Abstract
In this study we describe a methodology for diagnosing preclinical scrapie infection in hamsters from serum by a combination of Fourier-transform infrared (FT–IR) spectroscopy and chemometrics. Syrian hamsters (Mesocricetus auratus) were orally inoculated with the 263K scrapie agent, or mock-infected, and sera were obtained at 70, 100 and 130 days post infection (dpi) and at the terminal stage of scrapie (160 ± 10 dpi). The analysis of hamster sera by FT–IR spectroscopy and artificial neural networks (ANN) confirmed results from earlier studies which had indicated the existence of disease-related structural and compositional alterations in the sera of infected donors in the terminal stage of scrapie [Schmitt et al. (2002) Anal Chem 74:3865–3868]. Experimental data from sera of animals in the preclinical stages of scrapie revealed subtle but reproducible spectral variations that permitted the identification of a preclinical scrapie infection at 100 dpi and later, but not at 70 dpi. The IR spectral features that were discriminatory for the preclinical stages differed from those of the terminally ill individuals. In order to reliably identify scrapie-negative as well as preclinical (100 and 130 dpi) and terminal scrapie-positive animals, a hierarchical classification system of independent artificial neural networks (ANN) was established. A “toplevel” ANN was designed which discriminates between animals in the terminal stage of scrapie and preclinical scrapie-positive or control animals. Spectra identified by the “toplevel” ANN as preclinical or controls were then further analyzed by a second classifier, the “sublevel” ANN. Using independent external validation procedures, the toplevel classifier produced an overall classification accuracy of 98%, while the sublevel classifier yielded an accuracy of 93%, indicating that scrapie-specific serum markers were also present for preclinical disease stages. Possible spectral markers responsible for the discrimination capacity of the two different ANNs are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transmissible spongiform encephalopathies (TSEs) such as scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle and Creutzfeldt–Jacob disease (CJD) in humans are invariably fatal neurodegenerative disorders of the central nervous system [1]. After the emergence of BSE and variant Creutzfeldt–Jacob disease (vCJD) in 1986 and 1996, respectively, compelling evidence has gradually accumulated that the latter can most likely be attributed to transmissions, probably via contaminated food, of the BSE agent from cattle to man. Therefore, effective infection control measures for the surveillance and containment of the BSE epidemic have become a matter of crucial importance to public health. The countermeasures which have been implemented meanwhile in response to the BSE epidemic are expected to minimize the risk of new primary vCJD infections of humans directly originating from bovines.
However, further challenges in the area of infection control arise from the possibility of undetected preclinical BSE infections in the cattle population, possibly long incubation periods of TSE infections [2], and from the potentially very dangerous hazard of human-to-human transmission of vCJD via blood or blood products [3]. These issues could be effectively addressed if a test for the pre- or subclinical identification of TSEs in blood or blood components could be made available [4, 5].
In the last few years a number of diagnostic screening tests specifically targeting the causative agent of a TSE infection (the misfolded prion protein PrPsc, which is the TSE biomarker in blood or blood products) have been proposed [6–9]. Furthermore, studies are underway that use surrogate markers for the detection of a TSE infection in blood or serum [5, 10–13]. However, despite considerable research efforts, none of the proposed TSE blood tests has been developed into a practicable routine diagnostic method so far [4, 5]. Therefore, the development of new methods for the inexpensive, robust and rapid identification of pre- or subclinical BSE, vCJD and possible other TSEs from blood or its components constitutes a key challenge in the surveillance and control of TSEs.
Recently, we reported that Fourier-transform infrared (FT–IR) spectra from serum of clinically ill scrapie hamsters exhibit a variety of spectral features different from those observed in FT–IR spectra from scrapie-negative control hamsters [14]. These spectral differences reflect unknown molecular alterations in the sera of scrapie hamsters, and could be used, with the help of advanced computer-aided pattern recognition techniques (artificial neural networks, ANNs), to establish an experimental proof-of-concept for the rapid, inexpensive and reliable identification of TSE-affected animals.
Subsequently, the results from the feasibility study in hamsters were evaluated under field conditions with blinded serum samples from BSE-infected cattle [15]. This revealed, in two independent test series with bovine sera from more than 800 animals (including BSE-positive, healthy controls, and other animals suffering from conventional viral or bacterial infections), a test sensitivity and specificity of up to 92 and 96%, respectively. The results from these studies were confirmed by others using similarly designed approaches of FT–IR-based BSE serum testing [16]. Thus, the proof-of-concept for FT–IR based TSE serum testing originally established in the hamster model could be confirmed and expanded for clinical BSE in cattle.
Taken together, these findings strongly suggest that the molecular serum alterations detected by FT–IR spectroscopy and used for the identification of affected and healthy animals are specific for TSEs. In addition, they highlight scrapie-infected hamsters as a highly relevant animal model for the FT–IR-based TSE serum diagnostic in bovines, and possibly other animal species and humans. Here we report, for orally challenged hamsters, the preclinical detection of scrapie by FT–IR spectroscopic testing of serum.
Materials and methods
Donors and sampling of serum
In the present study, sera from orally (p.o.), intraperitoneally (i.p.) and mock-infected female and male Syrian hamsters were used. I.p. and p.o. infections with the scrapie agent were performed with 100 μl of a 10% 263K-scrapie brain homogenate containing a dose of 1–3×107 LD50 i.c., as described elsewhere [17]. Control animals were mock-infected with equivalent doses of normal hamster brain homogenate. Inocula were administered to the recipients at an age of 4–6 weeks. Orally infected hamsters were sacrificed at 70, 100, 130 dpi, or in the terminal stage of scrapie. Terminally infected animals showed mean incubation periods of 160 ± 10 (SD) days. In order to account for a catabolic type of metabolism in animals at the terminal stage of disease, some of the control animals were deprived of nutrition before euthanasia for 48h. An overview of the numbers of p.o. and i.p. scrapie-infected hamsters as well as of control animals is given in Table 1. Euthanasia with CO2, collection of blood and isolation of serum were carried out as previously described [14, 18].
FT–IR spectroscopy
For FT–IR transmission-type measurements, a serum volume of 2.6 μl was spread onto a ZnSe window of a sample wheel and air-dried for 15 minutes at 37°C. Subsequently, the multisample cuvette was sealed with a KBr window and transferred into an IFS 28/B spectrometer (Bruker Optik GmbH, Ettlingen, Germany). Infrared spectra were recorded between 500 and 4000 cm−1 by co-adding 128 scans for each individual spectrum. Nominal physical resolution was set to 4 cm−1, and a zero filling factor of 4 was employed, yielding a point spacing in the spectra of 1 cm−1. Spectra were collected using the data acquisition software OPUS supplied by Bruker. Each serum sample was measured in triplicate (three different times).
High-performance thin-layer chromatography (hpTLC)
HpTLC was carried out as described in [19, 20]. To perform semiquantitative analysis of serum lipid composition, 2 μl of serum were directly spotted onto the chromatography plates. The lipid standard was a kind gift from T.R. Appel.
Data analysis strategy
In this study, a hierarchical classification system of independent artificial neural networks (ANN) was used. The so-called toplevel ANN was designed to discriminate between spectra from animals in the terminal stage of scrapie infection and those from preclinical scrapie-infected and scrapie-negative control animals. Spectra identified by the toplevel ANN as preclinical or controls were then further subclassified by a second classifier, the sublevel ANN (see Fig. 1).
Schematic diagram of the hierarchical classifier developed for the identification of control hamsters, animals in the preclinical and those in the terminal stages of scrapie. A so-called toplevel artificial neural network (ANN) was specifically designed to identify IR serum spectra from terminally ill and preclinical or control hamsters. The discrimination between scrapie-negative controls and hamsters in the preclinical stage of scrapie is subsequently achieved by a second classifier, the sublevel ANN. Note that the top- and the sublevel ANNs are optimized for specific tasks and differ in terms of network architecture and data preprocessing
Data pretreatment
First, the raw spectral data were tested for an adequate signal-to-noise ratio (SNR), intensity thresholds and the presence of water vapor bands [21, 22]. All spectra that passed the tests for spectral quality were converted to second derivatives by applying a Savitzky–Golay algorithm with nine smoothing points. Subsequently, derivate spectra were vector-normalized utilizing the spectral range of 2820–2985 cm−1. In order to reduce the computational requirements, the effective point spacing was reduced by averaging three adjacent data points, resulting in 1206 new data points out of 3620.
The calculation of average and standard deviation spectra and univariate statistical tests (two sample t-tests of independent samples) were carried out by MATLAB-based software developed in-house (The MathWorks, Natick, MA, USA).
Spectral feature selection
The selection of appropriate spectral features is an essential step in the process of classification model development. Feature selection effectively diminishes redundancy of spectral information and thus reduces the complexity and dimensionality of the classification problem. In the present study, spectral feature selection was based on the covariances of the intensity values of preprocessed second-derivative spectra. Automatic and nonsubjective selection of the most discriminative IR features was carried out using spectral information contained in the following wavenumber regions: 700–1500 cm−1, 1690–1770 cm−1 and 2800–3400 cm−1 for toplevel classification, and 700–1500 cm−1, 1680–1700 cm−1 and 2800–3400 cm−1 for the sublevel classification problem.
Based on the covariance measure, a ranking of spectral features was established, and the best 85 (toplevel) or 75 (sublevel) spectral features were selected as input data for the ANNs.
Data analysis by ANNs
For network teaching and validation, infrared spectral data were split into subsets for teaching, internal and external validation. Spectra from the three independent measurements of the individual serum samples always appeared in only one of these three data sets. Table 2 gives an overview of the numbers of serum samples used to teach and validate the toplevel (second row) or the sublevel ANN (third row).
ANN analysis was carried out by utilizing Synthon’s NeuroDeveloper software v2.5 [23] (Synthon GmbH, Heidelberg, Germany), running under the Windows 2000 operating system. For toplevel classification, a three layer feedforward network with an 85-5-2 architecture (85 input, five hidden and two output units) was employed. At the sublevel, a 75-6-2 feedforward ANN was established. For both top- and sublevel ANNs, resilient backpropagation (Rprop) was used as the learning function [24].
A serum sample of an individual hamster was diagnosed as positive or negative when all or at least two of the corresponding spectra were classified as positive or as negative, respectively.
Results and discussion
To answer the question about the earliest time point at which a scrapie infection can be identified, we have systematically analyzed serum spectra from orally infected animals at 70, 100 and 130 dpi and compared these with the respective IR spectra from mock-infected and age-matched control hamsters. FT–IR serum analysis of the 70 dpi animals did not reveal any significant changes that would allow scrapie diagnosis as early as 70 dpi. On the other hand, we found strong indications for the existence of subtle, disease-related structural and compositional changes in the serum spectra of the 100 and 130 dpi scrapie-positive animals. These alterations differed quantitatively and qualitatively from those found for the animals in the terminal stages of scrapie. Most significantly, the amplitudes of the discriminative spectral features of the terminally infected individuals exceeded the amplitudes of features for animals in preclinical disease stages. Furthermore, a detailed analysis of scrapie-related spectral features yielded a high degree of similarity for the preclinical stages at 100 and 130 dpi. These features also differed qualitatively from features found for the terminal disease stage. The latter experimental findings, and the need to teach and validate classifiers with a statistically relevant number of samples per class, prompted us to define the following classes of sample spectra (see also Table 1):
-
i.
The class “control” of IR spectra obtained from serum samples of mock-infected control animals. As Table 1 shows, this class also contains spectra from uninfected individuals and from 50 control hamsters deprived of food.
-
ii.
The class “preclinical” scrapie contains spectra from p.o. infected animals at 100 and 130 dpi (spectra from the p.o. scrapie-infected animals at 70 dpi were not used for ANN teaching).
-
iii.
Spectra from the class “terminal” scrapie originated from the i.p. and p.o.-infected hamsters in the terminal stages of the disease (last two lines of Table 1).
Design of the ANN classifier
Classification by hierarchically organized classifiers offers a number of advantages. First, a complex multiclass classification problem is considerably simplified by converting it into separate two-class classification tasks [25]. Since each of the subclassifiers can be independently adapted to a specific classification problem, such classifiers frequently provide improved accuracy of prediction, particularly in cases with high class numbers [26]. Furthermore, this approach also offers enhanced flexibility in terms of extension with additional classes [27]. The most important advantage of modular-organized classifiers however is that this methodology permits to specifically optimized combinations of spectral features to be employed for each separate classification problem.
In the present study we have therefore utilized a hierarchically organized artificial neural network (ANN) classifier for the identification of control hamsters (i) and animals in the preclinical (ii) and in the terminal stages of a scrapie infection (iii). The upper level or toplevel ANN was specifically designed to identify IR spectra from terminally ill individuals and those of the remaining individuals, i.e., from preclinical or control hamsters. Identificaton of spectra from scrapie-negative controls and hamsters in the preclinical stages of infection was then achieved by a second classifier, the sublevel network (see Fig. 1).
ANN classification takes advantage of a high number of variables and free parameters. Since the number of teaching examples (spectra) is limited, this could result in ANN models with high complexity, which may be based on randomness in the data (overfitting). One of the strategies used to recognize overfitting is to challenge the classifier with independent test data, i.e., external validation spectra, which must not be used for network teaching and internal validation. For this reason, we created spectral subsets of data for teaching, internal and external validation. The process of model development for both the toplevel and the sublevel ANNs was carried out exclusively using the teaching and internal validation subsets. The prediction accuracy of the classifiers was then determined by challenging the classifiers with external validation data.
Toplevel classification
The results from toplevel classification i.e., the discrimination between serum spectra (external data set only) from terminally scrapie-infected animals and preclinical or control hamsters are summarized in Table 3. According to this table, 36 out of 37 terminally scrapie-infected animals gave positive test results. Furthermore, 380 out of 389 preclinical or control animals were correctly identified. Hence, the true positive rate (sensitivity) of toplevel classification was 97.7% and the true negative rate (specificity) was 97.3%. These classification results are comparable to earlier observations made on serum spectra of terminally scrapie-infected and control hamsters (sensitivity 97.5%, specificity: 100%, cf. [14]. Obviously, the additional presence of spectra from preclinical scrapie-positive animals and individuals with deprived nutrition status had no measurable impact on the classification results.
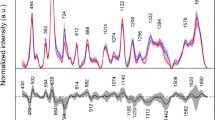
Mean absorbance and mean second-derivative spectra for the classes “control” plus “preclinical” scrapie and “terminal” scrapie are given in Fig. 2 (traces 1–4). Traces 2 and 4 were obtained from individual second derivative spectra which were vector-normalized over the complete spectral range. These mean spectra exhibit a very high degree of similarity, making it very difficult to identify the discriminative spectral information on which the toplevel classifier relies. In order to visualize these differences, Fig. 2 also displays the standard deviations of traces 2 and 4 (traces 5 and 6), as well as the difference spectrum between traces 4 and 2 (trace 7). Note that traces 5–7 are magnified by a factor of ten. The standard deviation spectra (which are also very similar) and the difference spectrum of trace 7 demonstrate that the difference between the class mean spectra is at many wavenumbers smaller than the intraclass standard deviation.
Discrimination between serum spectra of the classes “preclinical” scrapie or “control” (scrapie-negative) and the class “terminal” scrapie. Trace 1: mean infrared absorbance spectrum obtained from the combined classes of “preclinical” scrapie and scrapie-negative controls. Trace 2: mean second-derivative spectrum for the classes “preclinical” scrapie and the scrapie-negative controls (vector-normalized). Trace 3: mean absorbance spectrum for the class “terminal scrapie.” Trace 4: mean second-derivative spectrum for the class “terminal scrapie” (vector-normalized). Traces 5/6: standard deviations of traces 2 or 4, respectively (magnified by a factor of ten). Trace 7: difference between the vector-normalized second-derivative spectra (7=4−2, same scaling as traces 5 and 6). Trace 8: p-values obtained by independent t-tests using the values from vector-normalized second derivatives as inputs. Trace 9: spectral features in gray-marked regions of the IR spectra selected as input data for the toplevel ANN (see text for details)
We have furthermore performed t-tests to determine the statistical significance of distinct class means at each wavenumber. These t-tests were carried out on the basis of the intensities of vector-normalized second-derivative spectra. Trace 8 of Fig. 2 shows the p-values of the tests at each wavenumber on a logarithmic scale. Smaller p-values cast doubt on the null hypothesis that the class means are identical. Although the p-values are univariate criteria, which were not tested for statistical independence, it is likely that spectral regions with particularly small p-values denote real compositional differences in the sera. Interestingly, the smallest p-values are observed in the regions of 1730–1765 cm−1 (ν(C=O) ester) and 2800–3050 cm−1 (ν(C–H)). IR bands in the C–H stretching region can be primarily attributed to ν sy/as (>CH2) and ν sy/as (–CH3) vibrational modes of lipids. Vibrations observed at ∼1740 cm−1 indicate the presence of >C=O (ester) groups (e.g., of ester-bound fatty acids of triacylglycerols and phospholipids).
Trace 9 of Fig. 2 shows the infrared spectral features used as input data for the toplevel ANN. The heights of the bars indicate the relative importance of the individual toplevel feature obtained by the COVAR function of the NeuroDeveloper software [28]. Notice that the COVAR function is based on a multivariate approach (multiple covariance analysis) and a partial F-value. The aim of this function is to find and remove highly intercorrelated (redundant) spectral features. The results from feature extraction by the COVAR function may therefore substantially differ from those identified by univariate feature extraction criteria such as the p-values of the t-tests. Another important reason for varying spectral features lies in the fact that feature selection by the COVAR function is principally carried out by analyzing spectra from the teaching data subset only. Note that the t-test was performed on the complete data set. Gray boxes denote the spectral windows in which the COVAR feature extraction function was allowed to search.
Sublevel classification
Results from the sublevel classification are given in Table 4. The rate of true positive preclinical animals (sensitivity) in the external validation data subset was 27/31 (four false-negative classifications, sensitivity 87.1%). The corresponding true negative rate (specificity) was 93.7% (269 true negatives, 18 false positives). The weighted accuracy of prediction was 93.1%.
It was interesting to see that the false positive rate obtained separately for non-mock-infected animals (i.e., control hamsters not fed with normal brain homogenate) differed only slightly from the overall specificity. The specificity found for non-mock-infected control hamsters was 95.6% (three false positives and 65 true negatives), which is insignificantly higher than the corresponding value for the complete external validation subset (93.7%) of sublevel classification. This finding suggests that the supply of brain material does not induce nonspecific serum alterations on which scrapie identification could be based.
The classification results from the sublevel classifier demonstrate for the first time that scrapie markers can also be identified in the sera of hamsters in the preclinical stage of the disease. Since the identification of the molecular markers, or spectral features, on which the sublevel classification is based may provide new insights into the pathology of scrapie, we will focus on this particular issue in the following.
FT–IR spectroscopy of hamster sera from the classes “preclinical” scrapie and “control” produced spectra which are highly similar. This can be easily seen from traces 1–4 of Fig. 3, which show class mean absorbance spectra (traces 1 and 3) and mean spectra of vector-normalized second derivatives (traces 2 and 4). Furthermore, analogous to the example in Fig. 2, the standard deviations of traces 2 and 4 (second derivatives) and the difference spectrum for the class mean second derivative spectra are given (traces 5–7 of Fig. 3). These spectra exhibit, for most of the wavenumbers, low interclass and high intraclass variances. Additionally, a comparative analysis of the difference spectra in Fig. 2 (toplevel) and Fig. 3 (sublevel) revealed a number of surprising observations. First of all, at the top level, the C–H stretching region contributes significantly to the interclass variance. At the sublevel, however, we found exactly the opposite situation: the difference between the second derivative class mean spectra (“control” and “preclinical”) is much less pronounced. The latter statement also holds true for the region of 1730–1765 cm−1. It was found that the amplitudes in the >C=O (ester) stretching region of trace 7 in Fig. 2 (toplevel) exceed those in Fig. 3 (sublevel). We have not tested the statistical independence of the C–H and the C=O (ester) stretching regions, but compositional changes in serum lipids are likely to play a role at the terminal stage of scrapie, but not at preclinical stages. This conclusion is also supported by a comparison of the p-value plots of Figs. 2 and 3 (cf. trace 8 for each figure, respectively). Again, univariate testing by the t-test gave very low p-values at the toplevel in the “lipid” regions of 1730–1765 cm−1 and 2800–3050 cm−1. The respective data for preclinical serum samples indicated less pronounced differences for these spectral regions.
Discrimination between IR serum spectra of the classes “preclinical” scrapie and “control” (scrapie-negative) by the sublevel ANN. Trace 1: mean infrared absorbance spectrum obtained from individual IR spectra for the class “control.” Trace 2: mean second-derivative spectrum for the class scrapie-negative “controls” (vector-normalized). Trace 3: mean absorbance spectrum for the class “preclinical” scrapie. Trace 4: mean second-derivative spectrum for the class “preclinical” scrapie (vector-normalized). Traces 5/6: standard deviations of traces 2 or 4, respectively (magnified by a factor of ten). Trace 7: difference in vector-normalized second derivative spectra (7=4−2, same scaling as traces 5 and 6). Trace 8: p-values obtained by independent t-tests using the values from vector-normalized second derivatives as inputs. Trace 9: spectral features in gray-marked regions of the IR spectra selected as input data for the sublevel ANN (see text for details)
An overview of the ten most discriminative infrared spectral features for the top- and sublevel classification problems is given in Table 5. The data in this table were derived from the p-value vs. wavenumber curves given in Figs. 2 and 3 (trace 8 for each figure, respectively), and these show once again that the spectral information from the 1730–1765 cm−1 and the 2800–3050 cm−1 regions is discriminative at the toplevel, but not for the identification of preclinical disease stages.
Classification of scrapie-infected hamsters at 70 dpi
We stated earlier that spectra obtained from scrapie-infected hamsters 70 days after infection and spectra from age-matched controls cannot be reliably differentiated. On the other hand, it could not be excluded that IR patterns from 70 dpi scrapie-infected individuals represent an intermediate state between patterns from the control and the 100 and 130 dpi scrapie hamsters. For this reason, we have excluded all IR spectra from scrapie-infected hamsters at 70 dpi from the teaching subsets for both the top- and the sublevel ANN. On the other hand, the 70 dpi spectra of mock-infected animals were found to be suitable—as any control sample—for network teaching.
The results from toplevel classification of the 70 dpi data (24 mock-infected and 25 scrapie-infected animals) yielded a classification accuracy of 91.8%. This is below the general toplevel classification accuracy. Surprisingly, only one of the 70 dpi positive samples was misclassified, while the group of 70 dpi mock-infected hamsters contained three false positives. However, these differences are statistically not significant. Classification of the same data by the sublevel net yielded the following results: two of the 24 sera from mock-infected 70 dpi animals were misclassified as preclinical (specificity of 91.6%). From 25 scrapie-infected animals, 22 were classified as controls and only three of them as belonging to the class “preclinical” scrapie (sensitivity of 12%). These numbers support our findings, suggesting that a scrapie infection cannot be identified by the sublevel net as early as 70 days post infection.
High-performance thin-layer chromatography (hpTLC)
Spectral data from animals in the terminal stage of scrapie suggested reproducible compositional alterations of serum lipid constituents. However, the molecular identity of the particular lipid species cannot be revealed by FT–IR spectroscopy alone. The lipid composition of selected sera was therefore investigated by means of a specifically adapted high-performance thin-layer chromatography (hpTLC) method. The hpTLC technique enables separation and semiquantitative examination of natural lipid classes in serum, such as phospholipids, cholesterol, triglycerides and others [19, 20]. Using the hpTLC setup we have analyzed sera from five randomly selected p.o.-infected animals in the terminal disease stage and five mock-infected control hamsters. The results from the hpTLC experiment are displayed in Fig. 4. This chromatogram of serum lipids clearly shows that spots arising from triglycerides (TG) are more intense at even-numbered chromatography lanes (controls) and less intense at odd-numbered lanes (terminal scrapie). Furthermore, the relative abundance of cholesterol (CH) also seems to be associated with the disease status.
High-performance thin layer chromatography (hpTLC) of sera from hamsters orally infected with the scrapie agent 263K (terminal stage) or mock-infected controls. Spots of the individual serum lipids can be identified by comparison with a lipid standard. Lipid abbreviations: SQ, squalene; CL, cholesterol ester; TG, triacylglycerides; FA, mono fatty acid; CH, cholesterol; MG, monoacylglyceride; GC, galactocerebroside; SU, sulfatides; PG, phosphatidylglycerol; CA, cardiolipin; PI, phosphatidylinositol; PC, phosphatidylcholin; SM, sphingomyelin; LY, lysophospholipid (lyso-PC); GA, gangliosides
It cannot be excluded that the experimental findings of modified serum lipid patterns in clinically ill animals could be partly based on general metabolic processes in the very late stages of a scrapie infection. For example, it was observed that neurological and behavioral symptoms of an advanced 263K scrapie infection, such as head bobbing, generalized tremor and ataxia of gait, affect the ability to drink and to ingest food. Since the metabolism of triglycerides and other lipids is known to be associated with the actual nutrition status, animals in the terminal stages of scrapie may exhibit decreased lipid concentrations in serum. As a consequence, the specificity of some of the IR features in the lipid and also of the ester carbonyl region might be uncertain. A popular strategy for establishing classification systems with higher specificity is to modify the composition of the teaching subset such that the confounding variables are numerically equally represented in all predefined classes.
For this reason, the class “control” also comprised spectra from individuals exhibiting a catabolic type of metabolism (see Table 1). The catabolic serum patterns were obtained by depriving nutrition to healthy control hamsters for 48 h. Thus, the confounding variable “catabolic type of FT-IR serum pattern” was present in both classes of the toplevel ANN, i.e., in the classes “terminal” scrapie and “preclinical” or “control” animals. Interestingly, hpTLC investigations of sera from hamsters with deprived nutrition revealed lipid profiles that were similar to hamsters showing symptoms of a terminal scrapie infection (not shown).
Provided the toplevel classification was primarily based on nonspecific serum markers, one would expect misclassifications, particularly of control animals with an impaired nutrition status. The results from independent external validation given in Table 3 demonstrate, however, a high accuracy of prediction for catabolic control animals too, indicating that the discrimination of clinically ill individuals and animals showing no clinical signs of infection was undeniably not based on general changes in the metabolism. Moreover, this conclusion is backed up by the observation that only a surprisingly small number of toplevel ANN features found by the statistical t-test in the 1730–1765 cm−1 and the 2800–3050 cm−1 regions were confirmed by the COVAR function as reliable markers for toplevel identification (cf. traces 8 and 9 of Fig. 2). Obviously, nonspecific infrared spectral markers of a catabolic type of metabolism could be successfully eliminated from the set of features for ANN toplevel classification by extending the class of control spectra and applying advanced methods of feature selection.
Spectral features of the class “preclinical” scrapie
From a spectroscopic point of view it is interesting to note that two of the four most important sublevel ANN features are found in the amide I region (1620–1690 cm−1). The amide I band arises primarily from C=O stretching vibrations of the peptide backbone amide groups. Detailed analyses of this band have been the basis for countless IR studies examining secondary structures of proteins, since peptide backbone and side-chain infrared marker bands can be employed as conformation-sensitive indicators in order to derive structural parameters from protein spectra. For example, IR spectroscopy is particularly sensitive to β-pleated sheet structures. These secondary structural elements exhibit a strong amide I band component at ~1630 and a weaker band at 1685 cm−1. In addition, it is known that the causative agent of scrapie infection, the abnormally folded prion protein PrPsc, exhibits a number of remarkable structural differences compared with its normal cellular isoform PrPc. While PrPc comprises 42% α-helical structures and only 3% β-sheets [29–31], the content of β-sheets in the oligomeric PrPsc isoform is considerably increased. These findings notwithstanding, we do not believe that the discriminatory spectral features observed at 1630 cm−1 are due to a possible presence of PrPsc in the sera of preclinical hamsters. The analytical sensitivity of IR spectroscopy in the experimental set-up of whole serum analysis is doubtless too low to specifically detect subfemtomolar concentrations of PrPsc against a background of highly abundant proteins such as albumin, immunoglobulins and many others. It seems more plausible that compositional alterations of other as-yet unidentified serum proteins are responsible for the dissimilarities obtained in the sera of preclinical and control animals.
Interestingly, other important sublevel features are also attributable to functional groups characteristic of proteins or polypeptides. For example, the sublevel feature 3 at 3357.9 cm−1 can be assigned to the amide A band (N–H stretching vibration, amide linkage, see Table 5). Thus, the experimental findings from this study suggest that the composition of serum proteins, or polypeptides, is characteristically altered in clinically asymptomatic (preclinical) hamsters. The identification of these discriminatory molecular markers is essential for further improvements in the methodology in terms of classifier performance and will hopefully add also new insights to the pathology of TSEs. To address this problem, it is, however, necessary to improve the analytical sensitivity of the detection technique. We believe that a combination of separation techniques, such as HPLC in combination with a molecule-specific analytical technique such as mass spectroscopy, will be helpful in identifying these TSE-specific markers in serum.
Conclusions
In this study we have presented a methodology for diagnosing preclinical scrapie infections in hamsters from serum by a combination of Fourier-transform infrared (FT–IR) spectroscopy and chemometrics. The analysis of hamster sera by FT–IR spectroscopy and artificial neural networks (ANN) confirmed the results from previous studies which had demonstrated the presence of disease-related structural and compositional alterations in the sera of infected donors for the terminal stage of scrapie.
Furthermore, experimental data from sera from animals in asymptomatic (preclinical) stages of scrapie revealed subtle but reproducible spectral changes and permitted the identification of a preclinical scrapie infection at 100 dpi or later, but not at 70 dpi. The analysis of these IR spectroscopic differences provided experimental evidence for scrapie-associated compositional alterations of serum proteins. However, it appears to be highly unlikely that the spectral changes are attributable to the causative agent of a scrapie infection, the prion protein PrPsc. The identification of serum markers or spectral features on which the identification of preclinical animals is based is a key to obtaining new insights into the pathology of preclinical TSEs, and also to further improvements in the methodology in terms of accuracy of prediction.
Abbreviations
- ANN:
-
artificial neural networks
- BSE:
-
bovine spongiform encephalopathy
- CJD:
-
Creutzfeldt–Jakob Disease
- dpi:
-
days post infection
- FT-IR:
-
Fourier-transform infrared
- hpTLC:
-
high-performance thin-layer chromatography
- i.p.:
-
intraperitoneally
- p.o.:
-
orally
- PrP:
-
prion protein
- SNR:
-
signal-to-noise-ratio
- TSE:
-
transmissible spongiform encephalopathy
- vCJD:
-
variant Creutzfeldt–Jakob Disease
References
Prusiner SB (1998) Proc Natl Acad Sci USA 95:13363–13583
Collinge J, Whitfield J, McKintosh E, Beck J, Mead S et al (2006) Lancet 367(9528):2068–2074
Hunter N, Foster J, Chong A, McCutcheon S, Parnham D et al (2002) J Gen Virol 83(11):2897–2905
Cervenakova L, Brown P (2004) Expert Rev Anti Infect Ther 2(6):873–880
Parveen I, Moorby J, Allison G, Jackman R (2005) Vet Res 36(5–6):665–683
Trieschmann L, Navarrete Santos A, Kaschig K, Torkler S, Maas E et al (2005) BMC Biotechnol 4(5):26
Castilla J, Saa P, Soto C (2005) Nat Med 11(9):982–985
Yang WC, Yeung ES, Schmerr MJ (2005) Electrophoresis 26(9):1751–1759
Safar JG, Geschwind MD, Deering C, Didorenko S, Sattavat M et al (2005) Proc Natl Acad Sci USA 102(9):3501–3506
Otto M, Wiltfang J, Schütz E (1998) Br Med J 316:577–582
Maissen M, Roeckl C, Glatzel M, Goldmann W, Aguzzi A (2001) Lancet 357(9273):2026–2028
Schütz E, Urnovitz HB, Iakoubov L, Schulz-Schaeffer W, Wemheuer W et al (2005) Clin Diagn Lab Immunol 12(7):814–820
Lekishvili T, Sassoon J, Thompsett AR, Green A, Ironside JW, Brown DR (2004) Exp Neurol 190(1):233–244
Schmitt J, Beekes M, Brauer A, Udelhoven T, Lasch P, Naumann D (2002) Anal Chem 74:3865–3868
Lasch P, Schmitt J, Beekes M, Udelhoven T, Eiden M et al (2003) Anal Chem 75(23):6673–6678
Martin TC, Moecks J, Belooussov A, Cawthraw S, Dolenko B et al (2004) Analyst 129(10):897–901
Baldauf E, Beekes M, Diringer H (1997) J Gen Virol 78(5):1187–1197
Otto M, Beekes M, Wiltfang J, Bahn E, Poser S, Diringer H (1998) J Neurovirol 4(5):572–573
Klein TR, Kirsch D, Kaufmann R, Riesner D (1998) Biol Chem 379:655–666
Gellermann GP, Appel TR, Tannert A, Radestock A, Hortschansky P (2005) Proc Natl Acad Sci USA 102(18):6297–6302
Fabian H, Lasch P, Naumann D (2005) J Biomed Opt 10(3):031103
Naumann D, Lasch P, Fabian H (2006) Proc SPIE 6093:1–12
Synthon (2006) Company website. Synthon GmbH, Heidelberg, Germany (see http://www.synthon-analytics.de, last accessed 7th September 2006)
Riedmiller M, Braun HA (1993) ICNN-93, IEEE Intl Conf on Neural Networks, 28 March–1 April 1993, San Francisco, CA, pp 586–591
Marques de Sa JP (2001) Pattern recognition: concepts, methods and applications. Springer, Berlin Heidelberg New York
Gasser A, Kamel M (1998) J Intell Robot Syst 21:117–129
Udelhoven T, Naumann D, Schmitt J (2000) Appl Spectrosc 54(10):1471–1479
Schmitt J, Udelhoven T (2000) In: Gremlich HU, Yan B (eds) Infrared and Raman spectroscopy of biological materials. Marcel Dekker, New York
Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS (1991) Biochemistry 30(31):7672–7680
Gasset M, Baldwin MA, Lloyd DH, Gabriel JM, Holtzman DM et al (1992) Proc Natl Acad Sci USA 89(22):10940–10944
Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A et al (1993) Proc Natl Acad Sci USA 9d0(23):10962–10966
Acknowledgements
We thank Ramona Famulla, Stefanie Wolgast, Angelika Brauer (P25, Robert Koch-Institut) and Marion Joncic (P24, Robert Koch-Institut) for their excellent technical assistance. Furthermore we are grateful to Michael Eiden (Synthon GmbH, Heidelberg) and Elizabeth Baldauf for fruitful discussions and support. Thomas R. Appel (Fritz-Lipmann-Institut, Jena) is acknowledged for his help in performing thin-layer chromatography of the serum lipid constituents. This work was supported by the Bundesministerium für Bildung und Forschung (BMBF grant 0312727).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lasch, P., Beekes, M., Schmitt, J. et al. Detection of preclinical scrapie from serum by infrared spectroscopy and chemometrics. Anal Bioanal Chem 387, 1791–1800 (2007). https://doi.org/10.1007/s00216-006-0764-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-006-0764-z