Abstract
A new analytical method is proposed to determine more than 40 multiclass pesticides in different kinds of processed (whole, skimmed and powdered) and unprocessed (goat and human) milk samples using solid-phase microextraction (SPME). A comparative study between headspace (HS) and direct immersion (DI) was carried out. The effect of milk dilution and the use of acid to reduce the influence of the matrix in DI-SPME mode were also evaluated. DI of the SPME fiber into previously diluted and acidified milk samples achieved the best sensitivity results. Pesticides were determined using low-pressure gas chromatography-tandem mass spectrometry (LP-GC-MS/MS). Both of the selected techniques have been shown to be effective at reduce fat interference and can determine analytes present at very low concentrations (limits of quantification between 0.02 and 1.00 μg L−1). Performance characteristics such as linearity, recovery, precision, and lower limits, together with an estimation of the measurement uncertainty using validation data, are presented for each pesticide. All of the pesticides presented recovery rates of between 81 and 110% and precision values lower than 12% (expressed as the relative standard deviation). The overall uncertainty of the method was estimated at three different concentrations (10, 25 and 50 μg L−1) and was lower than 25.5% in all cases. The proposed analytical methodology was applied to the analysis of target pesticides in 35 samples: 15 commercial, 3 human and 17 goat milk samples. The metabolite p,p′-DDE was the compound most frequently found in both the breast and goat milk samples, at concentration levels <20 μg L−1. However, pesticide residues were not found in any of the other 15 commercial milk samples (skimmed, powdered and whole milk) analyzed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Due to its lipophilic nature, milk has been used to monitor the bioconcentration of environmentally persistent organic pollutants, such as organochlorinate pesticides. These compounds are commonly found in food products of animal origin like milk [1,2], mainly due to their use in the past but also due to the ongoing use of some of them in agricultural activities. Other pesticides considered to be less persistent or stable, like organophosphorus compounds, have also been found in different types of milk [3, 4], such as processed animal milk or unprocessed human milk. The reason for the contamination can vary, and may include [5]: (i) use of the compounds directly on the animal against disease vectors; (ii) hygienic treatments against insects in milk processing factories, and (iii) consumption of either foodstuffs containing pesticides or animal feed containing post-harvest plant materials treated during the growing season with pesticides.
The amount of pesticide in the milk depends on factors such as the stability of the pesticide, its metabolic fate in animals or humans, its mode of application or intake, and so on. Because pesticides are hazardous organic compounds, their determination in foods like milk is important to human health. Several organizations like the European Union [6] have therefore set maximum residue limits for parent pesticides.
The determination of pesticide residues in milk typically presents problems because fat is extracted along with the analytes of interest. Clean-up steps are often needed to eliminate interfering compounds from the matrix, which increases the amount of highly pure solvents used, the number of manual operations required, the uncertainty in the determination, the cost of the method and can result in loss of the analyte. Several methods have been proposed for the analysis of pesticides in milk, such as extraction with non-polar solvents [7], solid-matrix dispersion [5, 8], normal-phase liquid chromatography with column switching [9], liquid-liquid microextraction [10] and solid-phase microextraction (SPME) [1]. This last technique has proven its ability to analyze trace amounts of pesticides in various fluid samples like water [11], urine [12] or blood [13]. It simplifies the process of sample treatment before quantitative determination by gas or liquid chromatography, and avoids the use of organic solvents.
Pawliszyn and co-worker invented SPME in late 1989 [14, 15], and since then it has been proposed for use in environmental, botanical, clinical and forensic applications among others because it integrates sampling, extraction and concentration into a single step that does not need organic solvent use. SPME extraction is based on a multiphase equilibrium of the analytes between the sample, the gas above the sample and the coating of the SPME fiber. Once equilibrium is achieved, the amount extracted can be desorbed, either thermally into the hot GC injector or, in the case of liquid chromatography, eluted by the mobile phase. The theory of SPME has been extensively discussed by Pawliszyn [16].
After extraction, gas or liquid chromatographic determination can be carried out. Recently, low-pressure gas chromatography (LP-GC) has been proposed as an alternative to conventional capillary GC for faster chromatography [17, 18]. It can be successfully applied to the analysis of pesticide residues when mass spectrometry (MS) is used as the detection system. The reduction of analysis time using LP-GC columns is achieved through optimizing the velocity of helium under low-pressure conditions. Some characteristics of LP-GC are: faster analysis, 50–80 °C diminution in the elution temperature (reducing the thermal degradation of thermally labile compounds and the bleed of the analytical column), compatibility with standard injection techniques, high capacity, and finally, a wide range of film thicknesses and types are available.
In this work, we demonstrated the effectiveness of the on-line combination of SPME and LP-GC techniques at reducing fat interferences in the analysis of pesticides and metabolites in milks. This methodology has been fully validated (including estimating the uncertainty of the method) using in-house validation data [19]. Finally, the methodology was applied to evaluate the pesticide residue levels in commercial (powder and pasteurized) samples, as well as in both human breast and goat milk samples. The humans and goats were from an agricultural area (Almeria, Spain). The target pesticides studied here were chosen due to their use as pesticides in the same area.
Materials and methods
Reagents
Pesticide standards and the internal standard (IS), pentachlorobenzene, were obtained from Riedel-de-Haën (Seelze-Hannover, Germany); purities were always >99%. Pesticide-quality solvent (acetone) was supplied by Panreac (Barcelona, Spain). Stock standard solutions of individual compounds (with concentrations between 75 and 550 μg mL−1) were prepared by exact weighing and dissolution in acetone and were stored in a freezer (−30 °C). A multicompound working standard solution (2 μg mL−1 concentration of each compound) was prepared by appropriate dilution of the stock solutions with acetone, and was stored under refrigeration (4 °C). The sodium chloride used for residue analysis was obtained from Panreac. Formic acid and triethylamine (TEA) was obtained from Riedel-de-Haën.
Apparatus
GC-MS analysis was performed with a Varian 3800 gas chromatograph equipped with electronic flow control (EFC) and a Saturn 2000 ion-trap mass spectrometer (Varian Instruments, Sunnyvale, CA, USA) detector. Samples were injected using a Varian 8200 autosampler prepared for SPME analysis with agitation into an SPI/1079 split/splitless programmed-temperature injector. The autosampler carousel was an Autotherm system used as a thermostat for the SPME vials. A fused silica untreated capillary column 2 m×0.25 mm i.d. from Supelco (Bellefonte, PA, USA) was used as a guard column connected to a Rapid-MS (0.6 m×0.10 mm i.d. restriction coupled to a WCOT fused-silica CP-Sil 8 CB low bleed of 10 m×0.53 mm i.d.×0.25 μm film thickness) analytical column at the inlet end, from Varian Instruments, for high speed analysis. The mass spectrometer was operated in electron ionization mode (EI). The computer that controlled the system also held an EI-MS-MS library specially created to use with the target analytes in our experiment. Other EI-MS libraries were also available. The mass spectrometer was calibrated weekly with perfluorotributylamine. Helium (99.999%) at a flow rate of 1 mL min−1 was used as carrier and collision gas.
The syringe injector of the SPME unit (Supelco, Bellefonte, USA), equipped with 100 μm polydimethylsiloxane (PDMS) and 65 μm polydimethylsiloxane-divinylbenzene (PDMS-DVB) fibers (Supelco), was used for the extraction procedures. Fibers were conditioned before first use according to the supplier’s instructions.
Sample pretreatment
The human and goat milk samples were taken using a manual pump. They were collected in glass containers, sterilized and rapidly frozen at −20 °C. All of the milk samples were thoroughly mixed after thawing for spiking and analyzing procedures.
The powdered milk samples were dissolved in the amount of water (Milli-Q) recommended by the manufacturer and processed like liquid samples. Pesticide-free pasteurized milk was used as a blank for recovery studies and for preparing matrix-matched standards for calibration.
1 mL aliquots of milk were mixed with 500 μL of formic acid, 10 μL of TEA and 50 μL of 2 mg L−1 pentachlorbenzene in acetone solution. The mixture was centrifuged for 10 min at 3000 r.p.m. 1 mL of the superior layer was transferred to a test tube, where 2490 μL of milli-Q water was added and mixed. The sample was then centrifuged again for 2 min (3000 r.p.m.) and 1 mL of the superior layer was transferred to a GC vial for SPME extraction.
SPME conditions
Adsorption of the studied compounds in the fiber was achieved by placing them at 25 °C in a thermostated carousel for 55 min with agitation in order to reach the equilibrium. After that, the fiber was automatically introduced into the injection port of the GC, which was set at 250 °C (5 min) and then increased to 300 °C at 100 °C min−1, held for 10 min, in order to thermally desorb and transfer the analytes to the chromatographic column. The injector split ratio was initially 20:1. At 0.01 min the splitless mode was switched on and this was maintained until 9 minutes had elapsed. At 9.00 min the split ratio was 100:1 and at 15.00 min the split ratio was 50:1.
LP-GC-MS/MS conditions
The initial column temperature was set to 70 °C during injection (held for 9 min) and then increased at 50 °C min−1 to 95 °C, then at 5 °C min−1 to 165 °C, then at 20 °C min−1 to 190 °C, and then finally raised to 300 °C at 100 °C min−1, which was held for 5 min.
The ion-trap mass spectrometer was operated in EI-MS/MS. The transfer line, manifold and trap temperatures were 280, 50 and 200 °C, respectively. The Automatic Gain Control (AGC) was activated with an AGC-target of 5000 counts. The emission current for the ionization filament was set at 80 μA, generating electrons with energies of 70 eV. The axial modulation amplitude voltage was 3.0 V. The MS/MS process was carried out by Collision-Induced Dissociation (CID) with non-resonant excitation for all of the compounds studied, except for hexachlorobenzene and dieldrin which were dissociated by resonant excitation. The electron multiplier voltage was 1350 V (+200 V offset above the auto-tuning process). The scan rate and scanned mass range chosen depended on the number and type of compounds analyzed simultaneously. The specific MS/MS parameters used are shown in Table 1.
Results and discussion
Optimization of the chromatographic separation and MS/MS parameters
A LP-GC column was used to separate the compounds of the sample in a reduced time. Compared with conventional capillary columns, LP-GC is fast, it presents low elution temperatures and therefore it elutes higher boiling materials well and elutes thermally labile compounds at lower temperatures (lower by 50–80 °C), and it minimizes the bleed of the analytical column due to the lower elution temperature and higher sensitivity. The sample capacity of the column is higher than that of a narrow capillary analytical column. It also reduces potential milk interferences (from fats and proteins). Separation of the target pesticides was performed by testing several temperature programs and using electronic flow control for the carrier gas. After optimization, all of the compounds were eluted in a reasonably short time of 26 min.
The isolation, fragmentation and storage conditions of the ions obtained (the product ions) were optimized for each compound. In a first step, the precursor ion or an entire cluster of parent ions is isolated in the trap, and in a second stage the precursor ion or ions are dissociated by collisional activation, using helium as carrier gas. In general, the parent ion isolated was selected by finding the best compromise between both selectivity (the highest m/z ion) and sensitivity (the highest intensity ion). After this choice was made, it was necessary to set the excitation storage level before optimizing the CID step. The final values used in this study are shown in Table 1. Once the MS/MS conditions were optimized, the quantification ions were selected. The MS/MS spectra obtained under the final experimental conditions were stored in a MS/MS library created in-house.
Optimization of the extraction process
Headspace (HS) and direct immersion (DI) SPME techniques were evaluated for analyzing the target pesticides in milk. HS-SPME is generally used to analyze volatile compounds and DI-SPME for semivolatile compounds. The analysis of trace levels of pesticides by SPME in complex liquid samples like milk can be carried out using either technique, after solving certain limitations. On the one hand, HS-SPME avoids interferences from native compounds that milks contain (especially fats and proteins) that would be adsorbed on the fiber in DI-SPME mode. This mode would shorten the fiber lifetime and cause interferences in the resulting chromatograms. On the other hand, DI-SPME is simpler and potentially offers higher sensitivity when semivolatile compounds are SPME-sampled.
Some authors [19] have proposed the use of HS-SPME for analyzing pesticides in milk while heating samples at 100 °C. They have observed that vials have to be tightly sealed and the vial seals must be pierced with an injection needle before heating. Our experiments tested heating temperatures between 40 and 100 °C, but, even following the above mentioned recommendations, leak problems were observed at temperatures ≥70 °C. Predrilled commercially available SPME septa were also tested. However, leak problems still affected the repeatability of the test and so the heating temperature was set to 70 °C for HS-SPME experiments.
For HS-SPME, two coating fibers were evaluated: 100 μm polydimethylsiloxane, (PDMS) and 65 μm polydimethylsiloxane-divinylbenzene (PDMS-DVB). The influence of the adsorption time was studied by analyzing the extraction profiles for each pesticide between 15 and 75 min. To this end, aliquots of 1 mL of blank milk were spiked with the target pesticides at 10 μg L−1. The adsorption time was set to 40 and 45 min for PDMS and PDMS-DVB respectively; the minimum times at which saturation was reached for most compounds. Desorption time conditions were also studied, by evaluating several injection temperatures and heating times. The desorption temperature was selected to vaporize the analytes at their optimal temperatures, avoiding possible thermal degradations. The desorption time was varied from 5 to 20 min and was finally set to 15 min for both types of fiber; by this time the desorption was complete, even for the most persistent compounds (mirex and famfur). We checked for carryover effects by realizing blank injections between each experiment.
The influence of the ion strength in the sample was evaluated by adding different amounts of NaCl (from 0.05 to 0.20 g) to 1 mL aliquots of milk. We also found that better results were obtained when 0.1 g of salt was added. Better sensitivity was obtained using PDMS-DVB than PDMS, and for that reason it was chosen for the rest of the experiments.
DI-SPME analysis was also optimized using PDMS and PDMS-DVB fibers in the same way as done for HS-SPME. Direct immersion of the fiber into the milk samples was discarded after some preliminary experiments revealed a clear matrix effect on the calibration and that the results had poor repeatability and sensitivity. This was attributed to the high content of fat and proteins in the milk. Dilution of the milk samples with Milli-Q water was considered at various dilution levels (1:2, 1:3, 1:4 and 1:5 v/v). Obviously, this action would reduce the sensitivity of the method but increase the fiber lifetime and precision. A dilution of 1:4 v/v was finally selected, which sacrificed sensitivity but increased the precision and the fiber lifetime.
Later, in order to evaluate the effect of potential binding between the protein and the target pesticides, denaturation pretreatment of the milk was applied by following a previously-described procedure [2]. The protein denaturation method used increased the extraction efficiency, except for lindane, α-endosulfan, p,p′-DDE, o,p-DDT, dieldrin, methoxichlor and mirex. Nevertheless, the sensitivity achieved for these compounds was good enough. The addition of TEA during sample application masks strongly hydrophobic or polar sites from exposed silanol groups.
All of the experiments were carried out by agitating the fiber in order to increase sensitivity and to reduce equilibrium time. After optimization, the adsorption time was set at 40 and 55 min for PDMS and PDMS-DVB fibers. Desorption conditions were identical to those selected for HS-SPME, eliminating carryover effects. The influence of the ionic strength on the adsorption process was also evaluated by adding NaCl in amounts that varied from 0.05 to 0.80 g. In this case, the best DI-SPME results were obtained without adding salt. Finally, the PDMS-DVB fiber was selected for the DI-SPME experiments because it gave better sensitivity than PDMS.
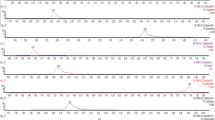
A final comparison between the HS and DI-SPME extractions was carried out by analyzing milk samples containing 1 μg L−1 of each pesticide. Figure 1 shows that a better sensitivity was generally achieved by immersing the fiber in the diluted milk than by using HS-SPME. Many of the studied compounds (organophosphorous pesticides) were not detected using HS-SPME at the tested concentration (parathion methyl, vinclozolin, malathion, fenthion, parathion ethyl, chlorfenvinphos, quinalphos, fenamiphos). The studied compounds were not volatile enough at 70 °C to compensate for lower distribution constants, and so DI-SPME adsorbed the analytes more efficiently, even when the milk samples were diluted. Using this method, a fiber could be used for at least 80–100 runs. Only a few compounds presented higher sensitivities with HS-SPME (aldrin, heptachlor and hexachlorobenzene).
Method validation
The DI-SPME-LP-GC-MS/MS method was validated in order to check its feasibility for the analysis of pesticide residues in milk samples. All validation parameters were assessed using pesticide-free milk.
Identification and confirmation of target analytes
The target pesticides were identified by searching in the appropriate retention time windows (RTWs). These were defined as the average retention time obtained when ten blank samples spiked at 20 μg L−1 with each compound were analyzed, ± three standard deviations of the retention time (Table 2). The confirmation of a compound previously identified by retention time only was done by comparing the MS/MS spectra obtained for the sample with those previously stored as reference spectra. The reference spectra were obtained and stored daily by analyzing blank milk spiked with the pesticide at the concentration of the second calibration level. The comparison results (FIT) are scaled to 1000 for the best match (identical spectra). To confirm the presence of a pesticide, the FIT value obtained must be equal or higher than the FIT threshold previously calculated for each pesticide. This FIT threshold is set by comparing the MS/MS spectra obtained from ten injections at 20 μg L−1, calculating an average FIT, and subtracting 250 units in order to compensate for small differences encountered during the analysis of real milk samples. These small differences are caused by variations in analyte concentrations in the standards and real samples and by differences in matrix compositions.
Limit of detection (LOD) and quantitation (LOQ)
The LODs and LOQs were taken to be the minimum analyte concentrations that can be detected with a response three and ten times greater than the noise level, respectively. To that end, we monitoring the quantitation ion using chromatograms. As can be seen in Table 2, LOD values ranged between 0.01–0.30 μg L−1, while the LOQ ranged from 0.02 to 1.0 μg L−1.
Linearity
The linearity of the method was studied in the range 1–50 μg L−1 by injecting spiked milk aliquots and applying the proposed SPME process. The matrix-matched calibration eliminated potential matrix effects in the analytical response. An internal standard calibration was performed by plotting the amount of analyte versus the ratio of peak area or height (analyte/IS). The results were improved slightly by using relative areas for all compounds. Table 2 summarizes the slopes, intercepts and the determination coefficient values for the calibration study. Three calibration levels and the origin (0,0) were used for calibration. The calibration curve was not forced through the origin. The calibration fit was plotted and inspected visually, avoiding reliance on correlation coefficients. The individual points did not deviate by more than ±20% from the calibration curve.
Trueness and precision
Recovery data were obtained by analyzing aliquots of an uncontaminated milk (n=10) spiked at 20 μg L−1. Mean recovery rates in the range of 81.3–110.0% (Table 3) were obtained for all of the target compounds. The precision (repeatability, expressed as the relative standard deviation, RSD) was <12% in all cases (Table 3).
Uncertainty
The overall uncertainty was estimated by considering each source of uncertainty and treating it separately to obtain the contribution from that source. It is often possible to evaluate the combined effect of several components as a global standard uncertainty [20]. Once the parameters and their associated uncertainties that contribute to the uncertainty of the method as a whole are listed, the individual uncertainties are combined in the uncertainty budget to give the relative global uncertainty Urel (Eq. 1). The expression includes the uncertainty derived from estimating the analyte concentration from the calibration curve, urel(CA), and the estimated uncertainty associated with the concentration of the diluted sample in the extract, urel(Fdil). The first term involves the uncertainty associated with transforming chromatographic signals to concentrations, preparing working standard solutions and the precision of the methodology. The other term covers the uncertainty derived from the steps followed to prepare the analyzable extract.
The expanded uncertainty, Urel, is obtained by multiplying urel by a coverage factor k, assuming a normal distribution of the measurand. k is usually 2, which provides an approximate level of confidence of 95%.
Table 3 shows the results obtained from estimating the uncertainty at three concentration levels. The uncertainty was never greater than 25.5% at the lowest calibration point for all concentration levels. The uncertainty values were very similar for all of the compounds. This can be explained if we bear in mind that standard solution preparation, the process of extraction and the rest of the analytical methodology were the same for all of them. In general, we observed that the most important contribution comes from estimating the concentration of the analyte in the sample (urel(CA)).
Application to the analysis of real samples
Once the analytical methodology was validated, it was applied to the analysis of real milk samples: commercial (powder and pasteurized), and goat and breast human samples. These analyses were performed in a laboratory certified by UNE-EN-ISO 9001:2000 for pesticide residue analysis in milks and other foods. Internal quality control procedures were routinely applied in order to check if the system was under control.
The quality control measures showed that the target compounds were properly identified; the laboratory reagent blank, laboratory matrix blank, and the laboratory spiked blank samples were analyzed at the same time as the set of samples. The laboratory reagent blank rejected any contamination from interference due to reagents during sample processing. The laboratory matrix blank extract avoided false positives due to possible contaminations of the extraction process or chemicals. We used the spiked blank to monitor the losses during the analytical procedure and to detect anomalies during the extraction step or due to instrumental causes. Sample analysis was performed if recovery rates (analyzing milk spiked with the pesticides at 1 μg L−1) were between 60% and 120%. Calibration curves were prepared daily to check both sensitivity and linearity within the working concentration range in order to avoid quantification mistakes caused by possible matrix effects or instrumental fluctuations (R2>0.99 was required).
The results showed that p,p′-DDE, metabolite of the insecticide p,p′-DDT, was found in two of the three human breast milk samples analyzed at concentration levels of 18.7 and 8.4 μg L–1, respectively (Fig. 2). Also, p,p′-DDE was found in five of the goat milk samples at levels of 1.2–11.0 μg L–1. However, pesticide residues were not found in any of the 15 analyzed commercial samples (four skimmed milk, three powdered milk and eight whole milk samples).
Conclusions
A rapid and simple mutiresidue method based on the DI-SPME technique has been developed in order to determine more than 40 pesticide residues in milk via LP-GC-MS/MS. The on-line combination of SPME and LP-GC-MS/MS gives a method with adequate sensitivity and selectivity, avoiding clean-up steps, reducing sample manipulation and saving solvents and time. To use the technique, the milk must be diluted with water and protein removed, and the SPME fiber should be immersed in the mixture. The method developed shows adequate validation parameters in terms of linearity, lower limits, recovery, precision and uncertainty.
Different kinds of milk samples were analyzed. Residues of p,p′-DDE were found in two of the three mother’s milk samples. This metabolite of p,p′-DDT was also determined in five goat milk samples at concentrations <11.0 μg L–1. However, the levels of target pesticides in commercial milk samples were lower than the detection limits of the proposed method. It is important to note that p,p′-DDE was found despite the fact that its parent compound has been totally banned due to its low biodegradability and high bioaccumulation and bioconcentration in lipid systems.
References
Röbrig L, Ulrich Meisch H (2000) Fresen J Anal Chem 366:106–111
Covaci A, Hura C, Shepens P (2001) Chromatographia 54:247–252
Bennett DA, Chung AC, Lee SM (1997) J AOAC Int 80:1065–1077
Sheridan RS, Meola JR (1999) J AOAC Int 82:982–987
Di Muccio A, Pelosi P, Camoni I, Barbini DA, Dommarco R, Generah T, Ausili A (1996) J Chromatogr A 754:497–507
EU (1993) European Union Council Directive n.93/57/CE of June 29, 1993; (1994) European Union Council Directive n.94/29/CE of June 23, 1994. EU, Brussels
Campoy C, Jiménez M, Olea-Serrano MF, Moreno Frias M, Cañabate F, Olea N, Bayés R, Molina Font JA (2001) Early Hum Dev 65:S183–S190
Di Muccio A, Pelosi P, Barbini DA, Generali T, Ausili A, Vergori F (1997) J Chromatogr A 765:51–60
René van der Hoff G, van Beuzekom AC, Th. Brinkman Udo A, Baumann RA, Van Zoonen P (1996) J Chromatogr A 754:487–496
Zhu L, Fe KH, Zhao L, Lee HK (2002) J Chromatogr A 963:335–343
Arrebola FJ, Córtes Aguado S, Sánchez Morito N, Garrido Frenich A, Martínez Vidal JL (2003) Anal Lett 37:99–117
Krämer Alcalde T, do Carmo Ruaro Peralba M, Alcaraz Zini C, Bastos Caramão E (2004) J Chromatogr A 1027:37–40
Guan F, Watanabe K, Ishii, H Seno A, Kumazawa T, Hattori H, Suzuki O (1998) J Chromatogr A 714:205–213
Belardi RP, Pawliszyn J (1989) Water Pollut Res J Can 24:179–191
Arthur CL, Pawliszyn J (1990) Anal Chem 62:2145–2148
Pawliszyn J (1997) Solid phase microextraction: theory and practice. Wiley, Chichester, UK
González Rodríguez MJ, Garrido Frenich A, Arrebola FJ, Martínez Vidal JL (2002) Rapid Commun Mass Spectrom 16:1216–1224
Arrebola FJ, Martínez Vidal JL, González Rodríguez MJ, Garrido Frenich A, Sánchez Morito N (2003) J Chromatogr A 105:131–141
Pauwels A, Wells DA, Covaci A, Schepens PJC (1999) J Chromatogr B 723:117–125
CITEC/EURACHEM (2000) Quantifying uncertainty in analytical measurement, 2nd edn. EURACHEM, Teddington, UK (download available at http://www.vtt.fi/ket/eurachem/quam2000-p1.pdf)
Acknowledgements
The authors are grateful to Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) (Project CAL03–087-C2–2) and FIAPA (Ce.-486/01) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
González-Rodríguez, M.J., Arrebola Liébanas, F.J., Garrido Frenich, A. et al. Determination of pesticides and some metabolites in different kinds of milk by solid-phase microextraction and low-pressure gas chromatography-tandem mass spectrometry. Anal Bioanal Chem 382, 164–172 (2005). https://doi.org/10.1007/s00216-005-3144-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-005-3144-1