The collection of in silico calculated fingerprints currently contains 499 amplicons but can easily be expanded by additional in silico data, such as other events, other restriction enzymes, or other screening elements, or even with experimentally determined fingerprints for events without such sequence information. Next, we examined whether the various events can be discriminated by comparing their in silico calculated fingerprints. An overview of the number of amplicons per event per Ad/AP combination shows that a fingerprint of a single GM event may contain ten to 20 amplicons per element/orientation or more than 40 amplicons if multiple screening elements are present (Table 1, columns).
Conversely, Table 1 (rows) also lists all amplicons per Ad/AP combination that may be simultaneously amplified in a product that contains multiple events. In such cases, the GM fingerprint becomes a composite fingerprint. For instance, the combination NcoI/p35S-R may detect amplicons of DAS-01507-1, DAS-59122-7, and MON-15985-7 (Table 1), while no amplicons are expected for the other events. To illustrate this in more detail, we plotted all possible amplicons for the combinations BfaI/p35S-F, XbaI/p35S-F, and NcoI/p35S-F (Fig. 5a–c). In general, using restriction enzymes with a four-base recognition sequence (tetra-cutter; BfaI, MboI), increases the chance to detect any event, but amplicons are less often unique for an event, and CE profiles are more likely to be complex (Fig. 5a). In contrast, using restriction enzymes with a six-base recognition sequence (hexa-cutter; BamHI, EcoRI, NcoI, XmaI, XbaI) decreases the chance to amplify a flanking amplicon (not all events contain the hexa-cutter restriction sites within 1,000–1,500 bp flanking the screening element) but increases the chance to generate a unique amplicon and to obtain a relatively simple profile (Fig. 5b, c). In practice, we therefore choose to use a combination of two tetra-cutters and five hexa-cutters to provide sufficient resolution and maintain a high chance to amplify unique amplicons from any unknown event (Table 1).
In silico calculated anchor-PCR amplicons of 13 known events are used to support interpretation of GM fingerprints. Experimentally determined amplicons derived from a novel GMO event (“unknown”; white squares) are indicated for comparison to amplicons of known events (black squares). All possible amplicons for a particular Ad/AP combination are plotted by increasing length to demonstrate discriminative power of the respective Ad/AP combinations. a Tetra-cutter BfaI in combination with the p35S-F anchor primer. b Hexa-cutter XbaI in combination with the p35S-F anchor primer. c Hexa-cutter NcoI in combination with the p35S-F anchor primer
Anchor-PCR fingerprinting of the novel event
Next, we performed the fluorescent anchor-PCR method on the novel GMO event. Because the anchor-PCR primers target the screening element sequence, these tests can be performed directly after positive detection of the screening element in the matrix approach. It does not require any additional sequence information of the unknown event. In this section, we will first confirm the method using the experimentally determined sequence of the insert. At the end of this section, we continue to describe how the obtained GM fingerprint is used to identify the unknown event in a blind sample.
Anchor-PCR was systematically performed for all possible Ad/AP combinations. The p35S and t-nos fingerprinting was performed in three and two independent experimental repetitions, respectively, each starting from separate restriction digestion/adapter ligation reactions. A total of 33 different amplicons flanking the p35S and t-nos screening elements were detected. A number of typical anchor-PCR profiles are presented in Fig. 6. Table 2 lists the number of observations over the number of independent experimental repetitions (listed per in silico calculated amplicon). These data show the reproducibility of the methodology including the digestion, ligation, and primary PCR steps. Reproducibility of independent amplification reactions is illustrated by the second nested fluorescent PCR. The similarity of profiles from different fluorescent labels shows good reproducibility, even in complex profiles with multiple amplicons (Fig. 6). To confirm the method, we compared the various detected amplicons to the in silico calculated fingerprint of the novel event (Table 2 and Fig. 7).
Fluorescent anchor-PCR profiles for the identification of GM events. For each panel, the relative position of the restriction sites is given with respect to the p35S screening element. a Simple fluorescent anchor-PCR profile containing a single anchor-PCR triplet obtained with either NcoI/p35S-F or NcoI/p35S-R combinations. b Complex fluorescent anchor-PCR profiles containing multiple triplets derived from two independent p35S elements, in combination with BfaI adapters. c Complex fluorescent anchor-PCR profile containing multiple triplets derived from two independent p35S elements in combination with MboI adapters. Simultaneous amplification of multiple amplicons from the same element (MboI a, b, d, e, g) indicates that adaptors are ligated at several adjacent MboI restriction sites. Amplification of amplicons from two different p35S elements (MboI a, b, d, e, g versus c, f; and h, i versus j) indicates that discriminative signals from independent screening elements can be simultaneously detected
Anchor-PCR fingerprinting for the identification of unknown events. a Position of restriction sites and anchor primers in the novel GMO insert. b Overall structure of the genetic elements in the novel GMO insert. c Anchor-PCR fingerprint on the p35S elements. d Anchor-PCR fingerprint on a t-nos element. All observed anchor-PCR amplicons are indicated at their relative position in the insert. If an observed amplicon corresponds to an amplicon present in the GM fingerprint collection, the amplicon is indicated with a solid line, and all candidate known events are indicated per amplicon. If no such amplicon exists in the collection, the observed amplicon is indicated with a dotted line and is labeled as UAA. These amplicons are sequenced to identify the unknown event
For the hexa-cutters, only a single amplicon is expected in some of the various combinations with the p35SF, p35SR, t-nos-F, and t-nos-R anchor primers. Examples of the single triplet structure obtained with NcoI/p35S-F or NcoI/p35S-R are given in Fig. 6a. Each of the amplicons expected for the hexa-cutters has been correctly detected (Table 2). The longest fragments that have been detected are the NcoI/t-nos-R fragment of 1,270 bp and the XmaI/p35S-R fragment of 1,271 bp.
For the tetra-cutters BfaI and MboI, multiple restriction sites are present within 100–1,500 bp from the screening elements. In most experiments, up to three amplicons were amplified in parallel. In one experiment, up to seven (MboI/p35S-F) and six (MboI/p35S-R) amplicons were detected (Table 2). Some restriction sites were close enough to one of the screening elements to be detected but too far away from another screening element. This explains the difference between the restriction sites that are detected in the respective p35S and t-nos fingerprints (Fig. 7b, c). Together, these data reveal that it is possible to amplify multiple amplicons in the same PCR leading to complex profiles (Fig. 6b, c). We illustrate this based on the MboI profiles. First, if an event carries multiple copies of the same screening element, as is often the case in the known events (Table 1; [35]), flanking regions from both copies can be amplified simultaneously. For instance, the MboI/p35S-R profile contains two triplets derived from the p35S promoter driving the expressed trait and a triplet derived from the p35S promoter driving the selection marker (Fig. 6c). This suggests that if a sample contains multiple events with a common screening element, amplicons from multiple events can also be amplified in parallel, allowing the detection of unique discriminative signals for each event. This observation is important because it suggests that detecting masked unknown events by direct observations may be possible in products with mixed GMO ingredients. Second, due to partial restriction digestion, a fraction of the gDNA molecules is not cut at the first restriction site neighboring the anchor primer, but adaptors are ligated at the second (or following) restriction sites (Fig. 6b, c). As a result, multiple amplicons covering the same flanking sequence but with increasing length are amplified simultaneously per Ad/AP combination, hence increasing the resolution of the fingerprint (Fig. 6b, c). The observed number of amplicons in a complex profile (Fig. 6b, c) is clearly less than the total number of possible amplicons stored in the collection (Table 1). In all cases, the shortest fragments are preferentially detected, as indicated by decreasing signal strength of longer amplicons (Table 2 and Fig. 6b, c). This may result from an efficient adapter ligation reaction (absence of longer fragments as template) or because short amplicons may have competitive advantage over relatively long amplicons during simultaneous PCR amplification.
In conclusion, all tested anchor primers, adapters, and reaction conditions are functional. The observed amplicon lengths were consistent with the in silico calculated amplicon lengths. In some cases, multiple amplicons are amplified in parallel, which may be derived from independent screening elements or are generated due to partial digestion. Partial digestion may occur due to sample impurities or may even be intentionally induced by dilution of the enzyme. The benefit of partial digestion might be exploited to increase the resolution of the fingerprint, even if it is difficult to control the degree. On the other hand, ruling out partial digestion would: (1) limit the method to the detection of the shortest fragment only, hence simplify fingerprints and increase the chance to detect amplicons from multiple events but (2) would lead to a potential loss of information on subsequent restriction sites (longer amplicons), which are still (or even more) informative for event identification.
In the second part of this section, we discuss how the obtained fingerprint may lead to the identification of an unknown event in a blind sample. Clearly, the unknown event may be identified by sequencing the full collection of amplicons. Comparison of individual sequenced amplicons to a DNA sequence database may be sufficient to reveal the identity of the corresponding event(s). For instance, amplicons obtained with combinations XmaI/p35S-F or BfaI/t-nos-R contain fragments of the expressed trait (rhIF) and would unambiguously identify the novel GMO. Alternatively, assembly of sequenced amplicons into a contiguous sequence could lead to a reconstruction of the insert of an unknown event. We aligned the observed amplicons with the experimentally determined sequence of the insert (Fig. 7). Together, these data reveal that the anchor-PCR amplicons cover a significant part of the construct, including the expressed trait (rhIF) and antibiotic resistance selection marker. Thus, it was possible to identify the novel unknown event with the respective data for p35S (Fig. 7c) or for t-nos (Fig. 7d). So, this procedure provides direct evidence of the identity of an unknown event, independent of any prior knowledge of the product composition.
Finally, we illustrate the use of the in silico calculated fingerprints for the interpretation of a GM fingerprint derived from a blind sample, using the data of the novel event. For each observed amplicon of the GM fingerprint, the size is determined using CE, giving a unique Ad/AP/size combination (Fig. 6a–c). Then, the collection is checked whether this amplicon is present in any of the known events (e.g., Fig. 5, Supplemental Table 2). In this example, a small size window (±5 bp) is used to account for minor inaccuracies in CE sizing. First, a list of amplicons that are not present in the collection is generated. Each represents a candidate unique signal derived from an unknown event and is characterized by subsequent sequencing of the amplicon. The novel GM fingerprint contains 20 such unique amplicons. For instance, a 181-bp NcoI/p35S-F fragment (Fig. 6a) does not have a related fragment in the collection (Fig. 5c) and is indicated as an “unknown anchor-PCR amplicon” (UAA) in the fingerprint (Fig. 7c). Conversely, the novel GMO fingerprint contains another 13 amplicons that are shared with one or more of eight known events. For instance, a 147-bp BfaI/p35S-F fragment (Fig. 6b) may be derived from MON-88017-3 or MON-89034-3 or indeed from an unknown event (Fig. 5a). Such amplicons are indicated with the candidate known events in the fingerprint (Fig. 7c, d). Amplicons may be shared between different events if they represent common fragments within the full p35S promoter and/or t-nos terminator sequence.
Conclusions
In this report, we illustrate that detection and identification of unknown events can be performed in a targeted or nontargeted fashion, depending on the available information on the distribution of GMO-derived products on the market, and/or on the genetic modifications in known and novel unknown GM crops. The choice between a targeted approach or a nontargeted approach will have to be made on a case-by-case basis.
First, we demonstrated that, if an effort is made to collect documented information on novel GMOs [11], targeted analytical confirmation of an unknown event can be performed (Fig. 1, steps 1 and 3). This case clearly reveals that a targeted discovery of products containing previously unknown events is possible based on documented evidence. So, integrating these steps into the sampling strategy will enrich the testing laboratory with potential high-risk samples. This, in turn, would enhance the efficient use of analytical resources dedicated to the detection of unauthorized events, by shifting from screening randomly selected blind samples to targeted analytical confirmation of products suspected to contain novel potentially unauthorized events.
In cases where information on the GMO content and/or DNA sequence level is not available, currently, the best strategy for the detection of unknown events is a nontargeted screening approach (Fig. 1, steps 2 and 4–14). However, three limitations are inherent to the screening approach: it yields only indirect evidence for unknown events; evidence is inconclusive in the absence of event identification, and unknown events may be masked by the presence of known authorized events in products with mixed GMO ingredients. To complement these limitations, an extended procedure, including a novel fluorescent anchor-PCR method, was developed for amplification of screening element flanking sequences.
As a proof of concept, we illustrated the approach on a previously uncharacterized novel GMO. These studies demonstrate that the procedure including anchor-PCR fingerprinting is useful for identifying an event after the screening approach identified a suspect sample. While these studies show the potential use of the methods, they have to be further evaluated, e.g., in processed food or feed or in samples with mixed GMO ingredients. Furthermore, the limit of detection will have to be evaluated in a range of products, taking into account the potential influences of DNA extract purity and DNA integrity. PCR methods that detect screening elements typically amplify relatively short (100 bp) fragments to avoid the negative influence of DNA degradation during food processing [39]. Using a DNA extract with high integrity, we were able to demonstrate that the established anchor-PCR method can amplify relatively long fragments of flanking sequences (up to 1,000–1,250 bp). Nevertheless, the method applicability is dependent on the level of DNA degradation in a given sample and may be limited in highly processed food or feed samples.
Importantly, the method generates a relatively large number of amplicons that can be sequenced to directly identify the causative event; hence, it yields conclusive evidence of the presence of an unknown and potentially unauthorized event. Sequencing all obtained anchor-PCR amplicons is a straightforward but labor-intensive and time-consuming approach, while not all amplicons may contain informative sequences. For instance, some sequences directly flanking the core screening element are present in multiple events and will not discriminate between known events and novel unknown events. To distinguish between those amplicons, a prototype reference collection of in silico calculated GM fingerprints of known events was established with three main purposes. First, we demonstrate its use for interpretation of experimentally generated GM fingerprints of blind samples. Unique discriminative signals for an unknown event are identified by comparison to fingerprints of known events. Thus, nondiscriminative signals can be excluded from sequencing analysis. Second, we used the in silico data to establish the level of resolution required to discriminate between events. The current resolution (two elements, two orientations, two tetra-cutters, and five hexa-cutters) is more than sufficient to discriminate between 13 commercialized events and an unknown event (Fig. 5, supplemental Table 2), and the system is fit for significant upscaling. For instance, about 120 different events are registered in the AgBios database [38]. Likewise, a recent study identified more than 90 novel events in advanced stages of development, which are expected to enter the market in the near future [2, 3]. If the anchor-PCR-based approach is to be used for GMO identification in the future, the corresponding fingerprints will have to be added to the collection to facilitate easier fingerprint interpretation. Such fingerprints can either be in silico calculated, as demonstrated here, or have to be experimentally determined for events without sequence information. Importantly, such in silico calculated fingerprints should ideally be based on sequence information of the integration locus rather than on the original construct used for transformation to account for possible rearrangements during the transformation process. Furthermore, in silico calculated fingerprints should be experimentally validated using appropriate reference material. Third, given the expected future expansion in the number of novel events, it is important to expand the screening platform itself with novel screening elements and consequently also with methods to identify their flanking sequences. Software applications that optimize the choice of analytical tests during routine screening are already under development to help manage the cost and workload [40]. The in silico calculated GM fingerprint data can be used to model the optimal choice of screening elements and adapters for a given set of (existing or expected) events. Using modeling, it can be predicted whether GM fingerprinting can still differentiate between all events (e.g., Fig. 5) or that additional combinations of screening elements and/or adapters are required to improve the resolution. This concept was illustrated using a limited set of known events (13) but can be expanded to cover all relevant events for a given jurisdiction.
Furthermore, because any amplicon that is not present in the fingerprint collection is a potential signal of an unknown event, it is important to suppress false-positive amplicons. In our setup, performing nested PCRs and using the highly specific triplet structure of pooled nested PCR products allows us to effectively counter-select false-positive amplicons. Finally, our nontargeted approach leads to the molecular identification of a novel GMO, independent of prior sequence information, thus transforming the GMO from an “unknown” to a “known” event. So, by following the procedure proposed in this work, sufficient sequence information is gathered to support the development of an event-specific method, which, in turn, can be incorporated in the targeted routine procedures. Screening for unknown and potentially unauthorized GMOs in the food and feed chain is important to protect environmental and public safety. At the same time, reporting the finding of unauthorized material may have huge consequences for international trade and public perception of the biotechnology industry. Therefore, it is equally important to avoid false alerts and to provide conclusive and unambiguous evidence that identifies the causative event(s).
References
James C (2009) ISAAA brief 39
Stein AJ, Rodríguez-Cerezo E (2009) European Communities, Luxembourg, JRC Technical Report EUR 23486 EN
Stein AJ, Rodríguez-Cerezo E (2009) European Communities, Luxembourg, JRC Technical Report EUR 23810 EN
Community Reference Laboratory for GM Food and Feed (2009) . Status of dossiers. http://gmo-crl.jrc.ec.europa.eu/statusofdoss.htm. Accessed 30 Sept 2009
Dorey E (2000) Nat Biotechnol 18:1136–1137
European Commission (2005) Off J Eur Union 101:14–16
European Commission (2006) Off J Eur Union 306:17–20
European Commission (2008) Off J Eur Union 96:29–34
Vermij P (2006) Nat Biotechnol 24:1301–1302
Holst-Jensen A (2008) Nat Biotechnol 26:858–859
Ruttink T, Morisset D, Van Droogenbroeck B, Lavrac N, Van Den Eede G, Zel J, De Loose M (2009) Anal Bioanal Chem. doi:10.1007/s00216-009-3218-6
ACP Pharma (2008) http://www.orfe.pl/wyniki_wyszukiwania.php?szukaj=acp_magazyn_03_2008.pdf Accessed 30 Sept 2009
Kuribara H, Shindo Y, Matsuoka T, Takubo K, Futo S, Aoki N, Hirao T, Akiyama H, Goda Y, Toyoda M, Hino A (2002) J AOAC Int 85:1077–1189
Hernandez M, Rodriguez-Lazaro D, Ferrando A (2005) Curr Anal Chem 1:203–221
Xu J, Miao H, Wu H, Huang W, Tang R, Qiu M, Wen J, Zhu S, Li Y (2006) Biosens Bioelectron 22:71–77
Holst-Jensen A, Rønning SB, Løvseth A, Berdal KG (2003) Anal Bioanal Chem 375:985–993
Leimanis S, Hernández M, Fernández S, Boyer F, Burns M, Bruderer S, Glouden T, Harris N, Kaeppeli O, Philipp P, Pla M, Puigdomènech P, Vaitilingom M, Bertheau Y, Remacle J (2006) Plant Mol Biol 61:123–139
Prins TW, van Dijk JP, Beenen HG, Van Hoef AA, Voorhuijzen MM, Schoen CD, Aarts HJ, Kok EJ (2008) BMC Genomics 9:584
Schmidt AM, Sahota R, Pope DS, Lawrence TS, Belton MP, Rott ME (2008) J Agric Food Chem 56:6791–6800
Chaouachi M, Chupeau G, Berard A, McKhann H, Romaniuk M, Giancola S, Laval V, Bertheau Y, Brunel D (2008) J Agric Food Chem 56:11596–11606
Randhawa GJ, Chhabra R, Singh M (2009) J Agric Food Chem 57:5167–5172
Mano J, Shigemitsu N, Futo S, Akiyama H, Teshima R, Hino A, Furui S, Kitta K (2009) J Agric Food Chem 57:26–37
Elenis DS, Kalogianni DP, Glynou K, Ioannou PC, Christopoulos TK (2008) Anal Bioanal Chem 392:347–354
Hamels S, Glouden T, Gillard K, Mazzara M, Debode F, Foti N, Sneyers M, Nuez TE, Pla M, Berben G, Moens W, Bertheau Y, Audeon C, Van den Eede G, Remacle J (2009) Eur Food Res Tech 228:531–543
Miraglia M, Berdal KG, Brera C, Corbisier P, Holst-Jensen A, Kok EJ, Marvin HJ, Schimmel H, Rentsch J, van Rie JP, Zagon J (2004) Food Chem Toxicol 42:1157–1180
GMO Compass (2009) http://www.gmo-compass.org. Accessed 30 Sept 2009
Cankar K, Chauvensy-Ancel V, Fortabat MN, Gruden K, Kobilinsky A, Žel J, Bertheau Y (2008) Anal Biochem 376:189–199
Raymond P, Gendron L, Paul S, Moreau ME, Coté MJ, Laberge S (2008) In: Book of abstracts. First Global Conference on GMO Analysis, Como, Italy, 24–27 June 2008
Bhat NS, Dibley K, Xie VRD, Partis L, Raymond P, Emslie KR (2008) In: Book of abstracts. First Global Conference on GMO Analysis, Como, Italy, 24–27 June 2008
Nesvold H, Kristoffersen AB, Holst-Jensen A, Berdal KG (2005) Bioinformatics 21:1917–1926
Querci M, Foti N, Bogni A, Kluga L, Broll H, Van den Eede G (2009) Food Anal. Methods. doi:10.1007/s12161-009-9093-0
Theuns I, Windels P, De Buck S, Depicker A, Van Bockstaele E, De Loose M (2002) Euphytica 123:75–84
Windels P, De Buck S, Van Bockstaele E, De Loose M, Depicker A (2003) Plant Physiol 133:2061–2068
Alonso JM et al (2003) Science 301:653–657
Dong W, Yang L, Shen K, Kim B, Kleter GA, Marvin HJP, Guo R, Liang W, Zhang D (2008) BMC Bioinformatics 9:260
Fedosov SN, Laursen NB, Nexø E, Moestrup SK, Petersen TE, Jensen EØ, Berglund L (2003) Eur J Biochem 270:3362–3367
Hvas AM, Buhl H, Laursen NB, Hesse B, Berglund L, Nexo E (2006) Haematologica 91:805–808
GM Database (2009) Agbios. http://www.agbios.com/dbase.php. Accessed 30 Sept 2009
Debode F, Janssen E, Berben G (2007) Eur Food Res Tech 226:273–280
GMOtrack (2009) http://kt.ijs.si/software/GMOtrack/. Accessed 30 Sept 2009
Acknowledgements
T. Ruttink is supported by the Belgian Federal Public Service for Health, Food Chain Safety, and Environment (contract no. RT-06/6 GMOdetec). R. Demeyer is indebted to the Institute for Agricultural and Fisheries Research (ILVO) for a predoctoral fellowship. I. Taverniers was involved in the 6th Framework Programme Integrated Project Co-Extra, Contract No. 007158. We are thankful for the excellent technical support of Annique Staelens, Cindy Merckaert, Mieke Dhondt, and Sabine van Glabeke.
Author information
Authors and Affiliations
Corresponding author
Additional information
T. Ruttink and R. Demeyer contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1957 kb)
Rights and permissions
About this article
Cite this article
Ruttink, T., Demeyer, R., Van Gulck, E. et al. Molecular toolbox for the identification of unknown genetically modified organisms. Anal Bioanal Chem 396, 2073–2089 (2010). https://doi.org/10.1007/s00216-009-3287-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-009-3287-6