Abstract
Silica particles containing immobilised peptidic templates have been used for the generation of hierarchically imprinted polymers. The pores of the silica mould were filled with a mixture of monomers/initiator and polymerised, followed by dissolution of the silica template. This method leaves behind imprinted polymers with binding sites located at the surface, which are capable of recognising larger molecules with the same immobilised epitope. All the products resulting from solid-phase synthesis of peptides were characterised by elemental analysis, FT-IR spectroscopy and fluorescence microscopy. The hierarchically imprinted polymers generated from these products were characterised by elemental analysis, FT-IR spectroscopy, fluorescence microscopy, scanning electron microscopy (SEM) and nitrogen adsorption, providing evidence concerning the reproducibility of each step. The chromatographic properties of the materials have been investigated and the advantages of the immobilisation method have been proven. The materials exhibit selectivity for their templates and other structurally related dipeptides. Furthermore, the polymers proved to be capable of recognising larger peptides containing the immobilised sequence.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chemically and mechanically stable materials which are able to mimic the recognition properties of biological structures, such as antibodies, can find numerous applications for the separation and analysis of a vast variety of biologically active or harmful substances in industry, health services and environmental monitoring. One technique that is being increasingly adopted for the generation of artificial macromolecular receptors is molecular imprinting in synthetic polymers [1].
Traditionally, molecularly imprinted polymers (MIPs) have been prepared as bulk polymer monoliths followed by mechanical grinding to obtain irregular, micrometre-sized particles. While this rather cumbersome and low-yielding method is still the most widely used to produce MIPs, some other specific applications require MIPs in defined physical forms. Therefore, there is a strong need for practical methods allowing the materials to be obtained in a bead format, with control of size and porosity independent from the generation of the templated sites.
Imprinted materials with a homogenous morphology have been produced by suspension or emulsion polymerisation [2], dispersion or precipitation polymerisation [3, 4]. The downside to these techniques is that the morphology of the resulting products is very sensitive to small changes in the synthesis conditions. Even under strictly controlled synthesis conditions, a simple change of template may require a complete reoptimisation of the conditions in order to achieve a given morphology. Furthermore, most of these procedures are limited with respect to the type of monomer and solvent that can be used for the polymerisation. One way to prevent these problems is by grafting MIPs to the surface of preformed support materials (e.g. silica or organic polymers supports) [5].
In another promising approach, hierarchical imprinting, surface-confined templated sites can be obtained with maintained control of the material’s morphology. The template molecule is attached to a pore structure directing agent (porogen), such as to the head group of surfactants forming an ordered surfactant system [6] or to the surface of a disposable porous solid [7, 8]. Polymerisation in the pores of the porogen followed by subsequent removal leads to the formation of a porous solid containing surface-confined templated sites (Fig. 1). In an approach independently developed by Yilmaz et al. [7] and Titirici et al. [9], porous silica is used as a mould to control the particle size, shape and porosity of the resulting imprinted polymer. The template can either be immobilised to the walls of the mould or can be simply dissolved in the monomer mixture. Here, the pores are filled with a given monomer/template/initiator mixture and, after the polymerisation in the pores, the silica is etched away and imprinted polymer beads are obtained exhibiting molecular recognition properties for the template and larger molecules containing the template substructure either in the batch [7] or chromatographic mode [8] (Fig. 1). The development of imprinted affinity phases for the separation of biological macromolecules (e.g. peptides and proteins) is an intriguing area. Most molecularly imprinted polymers are selective for low molecular mass compounds and the imprinting of oligopeptides and proteins for aqueous-based separations continues to be an important challenge. Those reports involving the protein itself in the polymerisation process [10, 11, 12] have been characterised by low selectivity and poor reproducibility. This may be related to the use of various solvents, temperatures, pH and ionic strengths causing partial denaturation of the protein. In order to circumvent these problems an epitope approach may be used. Here, a smaller peptide corresponding to a unique amino acid sequence of a target protein is used as a template to generate a site that can subsequently bind the larger target molecule [12].
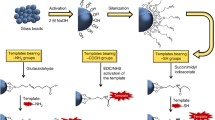
Expanding on this concept we recently used immobilized peptides as templates to generate surface-confined sites for larger peptides [13]. This was based on our general strategy to use crude solid-phase synthesis products as templates for hierarchical imprinting (Fig. 2). Herein we wish to present a more detailed account of this work. In the first step, the peptide epitopes were synthesized by standard solid-phase synthesis procedures on the surface of a porous silica support (Fig. 3). The immobilised peptides were then used as templates for the generation of hierarchically imprinted materials. These materials can be applied in HPLC and exhibit recognition for the immobilised peptidic templates as well as for larger peptides containing the immobilised sequence. The benefits of the immobilisation technique, where the sites are confined to the pore wall surface, are also demonstrated.
Solid-phase synthesis of peptide templates and preparation of corresponding hierarchically imprinted polymers. (i) BOC–Gly–OH, DCC/HOBt, DMF, 25°C, 12 h; (ii) Me3SiCl (1 M), C6H5OH (3 M), DMF, 25°C, 1 h; (iii) FMOC–Phe–OH, DCC/HOBt, DMF, 25°C,12 h; (iv) piperidine (20%), DMF, 25°C, 30 min; (v) (a) pore filling with EDMA (83%), MAA (17%) and AIBN (1% w/w) by repeated vacuum–nitrogen purge cycles;(b) Δ60°C, 24 h; (vi) (NH4)HF2 (aq), 12 h
Experimental
Chemicals
All the used peptides were purchased from Bachem (Heidelberg Germany). N,N′-Dicyclohexylcarbodiimide (DCC), 1-hydroxybenzotriazole hydrate (HOBt), ethylene glycol dimethacrylate (EDMA), methacrylic acid (MAA), (3-aminopropyl)triethoxysilane (APS), chlorotrimethylsilane (Me3SiCl), hexamethyldisilazane (HMDS), and phenol (PhOH) were obtained from Aldrich (Deisenhofen, Germany). EDMA and MAA were purified by distillation under reduced pressure; the other chemicals were used as received. Azo-N,N′-bisisobutyronitrile (AIBN), dimethylformamide (DMF), dichloromethane (DCM), toluene, methanol (all anhydrous), ammonium hydrogen difluoride, triethylamine, trifluoroacetic acid (TFA) and acetic acid (AcOH) biochemical grade were obtained from Acros Organics (Geel, Belgium). AIBN was recrystallised from methanol prior to use; triethylamine was distilled at atmospheric pressure prior to use. All other chemicals were used as received. 3-Aminoquinonile was purchased from Fluka (Steinhem, Germany). Acetonitrile (MeCN), methanol (MeOH), acetone, water (all HPLC grade) and hydrochloric acid (HCl fuming 37%) were purchased from Merck (Darmstadt, Germany). The porous silica gel (specific surface area (S)=350 m2 g−1, particle size (Ps)=10 µm, pore volume (V p)=1.083 mL g−1 and pore diameter (dp)=11.5 nm) was synthesised and characterised in our laboratories and subjected to calcination in air for a period of 6 h at 823 K (heating rate 1 K min−1).
Apparatus
The HPLC measurements were carried out on Hewlett–Packard HP 1050 instruments (Agilant Technologies, Waldbronn, Germany) equipped with a UV-DAD detector and an autosampler. The fluorescent labelled polymer particles were investigated using a LEICA DM R fluorescence microscope HC (Benzheim, Germany). The amounts of immobilized templates were estimated from elemental microanalysis obtained using a “CHN-rapid” HERAEUS analyzer (Hanau, Germany). The FT-IR spectra were recorded with a MATTSON 2030 Galaxy Serie FT-IR-Spectrophotometer (Madison, WI, USA). The particle morphology and size distribution were determined using a LEO 1530 “Gemini” scanning electron microscope (LEO Elektronemikroskopie GmbH, Oberkochen, Germany). The samples were deposited on a sample holder with an adhesive carbon foil. Nitrogen sorption measurements were performed on a Quantachrome Autosorb 6B (Quantachrome Corporation, Boynton Beach, FL, USA) automatic adsorption instrument. Before measurement, 100–150 mg of the samples were heated at 60°C under high vacuum (10−5 Pa) for at least 12 h.
Rehydroxylation of calcined silica
Calcined silica (20 g) was heated at reflux over night with stirring in 17% HCl in a 500-mL round-bottomed flask. The silica was filtered through a glass filter funnel and washed twice with 200 mL methanol. Finally, the silica was dried in a vacuum oven at 80°C for 12 h.
Silica surface modification with APS
The modification of silica with APS was performed according to the procedure described by Revillon et al. for obtaining initiator-modified support materials [14]. A batch of 20 g rehydroxylated silica was suspended in 160 mL dry toluene. According to the number of silanol groups on the silica surface (8 μmol m−2), the appropriate amount of APS (56 mmol, 12.39 g) was added to the mixture which was then heated at reflux (on a oil bath) with stirring for 12 h at 110°C. The product was filtered through a glass funnel and washed with 4×50-mL aliquots of toluene. The product was dried in a vacuum oven at 40°C for 12 h.
End-capping of the APTS-modified silica with HMDS
In order to react all the remaining silanol groups from the previous step, we used a procedure described in ref. [15]. Thus, 20 g of Si-APS were suspended in 180 mL dry DCM and 2.016 mL of HMDS in 20 mL dry DCM was added. The mixture was stirred at room temperature under nitrogen for 12 h and for a further 4 h at 40°C. The product was filtered through a glass funnel, washed with 200 mL methanol and 100 mL acetone to remove traces of unreacted HMDS and dried in a vacuum oven for 12 h at 40°C to afford 20 g of Si-APS.
Solid-phase synthesis of peptides using amino-modified silica
The peptides were synthesised using standard Merrifield chemistry [16]. Thus, in the first step, BOC–Gly–OH was coupled through DCC-catalysed amide bond formation. After deprotection, FMOC–Phe–OH was coupled to obtain the N-protected or, after deprotection, the free dipeptide coupled through its carboxy terminus to the support surface. The coverage of the silica surface with amino groups was determined from elemental analysis data (Table 1) and a two-fold excess of amino acids, DCC and HOBt were used.
Coupling BOC–Gly–OH onto the silica surface
Dry DMF (250 mL), BOC–Gly–OH (4.128 g), DCC (4.862 g) and HOBt (3.184 g) were placed in a 500-mL round-bottomed flask. These were stirred under a nitrogen atmosphere at room temperature. After 30 min, 10 g of the Si-APS were added to the mixture and the reaction was allowed to continue overnight. The product was filtered, suspended in 250 mL DMF and shaken for 35 min to remove the remaining dicyclohexylurea, filtered again, washed twice with 50-mL aliquots of DCM and MeOH and dried in a vacuum oven at 40°C for 6 h to afford 10 g of BOC–Gly–Si.
Deprotection of BOC–Gly–Si
As a deprotection agent for the N α-tert-butyloxycarbonyl group, the mild reagent containing 1 M chlorotrimethylsilane and 1 M phenol in DMF was used [17]. It should be mentioned that deprotection using TFA was tried, but a complete removal of the organic layer occurred. BOC–Gly–Si (7 g), prepared as described above, was shaken at room temperature for 10 min with 80 mL deprotection agent. The product was filtered and washed with 200 mL DCM and then suspended in 100 mL solution containing 4% water in DMF. After shaking for 5 min, the mixture was filtered, resuspended in 100 mL solution 10% triethylamine in DMF, shaken for another 5 min and finally washed with 3×100 mL DCM. The procedure was repeated once more, but the deprotection step was carried out for 30 min. The washing procedure was the same. The resulting hydrochloride of the peptide silica was neutralised by shaking with 100 mL of 10% triethylamine in DMF for 2 h at room temperature. Finally, the product was washed with 50 mL DCM and dried in a vacuum oven at 40°C for 6 h to afford 6.5 g of H–Gly–Si.
Coupling reaction between FMOC–Phe–OH and H–Gly–Si
Dry DMF (150 mL), FMOC–Phe–OH (4.555 g), DCC (2.956 g) and 1-HOBt (1.935 g) were introduced into a 250-mL round-bottomed flask. The mixture was stirred under a nitrogen atmosphere at room temperature. After 30 min, 4.5 g of the H–Gly–Si was added to the mixture and the reaction was allowed to continue overnight. The product was filtered, suspended in 250 mL DMF and shaken for 35 min to remove the remaining dicyclohexylurea, filtered again, washed with 50 mL DCM and dried in a vacuum oven at 40°C for 6 h. The washing procedure was the same as previously described and in the end the product was washed with 50 mL ethanol to afford 4.5 g of FMOC–Phe–Gly–Si.
Deprotection of FMOC–Phe–Gly–Si
FMOC–Phe–Gly–Si (2.5 g) was suspended in a 50-mL solution of 20% piperidine in DMF in a 100-mL round-bottomed flask, and shaken at room temperature for 6 h. The product was then washed with 50-mL aliquots of DMF and DCM and dried in a vacuum oven at 40°C for 6 h to afford 2 g of H–Phe–Gly–Si.
Preparation of the polymer–silica composites
A pre-polymerisation mixture consisting of 12 mmol MAA, 60 mmol EDMA and 0.129 g AIBN was prepared in a glass vial. According to the pore volume of silica (1.083 mL g−1), the appropriate amount of the pre-polymerisation mixture was calculated for a complete filling of the pores. Ninety percent of this amount was added to 2 g of peptide-modified silica in a flask and gently stirred with a steel spatula. Repeated nitrogen–vacuum cycles, each cycle lasting 30 min, were applied to force the monomer mixture into the silica pores. The flask was flushed with nitrogen at room temperature for 1 h and placed in a thermostatted water bath, where the polymerisation was thermally initiated at 60°C and allowed to continue for 24 h. The products were dried in a vacuum oven for 6 h at 40°C. Randomly imprinted polymers were also prepared in parallel by following the same procedure. In this case the template, FMOC–Phe–OH (1 mmol) was included in the pre-polymerisation mixture to obtain P(FMOC–Phe//Si). In all cases we obtained 3.8 g of polymer–silica composites, corresponding to 1.9 g polymer per g silica, from 2 g of peptide-modified silica and 1.94 mL of monomer mixture.
Preparation of molecularly imprinted polymers
A 2-g portion of the prepared silica–polymer composite (random and immobilised imprinted) and 50 mL of a 3 M aqueous NH4HF2 solution were placed in 50-mL a Teflon flask. The suspension was shaken at room temperature for 24 h and filtered through a glass funnel. The polymers were washed first with 100 mL water to remove unreacted and newly formed salts, then with 100 mL DMF to remove any traces of peptide and again with 100 mL acetone. The polymers were dried in a vacuum oven at 40°C for 12 h. In all cases, the weight loss following this treatment was around 50%.
HPLC measurements and evaluation
The particles were slurry packed into stainless steel columns (33×4 mm) using MeOH/H2O 80:20 (v/v) as pushing solvent. These were then evaluated using mobile phases of either pure MeCN or MeCN/H2O/AcOH (v/v/v) with varying contents of AcOH and water. The flow rate was 0.5 mL min−1 and 10-µL aliquots of a 1 mM solution of pure enantiomers were injected. The elution was monitored at 260 nm. Upon a change of the mobile phase, the column was washed with the new mobile phase until a stable baseline was obtained. The retention factors (k′, k′ l or k′ d ) and the separation factor (α) were calculated using the following formulae: k=(t R−t 0)/t 0,k′ l =(t l −t 0)/t 0, k′ d =(t d −t 0)/t 0 and α=k′ l /k d , where t R=the retention time of the injected molecule, t l =the retention time of the l enantiomer, t d =the retention time of the d enantiomer, and t 0 is the time of the void marker acetone.
Coupling of the fluorescence label
The obtained molecularly imprinted polymers or composites (0.04 g), HOBt and DCC were mixed in dry DCM (20 mL) and stirred for 30 min before a solution of 3-aminoquinoline (3AQ) (1.1 equivalents based on the theoretical amount of COOH groups of the polymer) in DCM was added dropwise. The solution was stirred for 24 h and filtered. The solid was then washed consecutively with 2×30-mL aliquots DCM and ethyl acetate. The particles were dried under vacuum at 40°C for 6 h.
Results and discussion
Characterisation of the solid-phase synthesis products
Each intermediate was characterised by elemental C, N microanalysis, infrared spectroscopy and fluorescent spectroscopy. From the change in carbon and nitrogen content, with reference to the starting material, the area density (D s) of the coupled ligand could be estimated, together with the associated coupling yield (Table 1). Assuming a maximum density of 8 µmol m−2 on the native silica gel, APS occupies approximately 50% of the available sites, which is in agreement with results reported in the literature [14]. The coupling of BOC–Gly–OH appeared quantitative and was accompanied by the appearance of strong characteristic amide bands in the IR spectrum (Fig. 4A). However, as indicated by the final area density of about 1 µmol m−2, the subsequent deprotection, even with a milder agent than TFA, still appears to remove part of the organic coating. The lower yield obtained by basing the calculation on the change in the carbon content indicates that the organic layer was removed from the support surface, thus including the APS part. The following steps appeared to occur in high yield and could, apart from the characteristic amide bands in the IR spectra, be followed visually by fluorescence microscopy (Fig. 5). Thus, coupling of FMOC–Phe–OH was accompanied by a strong particle fluorescence which completely disappeared upon deprotection. The area density of the final coupling products was 1–2 µmol m−2. Assuming a random ligand distribution, this corresponds to an average distance between ligands of 10–15 Å
Fluorescence micrographs (100x magnification) of A FMOC–Phe–Si and B the respective composite after the polymerisation. Note: after removal of the FMOC protecting group or the dissolution of silica, implying the removal of the molecular template, the samples no longer exhibit fluorescence properties
Characterisation of the molecularly imprinted polymers
Subsequent to the template immobilisation, the pores of the immobilized amino acid or peptide templates were filled with a mixture of MAA, EDMA and AIBN (Fig. 3). This mixture was thereafter thermally cured at 60°C. Dissolution of the silica mould by treatment with a solution of NH4HF2 (aq) resulted in organic polymer beads with a size and morphology reflecting those of the original silica mould (Figs. 6 and 7). The extent of removal of the silica and peptide template was revealed by the elemental analysis of the final polymer product (Table 1). Thus, the carbon and nitrogen contents indicated that more than 95% of the template was removed upon the fluoride treatment. Infrared spectra showed no peaks that could be assigned to remaining silica and were otherwise similar to those of the bulk material (Fig. 4B). The high surface area and mesoporosity (Table 1, Fig. 7) are also close to those of the precursor particles (350 m2 g−1). Particularly striking is the narrow pore size distribution observed, around 6–8 nm, which stands in stark contrast with the broad distribution observed for conventional bulk materials [18]. Furthermore, the low bulk swelling factors (<1.15 mL mL−1 in acetonitrile) imply that the materials have a relatively homogenous distribution of cross-links. This is also in contrast to their bulk-imprinted counterparts. To investigate the accessibility of the carboxylic acid functional groups on the surface, these were fluorescently labelled by reaction with 3AQ. The labelled particles were then studied with respect to the distribution of the fluorescence intensity between and within the particles. Only a weak intensity was observed after labelling the particles still containing the silica template (Fig. 6C), which efficiently blocks access to the carboxylic acid groups. However, particles no longer containing the silica template exhibited a strong fluorescence, indicative of a high accessibility of the surface carboxylic acid groups (Fig. 6D).
Characterisation of the polymer particles in the chromatographic mode
In order to investigate the effect of template immobilization, two materials prepared using free FMOC–Phe–OH (FMOC–Phe//Si) or FMOC–Phe–OH coupled to APS-Si (FMOC–Phe–Si) as templates (Table 1), were compared. As seen in Fig. 8A the polymer obtained using the latter template strongly retained N-protected phenylalanine derivatives, including the dipeptide FMOC–Phe–Gly–OH. Much larger retention factors (k′) (ca. 5 times) were observed on these phases compared to those prepared using soluble FMOC–Phe–OH as template. Meanwhile, phenylalanine derivatives containing free amino groups were retained similarly on both materials. This might be due to the fact that the N-protecting group FMOC also strongly contributes to the observed retention. In view of the similar template load used when preparing both materials, the enhanced retention factors are probably due to a higher accessibility of the surface-confined binding sites. This behaviour contrasted with the observed enantioselectivity of the materials, where P(FMOC–Phe//Si) exhibited a higher selectivity (α=1.5) than the surface-imprinted material P(FMOC–Phe–Si) (α=1.1) (Fig. 8B). Thus, embedded less accessible sites exhibit higher structural fidelity than surface-exposed sites of higher accessibility. This provides supporting evidence for the results reported by Gagne et al. on the relationship between site accessibility and fidelity in chiral bisnaphthol-imprinted materials [19].
Retention of amino acid derivatives and peptides injected (10 µL of 1 mM stock solutions) on columns (33×4 mm) packed with A P(FMOC–Phe–Si) and P(FMOC–Phe//Si) comparing surface and random imprinted polymers, B P(FMOC–Phe–Si) and P(FMOC–Phe//Si) comparing the enantioselectivity, C P(FMOC–Phe–Gly–Si), P(FMOC–Phe–Si), P(BOC–Gly–Phe–Si) and P(BOC–Gly–Si) using acetonitrile as a mobile phase at a flow rate of 0.5 mL min−1. D shows the elution profiles of FMOC–Phe–Gly–OH corresponding to the data shown in C
The dipeptide-imprinted materials exhibited an interesting retention behaviour. As seen in Fig. 8C and D, FMOC–Phe–Gly–OH is approximately twice as strongly retained on P(FMOC–Phe–Gly–Si) than on P(FMOC–Phe–Si) and about 15 times stronger than on P(BOC–Gly–Si). However, since similar relative retentions were seen for FMOC–Phe–OH on the three materials this indicates that the main contribution to the affinity is a different surface density of templated sites. Such difference can be expected from the different template coverage on the surface of the silica mould. Retention behaviour is the product of the amount of accessible sites and their affinity relative to the solute. In view of the approximately 2 times higher ligand coverage of FMOC–Phe–Gly–Si (1.81 µmol m−2) as compared with FMOC–Phe–Si (0.27 µmol m−2) (Table 1) and the concomitant doubling of the retention of FMOC–Phe–Gly–OH substrate (Fig. 8C and D), it seems that the site density is the controlling factor rather than the affinity of those sites towards the target. It is also clear that glycine contributes only weakly to the observed retention. Thus, BOC–Gly–OH, H–Gly–OEt and H–Gly–Phe–OH are retained weakly on all the above materials, but still about 2 times more retained on the polymers imprinted with the Gly motif P(BOC–Gly–Si) and P(BOC–Gly–Phe–Si) than on P(FMOC–Phe–Gly–Si) and P(FMOC–Phe–Si). Similar behaviour is observed also in water-containing mobile phases. For biological applications, the retention behaviour in aqueous mobile phases is important. With 5% water (Fig. 9) a pronounced selectivity for peptides containing the imprinted dipeptide motif is seen. This included also larger peptides containing the H–Phe–Gly motif at the N-terminus. Thus, H–Phe–Gly–Gly–Phe–OH is similarly retained to H–Phe–Gly–NH2, with a retention factor of almost 6 on P(FMOC–Phe–Gly–Si). This exceeds the retention observed on the exact complement P(H–Phe–Gly–Si). A plausible explanation for this behaviour may be found in the amount of immobilized ligands and how these are presented at the surface. The free amino groups resulting from the deprotection may interact with neighbouring silanol [16] groups, leading to a poor orientation of the ligand for the creation of high-fidelity templated sites; this would not occur for the protected counterpart. Further evidence for the presence of peptide-discriminating sites is provided by the retention behaviour of dipeptide H–Gly–Phe–OH with the inverse amino acid sequence. Opposite to the other dipeptides this is most strongly retained on the materials imprinted with the nearest complements used in this study, namely H–Gly–Si, BOC–Gly–Si and BOC–Gly–Phe–Si. Importantly, the retention exceeded those observed using the conventional bulk materials [20]. Finally, we showed that the heptadecapeptide nociceptin (H–FGGFTGARKSARKLANQ–OH), containing the H–Phe–Gly N-terminus was approximately 2 times more strongly retained on the dipeptide N-terminus complement P(H–Phe–Gly–Si) than on P(H–Gly–Si) and 1.2 times more retained on the P(FMOC–Phe–Gly–Si) than on P(BOC–Gly–Si) (Fig. 9)
Conclusions
Hierarchical imprinting allows the preparation of uniformly shaped and sized molecularly imprinted polymer particles with narrow pore size distributions and surface-confined binding sites. In an extension of this approach the crude products of solid-phase synthesis can be directly used as templates. This may lead to new recognition elements for biological macromolecules including, other than the peptide phases described here, nucleic acids and oligosaccharides. We are presently investigating the effect of the epitope size and the inorganic template porosity on the recognition properties of the imprinted polymers in order to extend the hierarchical imprinting technique to the recognition of other biologically active peptides.
References
Sellergren B (ed) (2001) Molecularly imprinted polymers. Man made mimics of antibodies and their applications in analytical chemistry. Elsevier, Amsterdam, vol 23; Wulff G (2002) Chem Rev 102(1):1
Perez N, Whitcombe MJ, Vulfson EN (2001) Macromolecules 34:30
Matsui J, Okada M, Tsuruoka M, Takeutchi T (1997) Anal Commun 34:85
Ye L, Cormack P, Mosbach K (1999) Anal Com 36:35; Tamayo FG, Casillas JL, Martin- Esteban A (2003) Anal Chim Acta 482(2):165
Sulitzky C, Rücket B, Hall AJ, Lanza F, Unger KK, Sellergren B (2002) Macromolecules 35(1):79; Sellergren B, Rückert B, Hall AJ (2002) Adv Mat14:1204
Dai S, Burleight MC, Ju YH, Gao HJ, Lin JS, Pennycook SJ, Barnes CE, Xue ZL (2000) J Am Chem Soc 122:992
Yilmaz E, Haupt K, Mosbach K (2000) Angew Chem Int Ed 39(12):2115
Sellergren B, Büchel G (1999) WO01/32760 A1
Titirici MM, Hall AJ, Sellergren B (2002) Chem Mater 14:21
Venton DL, Gudipati E (1996) Biochim Biophys Res Commun 227:419
Hjerten S, Liao J-L, Nakazato K, Wang Y, Zamaratskaia G, Zhang H-Y (1997) Chromatographia 44:227
Hirayama K, Burow M, Morikawa Y, Minoura N (1998) Chem Lett 731
Titirici MM, Hall AJ, Sellergren B (2003) Chem Mater 15:822
Carlier E, Guyot A, Revillon A (1991) React Polym 16:115
Vansant EF, Van Der Voort P, Vrancken KC (1995) Characterisation and chemical modification of the silica surface. Elsevier, Amsterdam, vol 93
Sherrington DC, Hodge P (1998) Synthesis and separation using functional polymers. Wiley, Chichester
Kaiser E, Picart F, Kubiak T, Tam JP, Merrifield RB (1993) J Org Chem 58:5167
Sellergren B, Shea KJ (1993) J Chromatogr 635:31
Gagne MR, Becker JJ, Brunkan NM (2000) Polym Prep 41:404
Rachckov A, Minoura N (2001) Biochem Biophys Acta 1544:255
Acknowledgements
We thank Gunnar Glasser (MPI für Polymerforschung, Mainz) for recording of the SEM micrographs. Financial contribution from the Deutsche Forschungsgemeinschaft (DFG) under Project 777/5–1 is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Titirici, M.M., Sellergren, B. Peptide recognition via hierarchical imprinting. Anal Bioanal Chem 378, 1913–1921 (2004). https://doi.org/10.1007/s00216-003-2445-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-003-2445-5