Abstract
The detection and identification of foodborne pathogens continue to rely on conventional culturing techniques. These are very elaborate, time-consuming, and have to be completed in a microbiology laboratory and are therefore not suitable for on-site monitoring. The need for a more rapid, reliable, specific, and sensitive method of detecting a target analyte, at low cost, is the focus of a great deal of research. Biosensor technology has the potential to speed up the detection, increase specificity and sensitivity, enable high-throughput analysis, and to be used for monitoring of critical control points in food production. This article reviews food pathogen detection methods based on electrochemical biosensors, specifically amperometric, potentiometric, and impedimetric biosensors. The underlying principles and application of these biosensors are discussed with special emphasis on new biorecognition elements, nanomaterials, and lab on a chip technology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The improvement of food and water safety and security depends on the ability to detect, identify, and trace food and water pathogens.
The European "Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents, Antimicrobial resistance and Foodborne Outbreaks in the European Union in 2005", published on 14 December 2006, reported a total of 5,311 foodborne outbreaks, involving 47,251 people and resulting in 5,330 hospitalizations and 24 deaths in the 23 member states during 2005. As in previous years, the most common agent reported in foodborne outbreaks was Salmonella followed by Campylobacter (64% and 9% of all reported outbreaks, respectively). Foodborne viruses (Calicivirus, including Norovirus) were reported to be the causative agent in 6% of all reported outbreaks. Other major causes of foodborne outbreaks in the European Union (EU) were Staphylococcus spp. (3% of all outbreaks), Clostridium spp. (2%), Bacillus spp. (1%), pathogenic E. coli (1%), and Shigella (1%) [1].
Foodborne illness occurring each year in Europe costs the European economy hundreds of millions of Euros in terms of medical costs, lost productivity, and product recalls. Food quality and safety is of major economic and societal importance in the EU policy, thus justifying its position as a priority area of research in the EU’s 7th Framework Programme [2, 3].
Similar trends were recorded in the USA, where the US Centers for Disease Control and Prevention (CDC) estimates that more than 36 million cases of illness occur annually because of foodborne and waterborne pathogens [4], and the market potential for detection and identification of bacterial and viral pathogens in the food safety area is estimated at around $150 million per year [5].
The increased awareness has prompted significant interest in the development of more effective methods for pathogen detection along the food processing chain. End-product testing alone is unable to assure safe food production. Thus, since its inception in the 1970s, hazard analysis critical control point (HACCP) methodology has evolved as the leading food safety strategy used by the food industry. HACCP identifies where potential contamination, time, and temperature problems can occur (the critical control points). However, key technologies needed to successfully implement any HACCP program are real-time microbial detection, traceability, and source identification [6].
The need for rapid, reliable, specific, and sensitive methods of detecting a target food pathogen, at low cost, is the focus of a great deal of research, as underlined by the huge amount of scientific literature on this field [7–15]. Lazcka et al. [16] recently gave an overview of the literature of the field of pathogen detection. Among the 2,005 scientific papers reported in literature over the last 20 years, 38% are related to food safety, 18% to clinical diagnosis, 16% to environmental monitoring, 27% to miscellaneous applications, and 1% to defense purposes.
To meet expectations of users, analytical methods for pathogen detection in food must have the specificity to distinguish between different bacteria, the adaptability to detect different analytes, and the sensitivity to detect bacteria on-line and directly in real samples without pre-enrichment [9]. The device must also be simple and inexpensive to design and manufacture. (Bio)sensor technology is claimed to satisfy these requirements [9].
This paper gives an overview of the field of food pathogen and toxin detection using electrochemical biosensors.
Overview of analytical methods in food pathogen detection
The major foodborne pathogens worldwide are Salmonella, E. coli 0157:H7, Listeria monocytogenes, Staphylococcus aureus, Campylobacter, Cryptosporidium, and Norwalk-like viruses (NLVs). Many west European countries propose the absence of pathogen in 0.01 g (100 colony forming units (CFU) per gram) for certain food products. The Environmental Protection Agency regulations specify the minimum frequency of water sampling and the maximum number of coliform organism allowed: treated drinking water should contain no coliforms in 100 mL [8].
Conventional methods for the detection and identification of pathogens mainly rely on specific microbiological and biochemical identification [7]. While these methods can be sensitive, inexpensive, and give both qualitative and quantitative information on the number and the nature of the microorganisms tested, they are greatly restricted by assay time, with initial enrichment needed in order to detect pathogens which typically occur in low numbers in food and water. Some standard methods, e.g., the DIN EN ISO 11290-1 method [17] for the detection of L. monocytogenes, can require up to 7 days to yield results, as they rely on the ability of microorganisms to multiply to visible colonies. Recent advances in technology allow the development of new methods that are often referred to as “rapid methods”, a subjective term used loosely to describe a vast array of tests that include: modified and automated conventional methods; bioluminescence, based on the measurement of bacterial adenosine triphosphate (ATP); cell counting methods, mainly based on direct epifluorescent microscopy; nucleic acid-based assays; and immunological methods [18].
The modification and automation of conventional methods in food microbiology allow the development of newer sample preparation methodologies and of newer plating techniques, such as the ALOA® method (AES laboratoire) [19], certified by AFNOR, the French standards body, which uses a chromogenic medium in conjunction with a Listeria monodisk for the detection of L. monocytogenes, and the development of several commercial identification kits based on biochemical tests (API systems (BioMerioux), MicroID (Remel), etc.). Even though these tests can reduce detection times down to a few days, this can still present difficulties in the quality control of semiperishable foods. In addition, viable bacterial strains in the environment can enter a dormancy state where they become nonculturable (viable but nonculturable, VBNC) which can subsequently lead to an underestimation of pathogen numbers or a failure to isolate a pathogen from a contaminated sample.
Bioluminescence method and the direct epifluorescent filter technique (DEFT), a direct method used for enumeration of microorganisms based on the binding properties of the fluorochrome acridine orange and fluorescent microscopic analysis, although not identifying specific species, can be used to rapidly enumerate the presence of total bacteria. The ATP measurements and the DEFT analysis have been proposed as very useful techniques in on-line hygiene monitoring in HACCP programs and some companies now produce some ATP hygiene monitoring kits. Nevertheless, these methods yield an unspecific measurement, and a significant challenge for researchers may be the provision of pathogen specificity to the ATP assay.
Genetic characterization methods lead to unequivocal species identification [13].
DNA probes (i.e., an oligonucleotide sequence immobilized on a fixed support able to hybridize the complementary strand present in solution) have been widely used for detection of specific RNA/DNA sequences of pathogenic bacteria. In this case, the probe is generally labeled with a radioisotope, usually 32P, or a fluorescent tag; these labeling and detection procedures require long and tedious steps. Moreover, when radioactive compounds are used, specific procedures for handling them must be adopted. Some kits based on this technology are now commercially available and are approved by AOAC Official Methods [20]. However, DNA probe technology requires a long culture-enrichment step, and 1–2 days are always needed to perform the analysis.
DNA chips are predicted to become the method of choice for species-specific identification of bacteria. DNA chips consists of oligonuleotide arrays bound to a solid support in a systematic manner, which enables hybridization results to be scored. DNA chips, however, face some of the same problems that DNA probes have experienced, such as the need for concentration of the relevant bacteria from the food matrix. Moreover, the use of DNA chips in analytical application is currently limited by the fact that the cost of the analysis is very high.
Detection of microorganisms by DNA amplification has been shown to be more suitable. The polymerase chain reaction (PCR) can be used to enhance the sensitivity of nucleic acid-based assays. PCR has distinct advantages over culture and other standard methods for the detection of microbial pathogens and offers the advantages of specificity, sensitivity, rapidity, accuracy, and capacity to detect small amounts of target nucleic acid in a sample. However, problems such as the sensitivity of the polymerase enzyme to environmental contaminants, difficulties in quantification, the generation of false positives through the detection of naked nucleic acids, nonviable microorganisms, or contamination of samples in the laboratory, may limit the use of PCR for the direct detection of microbial contamination. Development and application of extraction methods that concentrate the target organisms in a small volume, and which remove the target bacteria from the inhibitory substances, is therefore a high priority. An interesting approach is to capture the bacteria by antibody-coated paramagnetic beads using the immunomagnetic separation technique (IMS). Paramagnetic beads are coated with polyclonal antibodies, which can target and separate pathogen cells in a mixed suspension with no loss of viability, producing a normal isolate for further confirmation. IMS has been used as an alternative to selective enrichment broths for a variety of bacteria. IMS can reduce the time required for conventional methods by as much as 24 h. IMS has also been used in conjunction with rapid detection methods, including enzyme-linked immunosorbent assays (ELISAs), conductance microbiology, electrochemiluminescence, and, obviously, PCR. In the case of PCR, pathogens are specifically separated from the specimen, resulting in a useful sample for PCR with little or no nonspecific DNA or interfering factors.
Generally, detection of particular DNA sequences, amplified by PCR, is carried out using gel electrophoresis. Although it is simple and effective for detection of PCR products under research conditions, gel electrophoresis is not considered a suitable method for routine analysis; moreover gel electrophoresis cannot determine whether the sequence of the amplified target DNA is the same as that intended. In addition, ethidium bromide, which is a common staining agent, is carcinogenic. Amongst the different PCR variants, multiplex PCR is very useful as it allows the simultaneous detection of several organisms by introducing different primers to amplify DNA regions coding for specific genes of each bacterial strain targeted. Real-time PCR permits one to obtain quicker results without too much manipulation. This technique bases its detection in the fluorescent emission by a specific dye as it attaches itself to the targeted amplicon. One of the limitations of PCR techniques is that the user cannot discriminate between viable and nonviable cells because DNA is always present whether the cell is dead or alive. Reverse transcriptase PCR (RT-PCR) was developed in order to detect viable cells only [21]. RT is an enzyme able to synthesize single-stranded DNA from RNA in the 5′–3′ direction. Several genes specifically present during the bacteria’s growth phase can then be detected. Nucleic acid sequence-based amplification (NASBA) is another genetic technique considered to be an interesting alternative to RT-PCR, since it does not react with contaminating DNA. In recent years, quantitative NASBA assays have been developed for the detection of various viral and bacterial RNA in food samples [22, 23].
In summary, genetic methods are well developed and when applied as culture confirmation tests, they are reliable, fast, and sensitive. The ability to concentrate the target organisms from food, eliminating inhibitory substances, as well as the detection of the PCR product, in a simply and cheap way, still need to be improved before detection directly from processed food becomes a realistic goal and PCR can be used in routine analysis.
Immunological detection with antibodies is perhaps another technology that has been successfully employed for the detection of specific microorganisms and microbial toxins. The suitability of these antibodies depends mainly on their specificity [24, 25]. In the last year, immunological detection of microbial contamination has become more sensitive, specific, reproducible, and reliable with many commercial immunoassays available for the detection of a wide variety of microbes and their products. While nucleic acid-based detection may be more specific and sensitive than immunological-based detection, the latter is more robust and has the ability to detect not only contaminating organisms but also their biotoxins. ELISA and Western Blot are generally used for this purposes. Recently microarray immunoassays for pathogen detection were also reported [26].
Even though both antibody-based and nucleic acid-based detection have greatly decreased assay times compared with traditional culture techniques, they still lack the ability to detect microorganisms in “real time”. The need for a more rapid, reliable, specific, and sensitive method of detecting a target analyte, at low cost, is the focus of a great deal of research, especially for applications outside the laboratory environment.
Biosensor technology
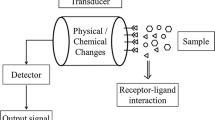
Sensors and mainly biosensors can be an exciting alternative to the more traditional methods for the detection of pathogens and toxins in food [5, 8, 9, 11, 14–16, 27]. Biosensors use a combination of biological receptor compounds (antibody, enzyme, nucleic acid, etc.) and the physical or physicochemical transducer directing, in most cases, real-time observation of a specific biological event (e.g., antibody–antigen interaction). Biosensors allow the detection of a broad spectrum of analytes in complex sample matrices, and have shown great promise in areas such as clinical diagnostics, food analysis, bioprocess and environmental monitoring. Biosensors may be divided into six basic groups, depending on the method of signal transduction: optical, mass, electrochemical, magnetic, micromechanical, and thermal sensors. In the recent years many papers appeared in the literature dealing with the use of biosensors in food safety. The sensitivity of each of the biosensor systems discussed in these papers may vary depending on the transducer’s properties and on the various biological elements. The main advantages of the use of biosensors, in comparison with other methods, is the short analysis time, low cost of analysis, the suitability to be integrated in automated assays, and the possibility to perform in situ real-time analysis. Nevertheless, in the field of biosensors for food safety, there is still a lack of portable, integrated systems. In this context, electrochemical detection allows simple integration with electronic system used for readout, making it a preferred choice if the sensitivity, miniaturization, and cost issues can be optimized.
Electrochemical biosensors
Electrochemical biosensors have some advantages over other analytical transducing systems, such as the possibility to operate in turbid media, comparable instrumental sensitivity, and possibility of miniaturization. As a consequence of miniaturization, small sample volume can be required. Modern electroanalytical techniques (square wave voltammetry, chronopotentiometry, chronoamperometry, differential pulse voltammetry) have very low detection limits (10−7–10−9 M). In situ or on-line measurements are both allowed. Furthermore the equipment required for electrochemical analysis is simple and cheap compared with most other analytical techniques. Basically electrochemical biosensors can be based on amperometric, potentiometric, conductimetric, or impedimetric transducers.
Amperometric biosensors
Amperometric biosensors rely on an electrochemically active analyte that can be oxidized or reduced at a working electrode. Typical electrode materials are platinum (Pt), gold (Au), and carbon. Nowadays some innovative techniques for electrode preparation, characterized by the possibility of mass-production and high reproducibility, have been proposed. Among these, the equipment needed for thick-film technology is less complex and costly and thus this is one of the most used techniques for sensor production. Thick-film technology consists of depositing inks on a substrate in a film of controlled pattern and thickness, mainly by screen-printing (Fig. 1). The inks may be printed on several kinds of substrates like glass, ceramic, or plastic. Many different types of ink are commercially available, differing in composition and electrical behavior. Screen-printing is a flexible and versatile technique, and one of its main advantages is the possibility of choosing shape and dimensions of the resulting sensor. Moreover, the interest in screen-printed sensors as electrochemical transducers is due to the possibility of making them disposable; this characteristic arises from the low cost and the mass-production of these systems. In electrochemistry a disposable sensor offers the advantage of not suffering from the electrode fouling that can result in loss of sensitivity and reproducibility. The avoidance of contamination among samples is another important advantage of using disposable sensors. The micro dimensions of these devices are important to satisfy the needs of decentralized testing. Finally, the high degree of reproducibility that is possible for these single-use electrodes eliminates the cumbersome requirement for repeated calibration.
Circuit pattern printing step: A the substrate (i.e., polyester) is mounted on a platform and is held in position by a vacuum chuck. B The ink is placed on one side of the screen and a squeegee crosses the screen under pressure, thereby bringing it into contact with the substrate and also forcing the ink through the open areas of the mesh. C The required circuit pattern is thus left on the substrate
Screen-printed electrochemical cells are, nowadays, a common choice in the development of an amperometric biosensor.
Amperometric or voltammetric biosensors typically rely on an enzyme system that catalytically converts electrochemically nonactive analytes into products that can be oxidized or reduced at a working electrode. Although these devices are the most commonly reported class of biosensor, they tend to have a small dynamic range due to saturation kinetics of the enzyme, and a large overpotential is required for oxidation of the analyte; this may lead to oxidation of interfering compounds as well (e.g., ascorbate in the detection of hydrogen peroxide). In addition to the use in enzyme-based biosensors, amperometric transducers have also been used to measure enzyme-labeled tracers for affinity-based biosensor (mainly immunosensors and genosensors). Enzymes which are commonly used include horse radishperoxidase (HRP) and alkaline phosphatase (AP). Limitations for amperometric and voltammetric transducer include potential interferences to the response if several electroactive compounds can generate false current values. These effects have been eliminated through the use of selective membranes which carefully control the molecular weight or the charge of compounds which have access to the electrode.
Amperometric biosensors for food pathogens are reviewed in the following sections as microbial metabolism-based, antibody-based (immunosensor), and DNA-based biosensors.
One of the problems still facing the production of biosensors for direct detection of bacteria is the sensitivity of the assay in real samples. For this reason, we selected from the literature examples of amperometric biosensors tested in real samples and we summarized their analytical performance and their application in food and water samples. The comparison between methods, reported in Table 1, cannot be used to determine the best method. The purpose of the table is simply to organize the relevant experiment for further study.
Table 1 lists also the analysis time. It should be noted that with the term analysis time some authors consider the entire analytical procedure (i.e., sample pretreatment, cultivation, extraction, etc., and detection) and some other only the detection step. We consider the analysis time as the time required for the entire analytical procedure, and where possible we specify both the detection time and the sample treatment time.
Microbial metabolism-based biosensor
Various combinations of biosensors based on the monitoring of microbial metabolism have been reported in the literature and summarized in some recent reviews [9]. These approaches can be based on direct measurements of a physical phenomena occurring during the biochemical reactions on a transducer surface. Parameters such as oxygen consumption can be measured by electrochemical transducers such as the amperometric Clark-type oxygen electrode [27, 28]. Recently, Ruan et al. [29] proposed an in situ method for monitoring of Salmonella typhimurium in a selective medium by measuring the cathodic peak current of oxygen in cyclic voltammograms during bacterial proliferation with an electrochemical voltammetric analyzer.
An interesting strategy to detect bacteria through their metabolic process was that based on the electrochemical detection of specific marker enzymes (mainly oxido-reductase), after incubation in an appropriate medium, such as the determination of the enzyme β-D-glucuronide glucuronosohydrolase (GUS) and of the enzyme β-d-galactosidase (β-GAL) for coliform detection in water samples. Conventional methods of E. coli detection using GUS or β-GAL, involve incorporation of chromogenic substrates into culturing media and spectrophotometric monitoring of the liberated chromophore. Conversion of the substrate p-nitrophenyl-β-D-glucuronide (PNPG) to p-nitrophenol (PNP) and D-glucuronic acid indicates the presence of GUS and hence E. coli. Mulchandani et al. [30] reported an electrooxidative method for GUS detection using a bacteria-based biosensor by immobilization of Moraxella species on a carbon paste electrode. Moraxella sp. degrades PNP and produces a more electroactive hydroquinone as an early intermediate. Hydroquinone was oxidized at a lower potential (+0.3 V) than PNP. Togo et al. [31] investigated an electrochemical method of GUS detection based on the production of PNP from PNPG and PNP degradation by a Moraxella sp. Pseudomonas putida JS444 was also used in initial studies because of its ability to degrade PNP faster than the Moraxella sp.
β-GAL has also been used for enumerating coliform in water media. Specifically, β-GAL catalyzes the breakdown of lactose into galactose and glucose. Perez et al. [32] described a rapid method for detection of viable E. coli in water samples, using an amperometric sensor for the detection of 4-aminophenol (4-AP) after hydrolysis of the substrate 4-aminophenyl-β-d-galactopyranoside (4-APGal) by the bacterial enzyme β-d-galactosidase. The bacteria were recovered by filtration and incubated in a selective medium, lauryl sulfate broth (LSB) supplemented with the substrate 4-APGal at 44.5 °C. The electrochemically active molecule 4-AP was produced after hydrolysis of 4-APGal by the enzyme β-galactosidase. 4-AP was measured by amperometry and was detected at a due concentration of E. coli. The time necessary for reaching that concentration was inversely related to the initial E. coli concentration of the sample. Serra et al. [33] reported a tyrosinase composite biosensor for improved amperometric detection of β-galactosidase activity. The method relies on the detection of phenol released after the hydrolysis of phenyl-D-galactopyranoside (PG) by β-galactosidase.
Immunosensors
Immunological detection with antibodies is perhaps another technology that has been successfully employed for the detection of specific microorganisms and microbial toxins. The suitability of these antibodies depends mainly on their specificity. Immunosensors for food pathogen are reviewed in the following sections as immunosensor based on antibodies immobilized directly on the electrode and on antibodies immobilized on magnetic beads.
Immunosensors based on antibodies immobilized directly on the electrode surface
Immobilization of antibodies directly on the surface of an electrode for rapid bacterial detection has been demonstrated for Salmonella and Staphylococcus [34, 35]. Croci et al [36] reported the application of a sandwich immunosensor assay against Salmonella in pork, chicken, and beef experimentally contaminated with different concentrations of salmonella. The activity of the label enzyme (horseradish peroxidase) was measured electrochemically using 3,3′,5,5′-tetramethylbenzidine as substrate. Brewster and Mazenko [37] developed an assay for rapid determination of E. coli O157:H7. The cells were labeled by incubation with an enzyme–antibody conjugate and captured by filtration of the sample/conjugate mixture through a 0.2-μm filter. The enzyme-labeled cells were detected by placing the filter on the surface of an electrode, incubating with enzyme substrate, and measuring the current produced by oxidation of the electroactive enzyme product; the method had a detection limit of 5 × 103 CFU mL−1 with a 25-min analysis time. The major obstacle to decreasing the lower detection limit was the nonspecific adsorption of the conjugate on the membrane. Adsorption is undoubtedly the simplest, quickest, and least reliable of the immobilization methods [16]. Since it consists in the random attachment of the antibodies onto the substrate, the correct orientation of the binding sites cannot be controlled. The exploitation of self-assembled monolayers (SAMs) to enhance biosensor performance has been widely reported. SAMs are structured oriented layers containing functional groups well ordered at the monolayer–liquid interface, and the binding of biological components through this functional group allows control of their orientation. Susmel et al. [38] demonstrated the feasibility of a one-step, specific, label-less, quantitative detection of food pathogens based on the measurement of the diffusion of a redox probe using screen-printed gold electrodes and thiol-based SAM for immobilization of antibodies on the electrode surface. Following antibody immobilization via the optimum SAM, the redox behavior and diffusion coefficient (D) of the potassium hexacyanoferrate(II) probe was monitored in the absence and presence of analyte. In the presence of analyte, a change in the apparent diffusion coefficient of the redox probe was observed, attributable to impedance of the diffusion of redox electrons to the electrode surface due to the formation of the antibody–bacteria immunocomplex. No change in the diffusion coefficient was observed when a nonspecific antibody (mouse IgG) was immobilized and antigen added. The system has been demonstrated with Listeria monocytogenes and Bacillus cereus.
In order to enhance the amperometric response of an immunosensor for detection of Vibrio parahaemolyticus (VP) based on a graphite screen-printed electrode (SPE) modified by VP antibody (HRP-anti-VP), a composite of agarose-doped nano-Au was prepared for immobilization of horseradish peroxidase (HRP)-labeled VP antibody [39]. The active site of HRP was shielded and the access of substrate molecules to HRP was partially shielded after HRP-anti-VP immunoreacted with VP to form immunocomplex during the incubation. The quantitative detection of VP based on the shift of reduction current showed excellent analytical performance (detection limit of 7 × 104 CFU mL−1) and good consistency with ELISA.
A different analytical application of immunosensors is that described by Mittelmann et al. [40]; these authors developed an amperometric immunosensor based on the activity of β-galactosidase for quantifying coliforms, represented by E. coli and Klebsiella pneumoniae. The experimental system allowed the simultaneous analysis of eight samples using disposable screen-printed electrodes. The specific detection of E. coli was achieved by using an antibody-coated electrode that specifically binds the target bacteria. The detection was based on the electrochemical measurement of β-galactosidase activity, using p-aminophenyl-β-D-galactopyranoside as substrate.
The same group [41] combined this method in a system involving a bacteriophage, a virus that recognizes, infects, and lyses only one bacterial species among mixed populations, thereby releasing intracellular enzymes that can be monitored by the amperometic measurement of enzymatic activity. The same electrochemical method developed for the identification of low concentrations of E. coli was thus applied to Bacillus cereus as a model for B. anthracis and Mycobacterium smegmatis as a model for M. tuberculosis [41]. Enzymatic activity was determined electrochemically using as substrate p-aminophenyl-α-D-glucopyranoside (p-AP-α-GLU) for B. cereus and p-aminophenyl-β-D-glucopyranoside (p-AP-β-GLU) for M. smegmatis. The product of the reaction, p-aminophenol (p-AP), was oxidized at the carbon anode at +220 mV vs. (Ag/AgCl) reference electrode. The detection procedure took less than 8 h.
Cyanobacteria are not strictly speaking food pathogens; however, their presence in water represents a serious problem because of the potent cyanotoxins that these algae may release. Cyanotoxins can also enter the food chain, as fish and birds may ingest and accumulate them during and after the occurrence of blooms. Consequently, the toxic compounds may be present not only in the algae, but also in the water and in other living organisms. Two electrochemical immunosensors for the detection of microcystin-LR (MC-LR) based on the affinity between this cyanotoxin and the corresponding monoclonal and polyclonal antibodies were reported in [42]. Disposable electrochemical immunosensors for the detection of another algal toxin, domoic acid (DA), were reported in [43, 44]. Electrochemical immunosensor for ochratoxin A and aflatoxin were reported in [45, 46].
Magnetic bead-based immunosensors
Different strategies have been proposed in the literature to selectively capture the microorganisms. Among these, antibody-coated superparamagnetic beads are very interesting. Magnetic beads are uniform microparticles comprising superparamagnetic material wrapped in a polymer shell. They have an even dispersion of magnetic material (Fe2O3 and Fe3O4) throughout the bead and are coated with a polymer that allows the adsorption or coupling of various molecules. The possibility of maintaining (by shaking or rotating) the beads in suspension ensures a rapid and efficient binding of the target analytes. Shape and size uniformity prevent “clumping” and nonspecific binding due to irregularly shaped particles. Their superparamagnetic properties allow the quantitative magnetic separation of the beads and ensure that they retain no residual magnetism when removed from the magnetic field. They have been used for the selective separation of bacteria and their quantization using different methods. Nowadays, well-defined functionalized nanoparticles and microparticles are commercial available. The main advantage in coupling these magnetic beads with electrochemical transducers is that nonspecific adsorption, which causes important errors in immunosensors, could be remarkably suppressed by antibody–antigen recognition and transduction at the surface of magnetic beads.
An enzyme-linked immunomagnetic electrochemical assay was reported in [47]. Salmonella typhimurrium was sandwiched between antibody-coated magnetic beads and an enzyme-conjugated antibody. With the aid of a magnet, the beads (with or without bound bacteria) were localized onto the surface of disposable graphite electrodes in a multiwell plate format. Enzyme substrate was added, and conversion of substrate to an electroactive product was measured. The electrochemical response was directly proportional to the number of captured bacteria. Using this technique, a minimum detectable level of 8 × 103 cells mL−1 of Salmonella typhimurium in buffer was achieved in ca. 80 min. Immunomagnetic separation followed by electrochemical analysis for the detection of E. coli was reported in [48].
An immunoelectrochemical biosensor utilizing immunomagnetic separation was developed for the detection of S. typhimurium in chicken carcass wash water in a time of 2.5 h [49]. Sampling involved the separation of antigen via magnetic beads coated with polyclonal antibody. After incubation, the sample was processed through a flow injection analysis cell for amperometric detection. An immunoelectrochemical method coupled with immunomagnetic separation was developed for rapid detection of E. coli O157:H7 in the same matrix [50].
An enzyme-linked immunoassay coupled with a tyrosinase-modified enzyme electrode for rapid detection of Campylobacter jejuni was reported in [51]. The immunomagnetic separation (IMS) method was investigated to achieve optimal isolation of C. jejuni cells. Eight types of beads with three different sizes and function groups were coated with anti-C. jejuni to isolate C. jejuni from the sample solution. This system was evaluated using C. jejuni pure culture and poultry samples inoculated with C. jejuni.
Immunomagnetic beads were used in an electrochemical enzyme-linked immunomagnetic electrochemistry (ELIME) assay to detect and quantify levels of S. aureus in broth cultures [52]. The assay was a modification of a “sandwich” format based on the use of common IgG as well as specific antibodies to bind protein A, an antigen localized in the cellular wall of S. aureus and partially extracted by boiling. Using this system the detection limit decreased about 2,000-fold.
DNA-based biosensors
In recent years various kinds of electrochemical biosensors based on identification of the bacterial nucleic acid have been developed. The development of DNA-based biosensors for the detection of specific nucleic acid sequences consists in the immobilization, onto the surface of a chosen transducer, of an oligonucleotide with a specific base sequence called the probe. The complementary sequence (target) present in the sample solution is recognized and captured by the probe through the hybridization reaction. The evaluation of the extent of the hybridization allows one to conclude whether the sample solution contains the complementary sequence of the probe or not. Electrochemical transducers have received considerable recent attention in connection with the detection of DNA hybridization. Some excellent reviews have summarized the recent progress in this field [53–58] and a few examples of pathogen detection using genosensors are cited. Mainly, DNA biosensors have been coupled to PCR as the specific detection method for the amplified base sequence.
In our recent paper [59], we described the simultaneous detection of different food pathogenic bacteria by means of a disposable electrochemical low density genosensor array. The analytical method relied on the use of screen-printed arrays of gold electrodes, modified using thiol-tethered oligonucleotide probes (Fig. 2). The samples identifying the bacteria of interest were obtained from the corresponding genomic DNAs through PCR amplification. These unmodified PCR products were captured at the electrode interface via sandwich hybridization with surface-tethered probes and biotinylated signaling probes. The resulting biotinylated hybrids were coupled with a streptavidin–alkaline phosphatase conjugate and then exposed to an α-naphthyl phosphate solution. Differential pulse voltammetry was finally used to detect the α-naphthol signal. The analytical strategy was based on the identification of toxins produced by the bacteria of interest. Thus, for each bacteria strain, the capture and signaling probes were selected within the sequence of a gene encoding a strain-specific toxin. This step was the most important in order to have a strain-specific assay. The genes were chosen considering the frequency of the presence of the expressed toxins in contaminated foods and the virulence of the different bacterial strains. We demonstrated the simultaneous analysis of Salmonella enterica, Lysteria monocytogenes, Staphylococcus aureus, E. coli 0157:H7 in less than 1 h .
Wang [57] developed genosensors for Cryptosporidium, E. coli, Giardia, and Microbacterium tubercolosis based on the immobilization of specific oligonucleotides onto a carbon paste electrode and chronopotentiometry for monitoring the hybridization events.
Lermo et al. [60] used a genomagnetic assay for the electrochemical detection of food pathogens based on in situ DNA amplification with magnetic primers. The performance of the genomagnetic assay was firstly demonstrated for a DNA synthetic target by its double hybridization with both a digoxigenin probe and a biotinylated capture probe, and further binding to streptavidin-modified magnetic beads. The DNA-sandwiched target bound on the magnetic beads was then separated by using a magneto electrode based on a graphite–epoxy composite. The electrochemical detection was finally achieved by an enzyme marker, anti-digoxigenin horseradish peroxidase (HRP). The novel strategy was used for the rapid and sensitive detection of polymerase chain reaction (PCR)-amplified samples. Promising resultants were also achieved for the DNA amplification directly performed on magnetic beads by using a novel magnetic primer, i.e., the up PCR primer bound to magnetic beads. The reliability of the assay was tested for Salmonella spp. in buffer solution.
Elsholz et al. [61] described a low-density electrical 16S rRNA specific oligonucleotide microarray and an automated analysis system for the identification and quantization of pathogens, such as E. coli, Pseudomonas aeruginosa, Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus epidermidis. Interdigitated gold array electrodes (IDA electrodes), which had structures in the nanometer range, were used for very sensitive analysis. Thiol-modified oligonucleotides were immobilized on the gold IDA as capture probes. They mediate the specific recognition of the target 16S rRNA by hybridization. Additionally three unlabeled oligonucleotides were hybridized in close proximity to the capturing site. They were supporting molecules, because they improve the RNA hybridization at the capturing site. A biotin-labeled detector oligonucleotide was also allowed to hybridize to the captured RNA sequence. The biotin labels enabled the binding of avidin alkaline phophatase conjugates. The phosphatase liberated the electrochemical mediator p-aminophenol from its electrically inactive phosphate derivative. The electrical signals were generated by amperometric redox cycling and detected by a multipotentiostat. The read out signals of the microarray were position-specific currents and changed over time in proportion to the analyte concentration. If two additional biotins were introduced into the affinity binding complex via the supporting oligonucleotides, the sensitivity of the assays increased more than 60%. The limit of detection of E. coli total RNA was determined to be 0.5 ng L−1. The control of fluidics for variable assay formats as well as the multichannel electrical read out and data handling were all fully automated. The fast and easy procedure did not require any amplification of the targeted nucleic acids by PCR.
Another example of a PCR-free method was proposed in [62], which described the development of a field-usable RNA biosensor for the specific, sensitive, and rapid detection of viable E. coli in water. Highly specific DNA probes hybridized with an E. coli mRNA sequence that was amplified using the isothermal NASBA technique. For sample preparation, a heat shock, was applied to the cells prior to disruption. mRNA was then extracted, purified, and finally amplified using the isothermal amplification NASBA technique. The amplified RNA was then quantified with the biosensor. The biosensor was a membrane-based DNA/RNA hybridization system using liposome amplification. The various biosensor components such as DNA probe sequences and concentration, buffers, and incubation times were optimized, and a detection limit of 5 fmol per sample was determined for a synthetic target sequence. An excellent correlation to a much more elaborate and expensive laboratory-based detection system was demonstrated, which can detect as few as 40 E. coli CFU mL−1.
Potentiometric, FET, and LAPS-based biosensors
The basic principle behind potentiometric measurements is the development of charge related to the analyte activity a 1 in the sample through the Nernst relationship:
where E 0 is the standard potential for a 1 = 1 mol L−1, R is the gas constant, F is the Faraday constant, T is the temperature in K, n is the total number of charges of ion i, and the signal + and − are for cations and anions, respectively. Typically, a reference electrode (inert) and one working electrode both in contact with the sample are required.
Potentiometric transducers are the least common in pathogen detection, but different strategies related to pH change or ion concentration monitoring could be possible. The main advantage of these devices is the wide concentration range for which ions can be detected, generally 10−6–10−1 mol L−1. Their continuous measurement capability is also an interesting possibility. The apparatus is inexpensive, portable, and is well suited for in situ measurements. The main disadvantage is that the limit of detection in some kinds of environmental samples can be rather high (10−5 mol L−1) and the selectivity can be rather poor.
Another approach uses suitably modified ion-selective field effect transistors (ISFETs) [63] which utilize the semiconductor field effect to detect biological recognition events. ISFETs use an electric field to create regions of excess charge in a semiconductor substrate in order to enhance or decrease local conductivity. They consist of a p-type silicon substrate with two n-doped regions known as source and drain, separated by a short distance (gate) covered by a layer of insulator. The gate insulator is typically SiO2 and it is covered by an ion-selective membrane which is selectively permeable to a certain ions, e.g., K+, Ca2+, F−. The application of these devices in the area of biosensors is reasonably new [16] and their use is not spreading as quickly as other electrochemical techniques due to, amongst others (a) problems related to production which include incompatibility of most biomolecule immobilization methods with the ISFET fabrication technology and difficult packaging and encapsulation at wafer level, (b) poor detection limits, linear range, and reproducibility, and (c) inadequate device stability. Evolving from ISFETs, a recent technology combines potentiometry and optical detection. It is known as light-addressable potentiometric sensor (LAPS) [64] and a commercial product, the Threshold Immunoassay System, is available and has successfully been applied to bacterial detection [65]. LAPS is based on the coupling of a transient photocurrent to an insulated n-doped or p-doped silicon thin layer in contact with an electrolyte. This transient photocurrent is induced by the application of transient illumination using an intensity-modulated light source such as light-emitting diodes (LEDs). The magnitude of the induced photocurrent depends on the potential applied to the silicon plate. It is even possible to detect different physicochemical phenomena by using different light sources on different spatial regions. If these regions are structurally different then the control of several different parameters on a single device is possible.
Gehring et al. [66] described the development of an immunoligand assay (ILA) in conjunction with a LAPS for the rapid detection of E. coli O157:H7 cells in buffer. The ILA protocol consisted of “sandwiching” bacterial analyte between biotinylated and fluoresceinated antibodies, indirect enzyme labeling of the bacteria with urease-labeled anti-fluorescein antibody, and active capture of the immune complex at a biotinylated bovine serum albumin blocked nitrocellulose filter membrane with streptavidin. Using live E. coli O157:H7, the efficiency of the ILA was compared using various ratios of the biotinylated and fluoresceinated antibodies. Simultaneous addition of equimolar biotinylated and fluoresceinated antibodies effected optimal urease labeling and subsequent active capture of the bacteria in the ILA. Equimolar concentrations of the antibodies were varied to achieve optimal LAPS detection response for the live bacteria. Using ILA with LAPS, a minimum detectable level of ca. 7.1 × 102 cells mL−1 of heat-killed or ca. 2.5 × 104 cells mL−1 of live E. coli O157:H7 bacteria was achieved in Tris-buffered saline in an assay time of ca. 45 min or ca. 30 min, respectively.
Ercole et al. [67] described the application of an antibody-based biosensor for the determination of E. coli cells in vegetable food. The presence of E. coli as a bioindicator of bacterial contamination—particularly faecal—was detected using the potentiometric alternating biosensing (PAB) system based on a LAPS transducing element, detecting pH variations due to NH3 production by a urease–E. coli antibody conjugate. Commercial samples of vegetable (lettuce, sliced carrots, and rucola) were washed with peptone water at pH 6.8, blended either in a stomacher or in a sonicator, to detach bacterial cells and to recover them in the liquid medium. This liquid phase was analyzed both by PAB system and conventional colony forming units (CFUs) methods. The proposed PAB system appears to be very sensitive and fast compared with conventional methods: a concentration of 10 cells mL−1 was detected in an assay time of ca. 1.5 h, i.e., the detection time was 10–20 times shorter than the conventional CFU procedure.
Examples of applications of LAPS biosensors to real matrices are reported in Table 2.
Impedimetric and conductimetric biosensor
Electrochemical impedance spectroscopy (EIS) is a powerful technique for the characterization of electrochemical systems. The fundamental approach of all impedance methods is to apply a small amplitude sinusoidal excitation signal to the system under investigation and measure the response (current or voltage or another signal of interest).
Impedance microbiology is one of the earliest physicochemical methods for the detection of bacteria in foods and has been developed, more than ten years ago, as a nonconventional method able to detect bacteria within 24 h. It is based on the measurement of changes in electrical impedance of a medium or a reaction solution resulting from the bacterial growth. Microbial metabolism usually results in an increase in both conductance and capacitance, while causing a decrease in impedance [9]. Therefore, impedance, conductance, capacitance, and resistance are merely different ways of monitoring the test system and are all interrelated [9]. The impedance method is accepted by the Association of Official Analytical Chemists (AOAC) as a method for the detection of Salmonella in food [20]. Several designs of impedimetric measurement methods were proposed in the literature [9, 16, 68].
An advantage of EIS compared with amperometry or potentiometry is that labels are no longer necessary, thus simplifying sensor preparation. However, the detection limits of EIS are still poor compared with traditional methods [16]. To increase the sensitivity Radke et al. [69] described a high density microelectrode array biosensor for the detection of Escherichia coli O157:H7. The biosensor was fabricated from (100) silicon with a 2-μm layer of thermal oxide as an insulating layer, an active area of 9.6 mm2, and consisted of an interdigitated gold electrode array. The sensor surface was functionalized for bacterial detection using heterobifunctional crosslinkers and immobilized polyclonal antibodies to create a biological sensing surface. Bacteria suspended in solution became attached to the immobilized antibodies when the biosensor was tested in liquid samples. The change in impedance caused by the bacteria was measured over a frequency range of 100 Hz–10 MHz. The biosensor was evaluated for E. coli O157:H7 detection in pure culture and inoculated food samples. The biosensor was able to discriminate between cellular concentrations of 104–107 CFU mL−1.
Muhhmmad-Tahir et al. [70] described the development of a conductometric biosensor for detecting foodborne pathogens. The biosensor consisted of two components: an immunosensor that was based on an electrochemical sandwich immunoassay, and a reader for signal measurement. The architecture of the immunosensor utilized a lateral flow system that allowed the liquid sample to move from one pad to another. The biosensor provides a specific, sensitive, low volume, and near-real-time detection mechanism. Results were presented to highlight the performance of the biosensor for enterohemorrhagic E. coli O157:H7 and Salmonella spp., which are of concern to biosecurity. The lower limit of detection was approximately 81 CFU mL−1 within a 10-min process. The ability to change the specificity of the antibodies enabled the biosensor to be used as a detection device for other types of foodborne pathogens.
Pal et al. [71] described the development of a direct charge transfer (DCT) biosensor for the detection of the foodborne pathogen Bacillus cereus. The biosensor was fabricated using antibodies as the sensing element and a polyaniline nanowire as the molecular electrical transducer. The sensor design consisted of four membrane pads, namely, sample application, conjugate, capture, and absorption pads. Two sets of polyclonal antibodies, secondary antibodies conjugated with polyaniline nanowires and capture antibodies, were applied to the conjugate and the capture pads of the biosensor, respectively. The detection technique was based on capillary flow action which allowed the liquid sample to move from one membrane to another. The working principle involved antigen–antibody interaction and direct electron charge flow to generate a resistance signal that was recorded. Detection from sample application to final results was completed in 6 min in a reagentless process. The biosensor sensitivity in pure cultures of B. cereus was found to be 101 to 102 CFU mL−1. The biosensor was also found to be specific in detecting the presence of B. cereus in a mixed culture of different Bacillus species and other foodborne pathogens.
Applications of impedimetric biosensors to real matrices are reported in Table 2.
New trends in biosensor development
Over recent years a lot of effort has gone into the study and development of biosensors for food pathogen detection. Even if it is true that their performance still needs improvement, biosensors have demonstrated potential for food microbial analysis, as reported in the sections above.
In order to become more attractive, biosensors first need to show that they are capable of reaching at least the same detection levels as traditional techniques (10–100 CFU mL−1) with a high level of reproducibility. The sensitivity of a biosensor depends on the (bio)recognition element and on transducers properties. The selection of more specific ligands (mainly by combinatorial techniques) and the development of new electrochemical platforms based on nanomaterials will give important benefits in the near future.
Moreover, emerging nanofabrication and microfabrication techniques will improve other important aspects such as sample treatment and the integration of biosensor into microdevices and nanodevices.
Biorecognition element
The two main factors important for every biorecognition element are affinity and specificity. Some developments related to biorecognition elements that appear to have potential for biosensor application in food pathogen detection are described below. First new ligands useful in genosensor development will be described (i.e., LNAs and PNAs). Other important affinity ligands such as aptamers and phage display peptides will then be reviewed.
Locked nucleic acid
A novel series of nucleotide analogs called locked nucleic acids (LNAs) have recently been reported [72]. A locked nucleic acid is a modified RNA nucleotide comprising a bicyclic ribonucleoside that is linked between the 2′-oxygen and the 4′-carbon atoms with a methylene unit (Fig. 3). LNAs can be used in any hybridization assay that requires high specificity and/or reproducibility. General properties of LNA oligonucleotides include highly stable base pairing with DNA and RNA [72], exceptionally high thermal stability, improved discrimination, compatibility with most enzymes, and predictable melting behavior. Application of LNAs has yielded good results in allele-specific PCR and mRNA sample preparation. There are enormous possibilities for developing LNA devices to detect various food pathogens by using the specific oligonucleotide probes.
Peptide nucleic acid (PNA)
Peptide nucleic acid (PNA) is a synthetic nucleic acid reported in the early 1990s [73] that has an achiral neutral polyamide backbone formed by repetitive units of N-(2-aminoethyl)glycine linked to nucleoside bases (Fig. 4). A PNA molecule that mimics DNA is advantageous as a probe molecule owing to superior hybridization characteristics and improved chemical and enzymatic stability relative to nucleic acids. Furthermore, its different molecular structure enables new modes of label-less detection, thereby contributing significantly towards the establishment of faster, more stable, and more reliable analytical processes. PNAs have been reported for various applications including hybridization biosensors and microarrays. Since PNAs are resistive to nuclease attack, they provide an extra edge over the use of conventional or naturally existing nucleic acids.
Aptamers
Aptamers are single-stranded DNA or RNA ligands which can be selected for different targets starting from a huge library of molecules containing randomly created sequences [74]. The selection process is called systematic evolution of ligands by exponential enrichment (SELEX), first reported in 1990 [74]. The SELEX process involves iterative cycles of selection and amplification starting from a large library of oligonucleotides with different sequences (generally 1015 different structures). After the incubation with the specific target and the partitioning of the binding from the nonbinding molecules, the oligonucleotides that are selected are amplified to create a new mixture enriched in those nucleic acid molecules having a higher affinity for the target. After several cycles of the selection process, the pool is enriched in the high affinity sequences at the expense of the low affinity binders.
The number of cycles required depends on the stringency conditions, but, once obtained and once the sequence is known, unlimited amounts of the aptamer can be easily prepared by chemical synthesis. Aptamers have other advantages over antibodies that make them very promising for analytical applications, in particular they avoid the use of animals or cell lines in their production. Antibodies against molecules that are not immunogenic are difficult to generate. In contrast, aptamers are isolated by in vitro methods that are independent of animals: an in vitro combinatorial library can be generated against any target. In addition, generation of antibodies in vivo means that it is the animal’s immune system that selects the sites on the target protein to which the antibodies bind. The in vivo parameters restrict the identification of antibodies that can recognize targets only under physiological conditions, thus limiting the extent to which the antibodies can be functionalized and applied. Moreover, the aptamer selection process can be manipulated to obtain aptamers that bind a specific region of the target and with specific binding properties under different binding conditions. After selection, aptamers are produced by chemical synthesis and purified to a very high degree by eliminating the batch-to-batch variation found when using antibodies. By chemical synthesis, modifications in the aptamer can be introduced to enhance the stability, affinity, and specificity of the molecules. Often the kinetic parameters of aptamer–target complex can be changed to achieve higher affinity or specificity. Another advantage over antibodies can be seen in the higher temperature stability of aptamers and they can recover their native active conformation after denaturation; in contrast, antibodies are large proteins that are sensitive to temperature and they can undergo irreversible denaturation.
Some aptamers have already been used for the detection of specific pathogens. Pan et al. [75] reported the first direct selection of aptamers for bacterial proteins involved in bacterial invasion. In particular, they focused on Salmonella enteric serovar Typhi. Another aptamer for pathogens has been developed for Campylobacter jejuni [76]. The selected DNA aptamer showed strong binding affinity towards Campylobacter with minimal cross-reactivity over other food pathogens. Another new type of assay aimed at the detection of bacterial pathogens was developed using a set of 25 unique DNA sequences that bind to Francisella tularensis [77]. Tularemia is a very rare disease that is caused by F. tularensis and humans most commonly contract this disease by handling or eating undercooked wild animal meat. A sandwich aptamer-linked immobilised sorbent assay (ALISA) was developed with the 25-aptamer cocktail and resulted in good sensitivity and high selectivity.
Phage display techniques
Phage display is a useful tool for the isolation of antibody fragments with desired specificities. The technique involves the display of a library of single-chain antibody (scFv) fragments on the surface of filamentous phages followed by selection of the desired recombinant phages by means of specific binding to an antigen of interest. Although phage display has advantages over conventional polyclonal and monoclonal antibody production, few reports have employed this technique for the selection of reagents for the detection of foodborne pathogens [78]. Phage display also provides several approaches for rational improvement of antibody affinity and selectivity. Given a set of phage clones with known affinity and selectivity profiles, selection strategies can be designed to isolate clones with optimal properties from the existing library of clones. By correlating affinity and selectivity data with DNA sequence information, it is feasible to design and construct novel sequences that express antibodies with the desired properties.
These techniques are evolving rapidly and appear to represent the future of recognition element selection technology [14].
Nanomaterials
Nowadays, nanomaterials (carbon nanotubes, metal nanoparticles, or metal oxide nanostructures) are exerting a big impact on the development of electrochemical biosensors [79]. Nanotechnology brings new possibilities for biosensor construction and for developing novel electrochemical bioassays. Nanoscale materials have been used to achieve direct wiring of enzymes to electrode surfaces, to promote electrochemical reactions, to impose nanobarcodes on biomaterials, and to amplify the signal from biorecognition events. Electrochemical nanobiosensors have been applied in areas such as cancer diagnostics and detection of infectious organisms, including food pathogens [79]. As an example, an electrochemical immunosensor for cholera toxin was developed based on poly(3,4-ethylenedioxythiophene)-coated carbon nanotubes [80]. The sensing interface consists of monoclonal antibody against the B subunit of cholera toxin that is linked to poly(3,4-ethylenedioxythiophene) coated on Nafion-supported multiwalled carbon nanotube casted film on a glassy carbon electrode. The cholera toxin (CT) was detected by a sandwich-type assay on the electronic transducers, where the toxin is first bound to the anti-CT antibody and then to the ganglioside-functionalized liposome loaded with potassium ferrocyanide for amplification.
As a general remark, electrochemical nanobiosensors will undoubtedly offer an important step toward the development of selective biorecognition devices for analytical applications with sensitivities down to a few target molecules. There is high expectation that such devices will develop toward reliable point-of-care diagnostics of cancer and other diseases, and as tools for food and environmental monitoring [79].
Advanced technology in sample treatment: from flow system to lab on a chip technology
A significant challenge for all biosensor systems is to achieve high assay sensitivity and specificity while minimizing sample preparation, requirements, operational complexity, and time (Fig. 5).
In many biosensor approaches, the separation and biosensing are performed in several steps with a manual transfer step in between. The multistep assay and manual transfer can result in sample loss and errors. The combination of separation with biosensor technology can dramatically improve speed, selectivity, and sensitivity of assays with minimal sample manipulation. The advantage of an on-line separation/biosensor system is that individual bacterial cells within a complex mixture are first purified and then detected by selective interactions with analysis times of minutes. Immunofiltration-based separations can offer an attractive approach [81], e.g., eliminating the need for pre-enrichment by the use of a flow-through immunofiltration assay combined with an amperometric sensor. Abdel-Hamid et al. reported a low detection limit of 50 cells mL−1 and an overall analysis time of 30 min but with a limited analytical range of 100/600 cells mL−1. The immunosensor consisted of a disposable antibody-modified filter membrane resting on top of a hollow carbon rod which acts as the working electrode. Another hollow carbon rod acts as a counter electrode, and a hollow Ag/AgCl disk as a reference electrode. The authors used a special device where the liquid flows from the inlet of the immunosensor, through the filter membrane, and then through the hollow channel formed in the working, reference, and counter electrodes, respectively. A sandwich scheme of immunoassay was employed. The activity of the peroxidase label (captured on the membrane surface) was measured using an amperometric technique with iodide ions in a phosphate buffer solution acting as the mediator. A polarization potential of 0.0 V versus Ag/AgCl was applied between the working and reference electrodes.
The approach proposed by Perez et al. [41], and developed in author’s lab, is based on an amperometric flow-injection analysis (FIA) system for the measurement of viable E. coli O157, measuring their respiratory activity. The selective immunological separation of E. coli was performed by using antibody-coated magnetic particles. The kinetics and the capacity parameters regarding the attachment of bacteria to the immunobeads were studied. The immunomagnetic separation was then used in conjunction with electrochemical detection to measure the concentration of viable bacteria. Electrochemical detection was carried out using redox mediators, potassium hexacyanoferrate(III) and 2,6-dichloroindophenol. The measurement was performed using an FIA system. A calibration curve of CFU against electrochemical response was obtained. The detection limit was 105 CFU mL−1, and the complete assay was performed in 2 h. This technique could easily be automated and the analysis can be performed quickly and continuously. The renewal of the immunosensor sensing surface was accomplished by removing the magnet and washing down the magnetic particles. The immunosensor was then ready for the injection of new antibody-modified magnetic particles for another cycle. Some advantages over ELISA methods are the direct detection of viable cells (and not total bacterial load) and the need for only one antibody (not enzyme-labeled), thus making the assay faster (only one washing step is necessary) and less expensive.
Sippy et al. [82] described a rapid and sensitive technique to detect low numbers of the model organism E. coli O55, combining lateral flow immunoassay (LFI) for capture and amperometry for sensitive detection. Nitrocellulose membranes were used as the solid phase for selective capture of the bacteria using antibodies to E. coli O55. Different concentrations of E. coli O55 in Ringer’s solution were applied to LFI strips and allowed to flow through the membrane to an absorbent pad. The capture region of the LFI strip was placed in close contact with the electrodes of a Clark cell poised at +0.7 V for the detection of hydrogen peroxide. Earlier research identified that the consumption of hydrogen peroxide by bacterial catalase provided a sensitive indicator of aerobic and facultative anaerobic microorganism numbers. Modification and application of this technique to the LFI strips demonstrated that the consumption of 8 mM hydrogen peroxide was correlated with the number of microorganisms presented to the LFI strips in the range of 2 × 101–2 × 107 CFU. Capture efficiency was dependent on the number of organisms applied and varied from 71% at 2 × 102 CFU to 25% at 2107 CFU. The procedure was completed in less than 10 min and could detect less than 10 CFU captured from a 200-mL sample applied to the LFI strip. The described method provided proof of principle for the basis of a new technological approach to the rapid, quantitative, and sensitive detection of bacteria that express catalase activity.
Other examples of intriguing miniaturized fluidics have been reported [69–71].
However, as already stressed, the combined use of microfabrication and nanofabrication techniques in the area of biosensors for food pathogens holds great promise especially in the field of sample treatment. Amongst the advantages of this smaller-scale approach are: (a) the possibility of mass production and reduced unit costs, (b) it allows the use of sample volumes in the range of nanolitres or less, which also implies that the cost of reagents is not too high, (c) microfluidics improve mixing rates and mass transport which is expected to result in much shorter analysis times, (d) the performance of multianalyte analysis is enabled in the same device, which also shortens analysis time, and (e) because the volumes manipulated are so tiny, these devices are safer and more environmentally friendly. Power consumption is extremely low and contamination associated with waste material may be easier to contain owing to the possibility of using tiny volumes and cartridge-like configurations.
Micro total analysis systems (μTAS) or lab on a chip (LOC) (term identifying devices that integrate (multiple) laboratory functions on a single chip of only millimeters to a few square centimeters in size and that are capable of handling extremely small fluid volumes down to less than picoliters) are the future to reduce reaction time, amount of reagents, labor, and cost.
A microfluidic biochip based on impedence measurement was proposed in [83] for detection of metabolic activity of Lysteria and E. coli.
An integrated portable genetic analysis microsystem including PCR amplification and capillary electrophoretic (CE) analysis was reported in [84].
A PCR-free microsystem was reported in [85] and [86]. A system for amperometric detection of E. coli based on the integration of microelectromechanical systems (MEMS), self-assembled monolayers (SAM), DNA hybridization, and enzyme amplification was reported in [85]. The analysis was performed with solution volumes of a few microliters and was completed in 40 min. The detection system was capable of detecting 1,000 E. coli cells without polymerase chain reaction with high specificity for E. coli vs. the bacteria Bordetella bronchiseptica.
A biosensor based on nucleic acid hybridization and liposome signal amplification with an integrated microfluidic system and a minipotentiostat for the quantification of Dengue virus RNA was reported in [86]. By combining microelectronics and microfluidics with the simple and effective liposome signal enhancement technology and an amperometric transducer, the authors designed a miniaturized electrochemical detection system (miniEC) that was easy to assemble and use. Physically, the miniEC had two parts. It consisted of a potentially disposable microfluidic sensor cartridge and the supporting instrumentation to power, measure, and display the sensor results. The amperometric transducer was placed directly into the microchannel of the cartridge. DNA and RNA molecules were quantified in this device based on a sandwich hybridization assay similar to the one previously described [87, 88], with the transduction mechanism being based on electrochemical rather than fluorescence detection.
Conclusions
In this paper we reviewed the scientific literature dealing with the use of electrochemical biosensors in food safety. Different approaches developed by various research groups to detect pathogens or their toxins are mentioned. The methods have different detection limits for the target organisms, different sample pretreatment requirements (extraction, amplification, preculture, etc.), and different assay complexities, degrees of automation, and required analysis times. Many of the described biosensor systems target the most important food pathogens, i.e., Salmonella, E. coli 0157:H7, Listeria monocytogenes, Staphylococcus aureus, Campylobacter. Although some of the reported examples are not targeted at the main important pathogens listed in the Introduction, these approaches are mentioned for illustration, since by using specific biomolecules they may be applied to more relevant targets.
From the analysis of the literature, biosensors have demonstrated huge potential for food microbial analysis. However, in terms of commercialization, one of the problems still facing the production of biosensors for direct detection of bacteria is the sensitivity of the assay in real sample. Analytical procedures must be able to provide a detection limit as low as a single coliform organism in 100 mL of potable water or a value between 10 and 100 CFU mL−1 in 0.01 g of food. As reported in Table 1 and in Table 2, only a few examples of biosensors for food pathogen detection seem to reach this important market requirement.
Specificity is another important issue: biosensors should have the same pathogen specificity as that of the plate culture method. The biosensor system must have the specificity to distinguish the target bacteria in a multiorganism matrix, the adaptability to detect different analytes, the sensitivity to detect bacteria on-line without pre-enrichment, and the rapidity to give real-time results. At the same time, biosensors must have relatively simple and inexpensive configurations [8, 9].
Obviously, enhancing the specificity of biosensor systems and incorporation of all the other features is a very complicated task. This is the main reason why penetration of biosensors into the market is so slow. Similar considerations were reported more or less ten years ago [8, 9], but the situation is still the same.
In our opinion the selection of more specific ligands (mainly by combinatorial techniques) will help in afford high specificity and reproducibility. For this reason we devoted a section to describe the important features of some innovative biorecognition elements such as LNAs, PNAs, aptamers, and phage display peptides that will help to define new biosensor designs.
Another important consideration is the stability of the biorecognition element. This issue is not of the same degree of relevance for all types of biomolecules; however, the use of innovative ligands can also help to overcome this problem. For instance PNAs and LNAs improved chemical and enzymatic stability relative to nucleic acids. Aptamers are considered to be more stable than antibodies since chemical synthesis allows modifications to be introduced into the aptamer molecules to enhance their stability, affinity, and specificity. Furthermore, aptamers exhibit higher temperature stability than antibodies and they can also recover their native active conformation after denaturation. In contrast, antibodies are large proteins that are sensitive to temperature and can undergo irreversible denaturation.
In the near future, pathogen detection, and biosensor technology itself, will undoubtedly benefit from the integration of biosensors into microdevices or nanodevices. Nanomaterials are claimed to improve electrochemical transducer sensitivity. In respect to other transducing principles, electrochemical techniques are much easier to use and allow the miniaturization for the integration in handheld devices, and thus electrochemical biosensors will greatly benefit from nanotechnology and μTAS technology.
References
European Food Safety Authority (2007) http://www.efsa.europa.eu. Accessed 13 Nov 2007
European Commission: 7th Framework Programme (2007) http://ec.europa.eu/research/fp7. Accessed 13 Nov 2007
Banati D (2003) Food Control 14:89–93
Centers for Disease Control and Prevention (2007) http://www.cdc.gov. Accessed 13 Nov 2007
Alocilja EC, Radke SM (2003) Biosens Bioelectron 18:841–846
Walker E, Pritchard C, Forsythe S (2003) Food Control 14:169–174
De Boer E, Beumer RR (1999) Int J Food Microbiol 50:119–130
Invitski D, Abdel-Hamid I, Atanasov P, Wilkins E (1999) Biosens Bioelectron 14:599–624
Invitski D, Abdel-Hamid I, Atanasov P, Wilkins E, Striker S (2000) Electroanalysis 12:317–325
Lim DV (2003) Proc IEEE 91:902–907
Leonard P, Hearty S, Brennan J, Dunne L, Quinn J, Chakraborty T, O’Kennedy R (2003) Enzyme Microb Tech 32:3–13
Siragusa GR, Cutter CN, Dorsa WJ, Koohmaraie M (1995) J Food Protect 58:770–775
Olsen JE (2000) Food Res Int 33:257–266
Rasooly A, Herold KE (2006) J AOAC Int 89:873–883
Arora K, Chand S, Malhotra BD (2006) Anal Chim Acta 568:259–274
Lazcka O, Del Campo FJ, Munoz FX (2007) Biosens Bioelectron 22:1205–1217
Molecular food safety testing (2007) Roche Diagnostics Corp.http://www.molecular-food-safety-testing.com. Accessed 13 Nov 2007
FDA Center for Food Safety and Applied Nutrition (2007) http://www.cfsan.fda.gov/ebam/bam-a1.html. Accessed 13 Nov 2007
Microbiology International (2007) http://www.microbiology-intl.com/ALOA/AFNOR.htm. Accessed 13 Nov 2007
AOAC Official Methods (1996) XVI edn
Yaron S (2002) J Appl Microbiol 92:633–640
Nugen SR, Leonard B, Baeumner AJ (2007) Biosens Bioelectron 22:2442–2448
Cook N (2003) J Microbiol Methods 1761:1–10
Cahill D, Roben P, Quinlan N, O’Kennedy R (1995) Production of antibodies. In: Townsend A (ed) Encyclopedia analytical science, vol IV. Academic, New York, pp 2057–2066
Fitzpatrick J, Fanning L, Hearty S, Leonard P, Manning BM, Quinn JG (2000) Anal Lett 33:2563–2609
Gehring AG, Albin BM (2007) J Rapid Methods Autom Microbiol 15:49–66
Patel PD (2002) TRACS 21:96–115
Suzuki H, Tamiya E, Karube I (1991) Electroanalysis 3:53
Ruan C, Yang L, Li Y (2002) J Electroanal Chem 519:33–38
Mulchandani P, Hangarter CM, Lei Y, Chen W, Mulchandani A (2005) Biosens Bioelectron 21:523–527
Togo CA, Wutor VC, Limson JL, Pletschke BI (2007) Biotechnol Lett (in press)
Perez FG, Tryland I, Mascini M, Fiksdal L (2001) Anal Chim Acta 427:149–154
Serra B, Morales MD, Zhang J, Julio Reviejo A, Hall EH, Pingarron JM (2005) Anal Chem 77:8115–8121
Brewster JD, Gehring AG, Mazenko RS, Van Houten LJ, Crawford CJ (1996) Anal Chem 68:4153–4159
Mirhabibollahi B, Brooks JL, Kroll RG (1990) Appl Microbiol Biotechonol 34:242
Croci L, Delibato E, Volpe G, Palleschi G (2001) Anal Lett 34:2597–2607
Brewster JD, Mazenko RS (1998) J Immunol Methods 211:1–18
Susmel S, Guilbault GG, O’Sullivan CK (2003) Biosens Bioelectron 18:881–889
Zhao G, Xing F, Deng S (2007) Electrochem Commun 9:1263–1268
Mittelmann AS, Ron EZ, Rishpon J (2002) Anal Chem 74:903–907
Yemini M, Yaron L, Ezra Y, Rishpon J (2007) Bioelectrochemistry 70:180–184
Campas M, Marty JL (2007) Biosens Bioelectron 22:1034–1040
Micheli L, Radoi A, Guarrina R, Massaud R, Bala C, Moscone D, Palleschi G (2004) Biosens Bioelectron 20:190–196
Kania M, Kreuzer M, Moore E, Pravda M, Hock B, Guilbault G (2003) Anal Lett 36:1851–1863
Sergio HA, Micheli L, Palleschi G, Compagnone D (2004) Anal Lett 37:1545–1558
Piermarini S, Micheli L, Ammida NHS, Palleschi G, Moscone D (2007) Biosens Bioelectron 22:1434–1440
Gehring AG, Crawford CG, Mazenko RS, Van Houten LJ, Brewster JD (1996) J Immunol Methods 195:15–25
Perez FG, Mascini M, Tothill IE, Turner APF (1998) Anal Chem 70:2380
Che Y, Li Y, Slavik M, Paul D (2000) J Food Prot 63:1043–1048
Ruan C, Wang H, Li Y (2002) Trans ASAE 45:249–255
Che Y, Li Y, Slavik M (2001) Biosens Bioelectron 791–797
Delibato E, Bancone M, Volpe G, De Medici D, Moscone D, Palleschi G, (2005) Anal Lett 38:1569–1586
Lucarelli F, Marrazza G, Turner APF, Mascini M (2004) Biosens Bioelectron 19:515–530
Kerman K, Kobayashi M, Tamiya E (2004) Meas Sci Technol 15:R1–R11
Palecek E (2002) Talanta 56:809–819
Palecek E, Fojta M (2001) Anal Chem 73:74A–83A
Wang J (2002) Anal Chim Acta 469:63–71
Pividori MI, Merkoci A, Alegret S (2000) Biosens Bioelectron 15:291–303
Farabullini F, Lucarelli F, Palchetti I, Marrazza G, Mascini M (2007) Biosens Bioelectron 22:1544–1549
Lermo A, Campoy S, Barbe J, Hernandez S, Alegret S, Pividori MI (2007) Biosens Bioelectron 22:2010–2017
Elsholz B, Wo R, Blohm L, Albers J, Feucht H, Grunwald T, Jurgen B, Schweder T, Hintsche R (2006) Anal Chem 78:4794–4802
Baeumner AJ, Cohen RN, Miksic V, Min J (2003) Biosens Bioelectron 18:405–413
Bergeveld P (2003) Sens Actuators B Chem 88:1–20
Hafeman DG, Parce JW, Mcconnell HM (1988) Science 240(4856):1182–1185
Dill K, Stanker L, Young CR (1999) J Biochem Biophys Methods 41:61–67
Gehring AG, Patterson DL, Tu SI (1998) Anal Biochem 258:293–298
Ercole C, Del Gallo M, Mosiello L, Baccella S, Lepidi A (2003) Sens Actuators B 91:163–168
Yang L, Li Y, Griffis CL, Johnson MG (2004) Biosens Bioelectron 19:1139–1147
Radke SM, Alocilja EC (2005) Biosens Bioelectron 20:1662–1667
Muhammad-Tahir Z, Alocilja EC (2004) Biosyst Eng 88:145–151
Pal S, Alocilja EC, Downes FP (2007) Biosens Bioelectron 22:2329–2336
Kaur H, Arora A, Wengel J, Maiti S (2006) Biochemistry 45:7347–7355
Nelson KE, Levy M, Miller SL (2000) Proc Nat Acad Sci USA 97:3868–3871
Tombelli S, Minunni M, Mascini M (2007) Biomol Eng 24:191–200
Pan Q, Zhang XL, Wu HY (2006) Antimic Agen Chemot 49:4052–4060
McMasters S, Stratis-Cullum DN (2006) Proc SPIE 6380
Vivekananda J, Kiel JL (2006) Lab Invest 86:610–618
Paoli GC, Chen CY, Brewster JD (2004) J Immunol Methods 289:147–155
Pumera M, Sanchez S, Ichinose I, Tang J (2007) Sens Actuators B 123:1195–1205
Viswanathan S, Wu LC, Huang MR, Ho JA (2006) Anal Chem 78:1115–1121
Abdel-Hamid I, Ivnitski D, Atanasov P, Wilkins E (1999) Biosens Bioelectron 14:309–316
Sippy N, Luxton R, Lewis RJ, Cowell DC (2003) Biosens Bioelectron 18:741–749
Gómez R, Bashir R, Bhunia AK (2002) Sens Actuators B Chem 86:198–208
Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, Sensabaugh GF, Riley LW, Mathies RA (2004) Anal Chem 76:3162–3170
Gau J Jr, Lan EH, Dunn B, Ho CM, Woo JCS (2001) Biosens Bioelectron 16:745–755
Kwakye S, Goral VN, Baeumner AJ (2006) Biosens Bioelectron 2:2217–2223
Kwakye S, Baeumner A (2003) Anal Bioanal Chem 376:1062–1068
Zaytseva NV, Montagna RA, Baeumner AJ (2005) Anal Chem 77:7520–7527
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Palchetti, I., Mascini, M. Electroanalytical biosensors and their potential for food pathogen and toxin detection. Anal Bioanal Chem 391, 455–471 (2008). https://doi.org/10.1007/s00216-008-1876-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-008-1876-4