Abstract
Infrared (IR) spectroscopic imaging coupled with microscopy has been used to investigate thin sections of cervix uteri encompassing normal tissue, precancerous structures, and squamous cell carcinoma. Methods for unsupervised distinction of tissue types based on IR spectroscopy were developed. One-hundred and twenty-two images of cervical tissue were recorded by an FTIR spectrometer with a 64×64 focal plane array detector. The 499,712 IR spectra obtained were grouped by an approach which used fuzzy C-means clustering followed by hierarchical cluster analysis. The resulting false color maps were correlated with the morphological characteristics of an adjacent section of hematoxylin and eosin-stained tissue. In the first step, cervical stroma, epithelium, inflammation, blood vessels, and mucus could be distinguished in IR images by analysis of the spectral fingerprint region (950–1480 cm−1). In the second step, analysis in the spectral window 1420–1480 cm−1 enables, for the first time, IR spectroscopic distinction between the basal layer, dysplastic lesions and squamous cell carcinoma within a particular sample. The joint application of IR microspectroscopic imaging and multivariate spectral processing combines diffraction-limited lateral optical resolution on the single cell level with highly specific and sensitive spectral classification on the molecular level. Compared with previous reports our approach constitutes a significant progress in the development of optical molecular spectroscopic techniques toward an additional diagnostic tool for the early histopathological characterization of cervical cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infrared (IR) spectroscopy relies on the absorption of radiation in the wavelength range 25 to 2.5 μm (corresponding to 400 to 4000 cm−1) which excites molecular vibrations. Biological macromolecules such as proteins, nucleic acids, and lipids have specific, fingerprint-like IR spectra in the wavenumber range 950–1800 cm−1. Because these spectral fingerprints are directly determined by molecular structures and cellular chemistry, IR spectroscopy has been shown to have promising potential to detect abnormal changes in cells and tissues [1]. Advantages of IR spectroscopic-based techniques are minimal or no sample preparation and objective results, because spectral data are collected and interpreted by computer controlled algorithms.
One field of research that received substantial attention in the last decade was the application of IR spectroscopy to gynecological screening for cervical dysplasia and malignancies (reviews in [1, 2]). Since pioneering work by Wong and coworkers [3], most studies have been performed on exfoliated cervical cells, and the main objective has been to improve the diagnostic accuracy of the Papanicolaou (Pap) smear. Cervical tissue sections have been examined in a few studies only [2, 4, 5, 6].
Standard histopathological assessment of carcinomas of the uterine cervix is conducted by means of hematoxylin and eosin (H&E) stained slides and the detected tumor is evaluated with regard to:
-
1
histological typing,
-
2
grading,
-
3
pattern of invasion (e.g. closed, finger-like, diffuse),
-
4
depth of infiltration into the cervical wall, and
-
5
lymphvascular space involvement [7].
Microscopic assessment on H&E-stained slides does not, however, reflect all the biological features characterizing the aggressiveness of a tumor and has only limited potential in terms of prognosis. Because of a substantial amount of subjectivity, evaluation of several morphological features has a restricted intra- and inter-observer correlation [8]. It is, furthermore, often difficult to diagnose whether a cervical intraepithelial neoplasia is accompanied by an early stroma invasion or to differentiate benign proliferative endocervical glandular lesions from a minimal deviation adenocarcinoma [9]. In addition, special cases, e.g. mesonephric carcinomas, are characterized inadequately by histology. Although supplementary examinations, for example special staining techniques (e.g. immunostaining for p16) or molecular biological analyses can elucidate the relationship between morphology, tumor biology, and pathogenesis, these methods usually focus on a special point and are expensive and time-consuming. Combination of IR spectroscopy with spatial coordinates is an alternative, less expensive, and time-saving approach, because it enables assessment of the chemical heterogeneity of a specimen in terms of spatial distribution of the system’s molecular constituents. This technique can, therefore, be used to improve the diagnostic accuracy and objectivity of the histopathological assessment, especially the characterization of cancer.
Advantage of imaging over mapping
The term “imaging” refers to the parallel detection of all pixels across the observed area by an array detector whereas “mapping” refers to sequential registration of all pixels by a single-channel detector. Array detectors comprise several thousands elements and thus enable correspondingly faster measurement. The effective spatial resolution of optical microspectrometers is limited by diffraction. Also, in mapping mode the lateral resolution of IR spectrometers is determined by the microscope aperture, which defines the area from which a spectrum is recorded, and by the step size of the raster grid with a spectrum recorded at each grid position. Such an aperture is set in conventional IR microspectrometers above 20×20 μm2, to achieve a reasonable signal-to-noise ratio in a short acquisition time. Special facilities offering IR synchrotron radiation with a brilliance advantage enable reduction of the aperture down to the diffraction limit of approximately 5×5 μm2 [10]. The total accumulation time depends on the number of spectra and the accumulation time per spectrum. A complete mapping measurement can take several hours. The imaging mode of IR spectrometers has a significant time advantage over the mapping mode because multichannel focal plane array (FPA) detectors instead of single-channel detectors enable recording of thousands of spatial points of a globally illuminated area in a single measurement which takes only few minutes [11]. In addition to the diffraction limit, the lateral resolution is determined by the pixel size of the FPA and the microscope magnification. First IR microspectroscopic imaging studies have been reported on the visualization of silicone gel in breast tissue [12], on collagen and proteoglycan in cartilage [13, 14], on the determination of CO3 2− in bone mineral [15], on breast tumor tissue [16], on germinal centers in the spleen [17], and on tissue microarrays of prostatic tissue [18]. These studies showed that the technique combines spatial localization at the single-cell level with the capability of chemical analysis.
To apply IR microspectroscopic imaging or mapping as an analytic tool for tissue evaluation, several mathematical procedures for analyzing the spectral data were developed. Univariate methods (also called functional group mapping) calculate peak positions, peak areas, or peak ratios of spectra to identify differences between different tissues. These methods do not, however, utilize the full content of the spectral information, which is usually distributed over a broad spectral region. Multivariate methods have therefore been applied for analysis like artificial neural networks [19], principal component analysis [20], linear discriminant analysis [21] and cluster analysis [2, 22]. Unsupervised methods do not require a-priori information. Supervised methods require the selection of training data to develop a classification model. In practice, the analysis results in a certain number of groups or clusters (in theory from one up to the overall number of pixels) which are assigned to a certain color so that, finally, a color-coded image can be assembled.
There have been only two reports of the examination of cervical tissue by IR microspectroscopic mapping with 20 to 30 μm spatial resolution; normal and precancerous areas were identified but no carcinomas [2, 5]. We expect IR microspectroscopic imaging to be a superior technique for sensitive and specific tissue diagnosis in the future, because of its advantages in spatial resolution and accumulation time. The discussion in this paper focuses on one sample of cervix uteri which encompasses squamous cell carcinoma, dysplasia, and normal tissue like stroma, epithelium, blood vessels, and mucus. The IR image comprises 499,712 IR spectra. They were analyzed in a two-step approach with initial fuzzy clustering for data reduction then hierarchical clustering for unsupervised classification. We succeeded for the first time in delimiting squamous cell carcinoma within a complex tissue section by IR microspectroscopy. This supervised analysis of data from one sample should establish a basis for the unsupervised classification of IR data from independent samples.
Material and methods
Tissue preparation
The sample originated from a patient with a squamous cell carcinoma of the uterine cervix who underwent a radical hysterectomy. The pathologic diagnoses were carcinoma extending beyond the cervix into the parametrium but not to pelvic sidewall (pT2b), regional/pelvic lymph nodes tumor-free (pN0), distant metastases cannot be stated (Mx), tumor well differentiated (G1), with lymph–vascular space involvement (L1), but without vascular space involvement (V0). Preparation of the cervix, fixation, and paraffin embedding have been described elsewhere [23].
Two adjacent sections 10 μm thick were prepared. One section was mounted on a conventional glass slide, stained with H&E and imaged by light microscopy. The second section was mounted on an IR-transparent calcium fluoride slide and imaged by IR spectroscopy in transmission mode. Both slides were deparaffinized by washing in xylol and ethanol. This approach enables comparison of the morphologies obtained. Photomicrographs were recorded using a digital camera fitted to a light microscope (Olympus IX70).
Data acquisition
IR spectroscopic images were recorded using a Bruker IFS66/S FTIR spectrometer coupled to a Hyperion microscope which was equipped with a 64×64 FPA detector (Bruker Optik, Ettlingen, Germany) and a 15× objective. The microscope was enclosed in a plastic box which was purged with dry air. Images of 4096 IR spectra at a spectral resolution of 8 cm−1 were acquired by the Opus software (Bruker) operating the FPA in continuous-scan mode by coadding 21 interferograms. Each image was recorded within 220 s, including 80 s data processing. Each FPA pixel refers to an area of 4.2×4.2 μm2 at 15× microscope magnification. A single microscopic image of 4096 pixels covers a sample area of 267×267 μm2. If the sample area exceeded 267×267 μm2, a series of IR images were recorded by moving the sample stage in increments of 267 μm. Background images were recorded after every six sample images to compensate for small fluctuations during the measurement series. Software written in-house was used to synchronize data acquisition and movement of the motorized stage.
Data preprocessing
Data pretreatment was performed by use of routines written in-house, in MatLab (The Mathworks, Natick, MA, USA). The treatment includes merging of different IR images to a single data set, filtering, baseline correction, and normalization. We used the spectral range from 950 to 1800 cm−1, and for special purposes the reduced ranges from 950 to 1480 cm−1 or from 1420 to 1480 cm−1 as fingerprint regions to discriminate between different tissue types. Different backgrounds and offsets in the spectra were corrected by subtracting a linear baseline over the considered ranges. The spectra were further scaled at the amide I band (1655 cm−1) to relative intensities between zero and unity by multiplying by a correction factor. This normalization is assumed to compensate for the effect of global intensity changes owing to slight variations in sample thickness. Low absorbance spectra with poor signal-to-noise ratios were removed from further analysis because they mainly originate from holes and fissures in tissue sections. The threshold criterion of the filter was the intensity of the strong amide I band of proteins near 1650 cm−1.
Nonhierachical fuzzy C-means cluster analysis
In the current context cluster analysis partitions the ensemble of spectra into groups (clusters) with distinct similarities within each cluster and differences between the clusters. Our implementation distributes the spectra over a fixed, pre-selected number of clusters. The probability that a selected spectrum belongs to one of the clusters is given by the so-called membership function with values between 0 (no class membership) and 1 (highest degree of cluster membership). The sum of the values of the membership function for one spectrum across all clusters is unity. We used the nonhierarchical fuzzy C-means (FCM) iterative algorithm to calculate the membership functions. Stop criteria were the maximum number of iterations (300) or the minimum incremental improvement (0.00001). Mean spectra for each cluster, so called cluster centroids, were calculated as weighted means over the IR spectra of one cluster with the spectra weighted by the respective values of the membership function. Finally, each IR spectrum is assigned to the cluster for which the value of the membership function is maximum. The program was optimized for parallel computing using several processors to analyze large data sets of more than 100,000 spectra.
Hierarchical cluster analysis.
The hierarchical cluster analysis (HCA) algorithm calculates the symmetric distance matrix (size n×n) between all considered spectra (number n) as a measure of their pairwise similarity. The algorithm then searches for the minimum distance, collects the two most similar spectra into a first cluster and recalculates spectral distances between all remaining spectra and the first cluster. In the next step the algorithm performs a new search for the most similar objects, which now can be spectra or clusters. The two most similar objects are clustered again and the distance values of the newly formed cluster are recalculated. This iterative procedure is repeated n−1 times until all spectra have been merged into one cluster. The hierarchical clustering classifies the spectra in terms of a two-dimensional dendrogram in which one axis refers to the reduction of clusters with increasing number of iterations and the other axis to the respective spectral distances. This study uses the cluster analysis implemented in the PLS toolbox (Eigenvector Research, Manson, WA, USA). Accordingly, the HCA algorithm provides two measures which can be used for visualization, namely the number of clusters at fixed spectral distance or, alternatively, the spectral distances for a fixed number of clusters. If the latter is used the number of clusters was usually adjusted to correspond to the classification of tissue types suggested after pathological inspection of the microscopic images.
Combining fuzzy C-means and hierarchical cluster analysis
FCM clustering is advantageous because of its capability to handle large spectral data sets within the framework of a limited and therefore concise number of clusters. Its disadvantages are that the exact number of clusters is usually not available and the result may depend on the random initial state. HCA clustering classifies the spectra within the data set in the form of a dendrogram, which reveals differences and similarities between all spectra in a highly detailed and stable fashion. Problems occur if the size of the distance matrix, the square of the number of objects, is large. We developed a two-step approach which combines the advantages of both clustering methods to distinctly enlarge the tissue area to be characterized at once by IR spectroscopic imaging. FCM clustering was performed in the first step so that every tissue type in the tissue sections is represented by at least one FCM cluster. The respective cluster centroids provide the starting point for subsequent HCA. This procedure reduces the number of potential HCA clusters to less than 100 which is a manageable size for computation. Note that this approach also improves the signal-to-noise ratio, because each FCM cluster centroid is the average over a certain set of IR spectra. Another means of obtaining a subset of a large number of image data for HCA is to generate this set randomly [24]. There is, however, a real danger of missing a small cluster.
Results
H&E stained image: Overview
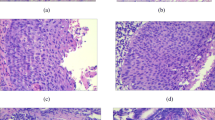
Figure 1 shows a 16×16 mm2 H&E-stained section of cervix uteri which is directly adjacent to the unstained tissue section studied by IR spectroscopic imaging. We selected seven representative areas for IR analysis encompassing the major tissue types present in this section (Fig. 1). Table 1 summarizes the size of each tissue area selected, the number of IR spectra, and a description of the tissue. A total of 499,712 IR spectra covering an area of 8.7 mm2 were selected for this study. The histopathological architecture in Fig. 1 can be divided into five major compartments—cervical stroma, squamous epithelium, squamous cell carcinoma, dysplasia, and peritumoral inflammatory response. In addition, bright areas correspond to glandular tissue and red spots surrounded by a thin ring correspond to blood vessels.
H&E-stained tissue section of the cervix uteri. The areas for IR spectroscopic imaging of an adjacent unstained tissue sections are depicted: cervical stroma (1), squamous epithelium (2), dysplasia (3, 4, 5), and squamous cell carcinoma (6, 7). See Table 1 for detailed descriptions
IR spectroscopic imaging of cervical stroma and epithelium
As described in the material and methods section, we combined FCM clustering and HCA to take advantage of the capacity of FCM clustering to analyze large data sets and because HCA can display similarities between IR spectra in detail. The dendrogram in Fig. 2c plots the distances between 35 cluster centroids which were obtained after FCM clustering of approximately 45,000 spectra in the wavenumber range 950 to 1480 cm−1 referring to area 2 of Fig. 1. The dashed vertical line at the distance value 5.5 groups the FCM cluster centroids into ten HCA clusters. This means cluster centroids with distances smaller than 5.5 were merged. After comparison with the H&E-stained tissue section (Fig. 2a), the color coded clusters in Fig. 2b could be assigned to histological features. The left part (pink) is dominated by stroma. At the border to the basal cell layer (red) a thin layer of cells can be found which is typical of the area of inflammatory response (blue). A further cluster (cyan) corresponds to the transition between cervical stroma and the area of inflammatory response, and so is tentatively attributed to mild inflammation.
IR image of the transition from cervical stroma to squamous epithelium. a. H&E-stained tissue section of cervical stroma and epithelium. b. Ten-color-coded result of cluster analysis of IR spectroscopic images from area 2 of Fig. 1. Cervical stroma (pink), inflammation (cyan, blue), basal cell layer (red), transition from basal to parabasal cell layer (light green, dark green), parabasal cell layer (yellow), intermediate cell layer (light orange, dark orange), superficial cell layer (brown). c. Dendrogram of hierachical cluster analysis with 35 centroid spectra of fuzzy C-means clustering as input. The dashed line indicates the histology-based decision of the number of clusters chosen
The overlaid cluster centroid IR spectra in Fig. 3a confirm this observation. The spectrum corresponding to the transition cluster (cyan) lies between the spectra of cervical stroma (pink) and the main area of inflammatory response (blue). All three spectra contain similar bands near 1656, 1535, 1452, 1400, 1336, and 1232 cm−1 which can mainly be assigned to proteins as the major constituent of these tissues. An increase at 1082 cm−1 is the most significantly difference between the spectra of Fig. 3a and is, therefore, regarded as an indicator of inflammatory response. The band near 1232 cm−1 can be assigned to collagen contained in cervical stroma. The higher relative amount of collagen in the tissue types of Fig. 3a correlates with the lower relative intensity of this band in the spectra of epithelium in Figs. 3b,c. The dendrogram (Fig. 2c) shows that the distance between cervical stroma (pink) and the two forms of inflammation (cyan, blue) is smaller than between cervical stroma (pink) and epithelium (red through brown). This result suggests that this inflammation pertains to cervical stroma.
IR spectra from 950 to 1800 cm−1 representing cluster averages in Fig. 2 after baseline correction and normalization to amide I band intensity. a. Overlay of cervical stroma (magenta), mild inflammation (cyan), and severe inflammation (blue). b. Overlay of basal layer (red), transition to parabasal layer (light green, dark green) and parabasal cell layer (black=yellow-colored cluster in Fig. 2). c: Overlay of intermediate cell layer (light orange, dark orange) and superficial layer (brown)
Increasing the spectral distance, e.g. by shifting the dashed line in Fig. 2a to 20, results in only two clusters referring to cervical stroma and epithelium. The differences between the IR spectra of epithelium in Figs. 3b,c and cervical stroma in Fig. 3a correlate with distances in the dendrogram. In particular, if one compares both groups, the bands at 1537, 1460, 1394, and 1239 cm−1 decrease in intensity and shift their positions by more than two wavenumbers.
Two narrow clusters (light green, dark green) in Fig. 2b indicate a transition zone from basal (red) to parabasal tissue (yellow). The respective spectra in Fig. 3b are characterized by slightly increased absorbance near 1028 cm−1 with minimum absorbance for basal tissue. The basal layer is the germ line for the epithelium. It consists of progenitor cells that tend to be one cell layer thick. This is consistent with the width of the red colored area which ranges from 15 to 30 μm.
The clusters next to the transition zone are assigned to the parabasal cell layer (yellow), the intermediate cell layer (light and dark orange), and the superficial cell layer (brown). Their spectra in Fig. 3c change in a fashion similar to the spectra of the transition zone. In particular, the band 1028 cm−1 further increases accompanied by an increase of the less intense bands at 1081 and 1153 cm−1 which were not fully resolved in Fig. 3b because of low intensities and strong overlap with other bands. The spectral signature of bands at 1028, 1081, and 1153 cm−1 is a marker for an increase in the concentration of glycogen in the tissue. This trend seems plausible, because glycogen usually accumulates on maturation of epithelial cells [6].
The distinction of tissue types did not improve when the cluster analysis was extended from the region 950–1480 cm−1 to 950–1800 cm−1. The major bands in the wavenumber region 1480–1800 cm−1 are assigned to proteins (amide II near 1545 cm−1, amide I near 1655 cm−1), and to the C=O ester groups of lipids near 1740 cm−1. These bands do not enable clear differentiation between the discussed epithelial layers because variances such as band positions, band widths, or relative intensities are too small in this region, as is evident from the overlay of the normalized spectra (Fig. 3).
IR spectroscopic imaging of all tissue areas
In Fig. 4 the photomicrographs of the H&E-stained areas (left row) are compared with the results from cluster analysis of the IR spectroscopic images from all considered areas 1 to 7 in the spectral range 950–1480 cm−1 (middle row). Altogether the data set consists of 499,712 spectra. Note that the code of ten colors is slightly different from that used in Fig. 2, because of different tissue properties. Regions where the preprocessing procedure removed the spectra are shaded gray. The imperfect coincidence between the IR spectroscopic image and the H&E stained photograph—in particular for area 2—is because the sections are just adjacent, not identical.
H&E-stained tissue areas 1 to 7 of Fig. 1 (left row), pooled cluster analysis of all IR spectroscopic images in the spectral range 950–1480 cm−1 (middle row), and selected cluster analysis of those IR spectra assigned in the pooled cluster analysis to the nucleus-dense areas in the spectral range 1420−1480 cm−1 (right row). Bar=200 μm. Coloring scheme in middle row: nucleus-dense areas of basal cell layer, dysplasia, squamous cell carcinoma (red), parabasal cell layer (light and dark orange), cervical stroma (pink, magenta), inflammation (cyan, blue), mucus (yellow), blood vessel (green). Coloring scheme in right row: basal cell layer (yellow), dysplasia (orange), squamous cell carcinoma (red), non-basal cell layer and non-malignant tissue (blue)
Analysis of the pooled tissue areas gives results for area 2 (Fig. 4, middle row) almost identical with those from previous analysis of the isolated area (Fig. 2), although with a slightly reduced number of clusters. The four clusters forming the zone above the basement membrane (red, light green, dark green, yellow clusters in Fig. 2b) are now integrated with the basal and the parabasal cell layers (red and light orange clusters, respectively). The area of mild inflammatory response (cyan in Fig. 2b) is included with cervical stroma (pink). Good agreement between both cluster analyses is obtained for the intermediate (light and dark orange) and superficial layers (brown). A few spectra of cervical stroma make up a separate cluster (magenta). This result demonstrates the highly reproducible distinction of the main tissue components cervical stroma, basal layer, and the upper layers of the epithelium.
Tissue area 1 consists of cervical stroma indicated by pink and magenta colors. The blood vessel in the upper part of area 1 is clearly distinguished from the surrounding tissue and displayed by the green cluster. As is evident from Fig. 1, this area is located far from tumor, dysplasia, and epithelium. The low frequency of colors other than pink, magenta, and green shows that fewer than 0.1% of the spectra were grouped into other clusters.
The lower part of area 3 is similar to area 2. It shows that inflammatory infiltration and basal cell layer (red) are located between the cervical stroma and the upper layers of the epithelium. In the upper part of area 3, however, the inflammation zone (blue) broadens and in addition, dysplastic tissue (red) is found. The red coloring for dysplasia and basal cell layer indicates that this approach cannot yet distinguish normal from premalignant epithelium. Analogous problems occur for areas 4 to 7 with the red colored squamous cell carcinoma. The strong overlap in the red cluster can be morphologically attributed to the higher nucleus density in relation to the other tissue areas. Therefore, the red cluster was designated as nucleus-dense areas. To improve the results of delimitation, a more advanced approach will be introduced in the next section.
Area 4 is dominated by dysplasia (red) surrounded by inflammatory infiltration of the cervical stroma (blue/cyan). Almost no inconspicuous cervical stroma (pink/magenta) or epithelium (orange) is detected. A few spectra are grouped in a new cluster (yellow). This additional feature is assigned to mucus which is also present in areas 5, 6, and 7.
In area 5 some cervical stroma at the bottom indicates the transition from normal (pink) to dysplastic and cancerous tissue (red). Extended areas of inflammation (cyan, blue) exist at the interface.
Analogous to area 1, the green cluster in area 6 correlates with a blood vessel. Interestingly, the blood vessel seems to be embedded in cervical stroma (pink) and inflammatory tissue (cyan, blue). The top and the bottom parts consist of tumor tissue and dysplasia, respectively.
In area 7 the transition from cervical stroma (pink, magenta) to tumor tissue (red) is apparent. Although in area 5 the inflammatory zone (cyan, blue) forms a continuous layer between normal and malignant tissue, this separation is inhomogeneous in area 7. Although endocervical glands can be identified by histology, IR spectroscopic segmentation attributed them to inflammatory cells. This problem shows that in this aspect the current approach needs improvement.
Improved cluster analysis of the nucleus-dense areas
Figure 5 shows IR spectra of basal cell layer, dysplasia, and squamous cell carcinoma from 950 to 1480 cm−1 after baseline correction and normalization, as described in the experimental section. The different spectra are virtually identical in the spectral range 950–1350 cm−1. At higher wavenumbers some of the bands change characteristically between different tissues. For example, the position of the band due to the deformation vibration of the CH2 and CH3 groups shifts from 1460 to 1452 cm−1 as a function of malignancy. This trend is accompanied by the decrease of intensity which is maximum for basal cell layer, medium for dysplastic tissue, and minimum for squamous cell carcinoma.
These spectral characteristics enable us to distinguish basal cell layer, dysplastic tissue, and squamous cell carcinoma by cluster analysis which uses the relevant spectral range from 1420 to 1480 cm−1 and which starts with the red clusters from Fig. 4, middle row. All clusters from Fig. 4 (middle row) assigned to cervical stroma, epithelium, mucus, blood vessels, and inflammatory response are now excluded from further analysis. Figure 4, right row, shows the results after FCM clustering of the red clusters of Fig. 4, middle row, into ten cluster centroids and subsequent HCA, providing four relevant clusters. This algorithm correctly assigns normal basal cell layer (yellow) in areas 2 and 3. It also detects dysplastic tissue (orange) in the upper part of area 3, throughout area 4 and at the bottom of area 6. Moreover, squamous cell carcinoma (red) dominates in the areas 5, 6 (top), and 7. On one hand, only very few pixels representing normal basal cells (yellow) are detected in areas 4, 5, 6, and 7. On the other hand, very few dysplastic and tumor cells (orange and red pixels) in area 2 and in the lower part of area 3 (right row) are misclassified within the basal cell layer, and few basal cells (yellow pixels) in the upper part of area 3 (right row) are misclassified within the dysplasia. The morphological assignments for areas 4 and 5 are ambiguous which is reflected for area 5 (right row) by a melange of red and orange pixels for squamous cell carcinoma and dysplasia, respectively. Although a correct distinction was obtained for area 6 with few exceptions only, the dysplastic regions mainly at the bottom and at the top of area 7 should be tumor after morphological inspection. The tissue distinction for areas 6 and 7 was, furthermore, correct with few exceptions only, e.g. the morphologically unambiguous regions of cancer at the top of both areas contains a few dysplastic parts (orange pixels).
Another cluster (blue) appears in all areas whose spectral signature was not assigned to malignant tissue or basal cell layer. For example, all spectra in area 1 which were first grouped in the red cluster are now attributed to the blue cluster. This cluster reveals the accuracy of the advanced clustering approach which is controlled by the biological variance within the tissue section. Although clustering of the complete set of spectra resulted in some incorrect classifications, restriction to the data set of nucleus-dense areas could compensate for this error.
Discussion
A recent study compares results from cluster analyses of IR spectroscopic mapping data of a colorectal adenocarcinoma section with photomicrographs of the respective H&E-stained slide in terms of image quality and consistency [25]. The authors show that hierarchical clustering (Ward’s technique) is a superior algorithm to fuzzy C-means clustering and k-means clustering. Spectral tissue structure differentiation agreed satisfactorily with histological tissue architecture (“supracellular level”), but some of the colored image areas could not be assigned unequivocally to histologically relevant tissue types. Although hierarchical clustering was the best method, its computational demands increase dramatically with increasing number of spectra. We therefore developed a modified clustering approach to analyze IR spectroscopic images from a thin section of cervical tissue.
To handle hundreds of thousands of spectra—corresponding to hundreds of thousands of pixels or cells—a two-step algorithm was developed. Fuzzy C-means (FCM) clustering was used for data reduction then hierarchical cluster analysis (HCA) for classification based on spectral similarities. The results were compared with those from an adjacent tissue section stained with H&E. Cluster analysis in the fingerprint region 950–1480 cm−1 enabled distinction between cervical stroma, layers of squamous epithelium above the basal cells, inflammation, mucus, blood vessels, and nucleus-dense areas such as the basal cell layer of squamous epithelium, dysplastic epithelium, and squamous cell carcinoma. The progression from normal tissue through dysplasia to squamous cell carcinoma could be clearly identified by cluster analysis of the IR spectra in the range 1420–1480 cm−1, a specific marker for differentiation of the nucleus-dense areas. This spectral range was found by pattern-recognition techniques and is consistent with chemical heterogeneity in benign, dysplastic, and neoplastic regions. In a very simplistic view of cell biology, all cells, benign or otherwise, are made of the same basic components, for example protein, nucleic acid, lipids, fats, carbohydrates etc. Tumorgenesis (benign or malignant) is reflected in the morphology of cells by changes in the relative proportions of these components. An assignment of a single signal near 1450 to 1460 cm−1 to a specific compound is extremely difficult, in particular after changing the chemical composition by the washing procedure to remove paraffin. Such an assignment is not important for the IR-based approach, however, if we consider this spectral feature as a fingerprint for squamous cell carcinoma.
The high lateral resolution in the IR images revealed new details which have not been identified before by mapping IR spectroscopy. The stromal infiltration of inflammatory cells is located below the normal epithelium and is especially present at the interface with dysplasia and squamous cell carcinoma. Normal and abnormal tissue could also been differentiated in great detail, reflecting the chemical changes during carcinogenesis. These results clearly demonstrate the progress of our approach for IR spectroscopy-based tissue diagnosis.
Previous studies reported IR spectral differences between normal and abnormal tissue samples from cervix uteri which were related to changes in glycogen, nucleic acid, and protein content. Further studies demonstrated that the observed spectral changes might not have been related to the presence of dysplastic cells per se, but rather to nonspecific factors such as localized inflammatory effects [6, 26], the cell type being measured [27], and nonspecific depletion of cytoplasmic glycogen. For instance, glycogen concentration varies dramatically when samples are collected at different stages of the menstrual cycle [28] and nucleic acid concentration changes as a function of the cell cycle [29]. It has, therefore, been suggested that IR spectroscopy does not identify cancerous samples directly by detection of specific tumor markers but only indirectly by detecting differences between their average cell cycle states [30]. This might also explain why the distinction of normal basal cell layer from dysplasia and squamous cell carcinoma was much more difficult than distinction of the different tissue types such as cervical stroma or upper layers of the epithelium. The reason for the difficulties could be that normal and neoplastic basal cells go through the same cell cycle stages at approximately the same rate. The key difference is that neoplastic cells do not regulate this process which is a hallmark of cancer [31]. As a consequence, the cytoplasm consists of similar chemical compositions which all give high cell densities and similar patterns after H&E staining. As far as we are aware we have succeeded for the first time in distinguishing basal cells, dysplasia, and tumor in a single sample by IR spectroscopy.
In summary, this study demonstrated that IR spectroscopic imaging in combination with multivariate algorithms is capable of differentiation of cervical tissue which is comparable with H&E staining as the gold standard. IR spectroscopy can complement established methods, because it probes molecular fingerprints as an independent property; this technique has not previously been used for diagnostic purposes. The next task is to use this approach to classify normal and abnormal tissue in thin sections from different patients. We expect that this task to be feasible, because a previous study demonstrated that patient-to-patient variations are smaller than those encountered between different tissue types and stages of disease [32]. The final objective of our work is to develop a rapid and objective algorithm for automatic classification of cervical tissue sections on the basis of IR spectroscopy alone.
Abbreviations
- IR:
-
Infrared
- Pap:
-
Papanicolaou
- H&E:
-
Hematoxylin and eosin
- FPA:
-
Focal plane array
- FT:
-
Fourier transform
- FCM:
-
Fuzzy C-means
- HCA:
-
Hierarchical cluster analysis
References
Dukor RK (2002) In: Chalmers JM, Griffiths PR (eds.) Handbook of Vibrational Spectroscopy. John Wiley and Sons Ltd., New York 3335–3361
Wood BR, Chiriboga L, Yee H, Quinn MA, McNaughton D, Diem M (2004) Gynecol Oncol 93:59–68
Wong PTT, Lacelle S, Fung Kee Fung M, Sentermann M, Mikhael NZ (1995) Biospectroscopy 1:357–364
Mordechai S, Sahu RK, Hammody Z, Mark S, Kantarovich K, Guterman H, Podshyvalov A, Goldstein J, Argov S (2004) J Microsc 215:86–91
Chang JI, Huang YB, Wu PC, Chen CC, Huang SC, Tsai YH (2003) Gynecol Oncol 91:577–583
Chiriboga L, Xie P, Yee H, Zarou D, Zakim D, Diem M (1998) Cell Mol Biol 44:219–229
Scully RE, Bonfiglio TA, Kurman RJ, Silverberg SG, Wilkinson EJ (1994) WHO—Histological typing of female genital tract tumors. Springer, Berlin Heidelberg New York
Zaino RJ, Ward S, Delgado G, Bundy B, Gore H, Fetter G, Ganjei P, Frauenhoffer E (1992) Cancer 69:1750–1758
Tsuda H, Mikami Y, Kaku T, Akiyama F, Hasegawa T, Okada S, Hayashi I, Kasamatsu T (2003) Pathol Int 53:440–449
Dumas P, Jamin N, Teillaud JL, Miller LM, Beccard B (2004) Faraday Discuss 126:289–302
Lewis EN, Treado PJ, Reeder RC, Story GM, Dowrey AE, Marcott C, Levin IW (1995) Anal Chem 67:3377–3381
Kidder LH, Kalasinsky VF, Luke JL, Levin IW, Lewis EN (1997) Nature Medicine 3:235–237
Potter K, Kidder LH, Levin IW, Lewis EN, Spencer RGS (2001) Arthritis and Rheumatism 44:846–855
Camacho NP, West P, Torzilli PA, Mendelsohn R (2001) Biopolymers 62:1–8
Ou-Yang H, Paschalis EP, Mayo WE, Boskey AL, Mendelsohn R (2001) J Bone Miner Res 16:893-900
Fabian H, Lasch P, Boese M, Haensch W (2002) Biopolymers 67:354–357
Krafft C, Salzer R, Soff G, Meyer-Hermann M (2005) Cytometry A 64A:53–61
Fernandez DC, Bhargava R, Hewitt SM, Levin IW (2005) Nat Biotech 23:469–474
Romeo M, Burden FR, Wood BR, Quinn MA, Tait B, McNaughton D (1998) Cell Mol Biol 44:179–187
Cohenford MA, Godwin TA, Cahn F, Bhandare P, Caputo TA, Rigas B (1997) Gynecol Oncol 66:59–65
Shaw RA, Guijon FB, Paraskevas M, Ying SL, Mantsch HH (1999) Anal Quant Cytol Histol 21:292–302
Lasch P, Naumann D (1998) Cell Mol Biol 44:189–202
Horn LC, Fischer U, Bilek K (2001) Zentralbl Gynakol 123:255–265
Tran TN, Wehrens R, Buydens LMC (2005) Chem Intell Lab Systems 77:3–17
Lasch P, Haensch W, Naumann D, Diem M (2004) Biochim Biophys Acta 1688:176–186
Wood BR, Quinn MA, Tait B, Romeo M (1998) Biospectroscopy 4:75–91
Chiriboga L, Xie P, Yee H, Vigorita V, Zarou D, Zakim D, Diem M (1998) Biospectroscopy 4:47–53
Romeo M, Wood BR, McNaughton D (2002) Vib Spectrosc 28:167–175
Boydston-White S, Gopen T, Houser S, Bargonetti J, Diem M (1999) Biospectroscopy 5:219–227
Diem M, Boydston-White S, Chiriboga L (1999) Applied Spectroscopy 53:148A–161A
Hanahan D, Weinberg RA (2000) Cell 100:57–70
Lasch P, Naumann D (1998) Cell Mol Biol 44:189–202
Acknowledgement
W. Steller and C. Krafft are supported by the Volkswagen Foundation within the project “Molecular Endospectroscopy” of the program “Junior Research Groups at Universities”. U.-D. Braumann and H. Binder acknowledge financial support of the Deutsche Forschungsgemeinschaft under grant no. BIZ 6/1-2.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Steller, W., Einenkel, J., Horn, LC. et al. Delimitation of squamous cell cervical carcinoma using infrared microspectroscopic imaging. Anal Bioanal Chem 384, 145–154 (2006). https://doi.org/10.1007/s00216-005-0124-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-005-0124-4