Abstract
We examine vibrational energy transfer across the heme–protein and protein–solvent interfaces of cytochrome c, using, as appropriate, classical, semiclassical, and quantum approaches. To characterize energy flow across the interface between the heme and the rest of cytochrome c, we calculate communication maps for the protein in its native structure as well as two structures with Met80 dissociated from the heme at 300 K. The response to excess energy in the heme is mediated by covalent and hydrogen bonds to the heme, as well as several through-space interactions, including those involving the dissociated Met80. This observation suggests no energy flow bottleneck between the heme and Met80 that would impede rebinding kinetics at 300 K. We examine the possibility of additional bottlenecks to energy flow by calculating the temperature dependence of the ergodicity threshold in an imidazole-ligated Fe-porphyrin system that constitutes the core of the heme–histidine complex. The ergodic threshold, which we calculate quantum mechanically, corresponds to a temperature of about 140 K. We also address the flow of excess vibrational energy from the protein to the solvent. We calculate the thermal boundary conductance between cytochrome c and water semiclassically over a range of temperatures and find that the protein–water interface poses no greater resistance to thermal flow than the protein itself.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Pathways and bottlenecks to energy flow in proteins mediate chemical reaction kinetics including allosteric transitions, charge transfer, and ligand binding and dissociation [1–9]. For this reason, there have been numerous experimental and computational studies of vibrational energy relaxation in heme proteins [10–22]. Cytochrome c, a heme protein consisting of a single polypeptide chain containing 104 amino acid residues organized into a series of five α helices and six β turns, is an important participant in electron transfer for ATP synthesis in mitochondria and as signal amplifier in cell apoptosis [23]. The heme active site in cytochrome c has a six-coordinate low-spin iron, with His18 and Met80 bound as internal ligands. A number of experimental studies [24–26] have elucidated its ultrafast dynamics following photoexcitation, sometimes leading to ligand dissociation and recombination. Recent computational work has explored the non-adiabatic transitions involved in ligand recombination using ab initio potential energy surfaces [27] revealing heterogeneity in the rebinding kinetics determined by protein and solvent structure and temperature [28]. The rebinding kinetics can also be influenced by energy flow. Bu and Straub reported detailed molecular dynamics (MD) simulations of vibrational energy relaxation from the heme of cytochrome c in its native state [29]. In this article, we further study vibrational energy flow in cytochrome c, both in its native structure and in two structures with Met80 dissociated from the heme. We examine, using classical, semiclassical, and quantum methods, vibrational energy transfer across two interfaces: (1) between the heme and the rest of cytochrome c when Met80 is bonded and dissociated from the heme, and (2) the interface between cytochrome c and water.
Bottlenecks to vibrational energy transfer in a molecule as large as a protein can occur on different length scales. On the scale of a residue or cofactor, energy redistribution among vibrational modes within such a subspace of the protein depends on anharmonic coupling and a local density of states made up of these modes. Quantum mechanical first-order time-dependent perturbation theory can be adopted to calculate energy transfer by anharmonic coupling [30–32], an approach that becomes valid when the product of the anharmonic coupling and local density of states is at least of order 1 [33, 34]. When this criterion is not met, energy may still flow but the dynamics can be quite complex in molecules of even modest size [35–38]. On larger length scales, we may be interested in energy flow through residues of the protein, for instance, how bonds or other interactions between residues influence redistribution of excess energy in different parts of the protein [29, 39]. At even larger length scales, we can consider vibrational energy flow between the protein as a whole and the solvent, e.g., to address how excess energy already distributed within the protein flows into the solvent environment and whether additional resistance to energy redistribution is encountered at the protein–solvent interface [40–42].
In our study of energy flow across the heme–protein interface, we calculate communication maps [43], which we use to determine the response of the protein to excess energy in the heme at 300 K when Met80 is bound to and dissociated from the heme. Met80-heme rebinding kinetics depends on whether the Met80 orientation to the heme is upward or downward, i.e., the upward structure does not bind to the heme and must isomerize to the downward to bind [27, 28]. Isomerization kinetics can be mediated by energy flow in each structure as well as the barrier between structures [44–50], so we examine if an energy flow bottleneck is introduced when Met80 is dissociated from the heme. In the native structure, Met80 responds more rapidly than any other residue to excess energy in the heme. We find the response time to be diminished but still quite rapid when Met80 is dissociated from the heme, so that, at least at temperatures near 300 K, there appears to be no energy flow bottleneck between the heme and Met80. Dissociation of Met80 from the heme does not affect the response of other parts of the protein to excess vibrational energy in the heme.
Another way in which energy flow could influence the rate constant for Met80-heme rebinding involves limited or slow energy relaxation within the heme itself, which might occur at low temperature. We can think of the binding region as a system of many coupled nonlinear oscillators, which may exhibit ergodicity only above an energy threshold [51–55]. We examine the energy and temperature dependence of ergodicity quantum mechanically in an imidazole-ligated Fe-porphyrin system that forms the binding region of the heme. Vibrational relaxation in that system was studied by Straub and coworkers using quantum mechanical time-dependent perturbation theory [31]. We use the vibrational frequencies calculated in that work to estimate the temperature-dependent ergodicity threshold, which we find to be near 140 K. We thus expect limited or sluggish energy flow within the binding region at lower temperatures.
Having examined how excess energy flows from the heme into the rest of the protein, we consider energy flow across the boundary between cytochrome c and solvent. We coarse grain the system beyond the level of the residues and address thermal transport across the interface of the protein and bulk water. Thermal transport within proteins has been studied by MD simulations [56–58], computational approaches based on the vibrational modes of the system [45, 59, 60], and experimentally by time-resolved spectroscopy [61–64]. Below we adopt a semiclassical approach [65] to calculate the resistance to thermal flow across the cytochrome c–water interface and compare the result with the resistance to thermal flow in proteins computed earlier [60] to determine whether the flow of excess vibrational energy in cytochrome c is impeded more by the protein itself or its interface with water.
In the following section, we summarize the calculation of communication maps to describe the response of cytochrome c to excess vibrational energy in the heme and thermal boundary conductance at the cytochrome c–water interface. In Sect. 3, we present and discuss our results. Concluding remarks are given in Sect. 4.
2 Methods
2.1 Communication maps and molecular simulations
Protein structures and calculation of normal modes, which was carried out using the CHARMM simulation package with the CHARMM28 force field, have been described in previous work by two of us [27, 28]. The sequence of 1HRC from the PDB was used to define the human cytochrome c system, and the crystal structure was taken as the initial configuration. Using the normal modes for cytochrome c in the native structure and two dissociated structures, i.e., upward and downward [27, 28], we computed frequency-resolved communication maps. The calculation of communication maps has been detailed elsewhere [43], and in this subsection, we summarize the method. Thermal transport coefficients have been computed for molecules and other nanoscale objects using a number of approaches [66–68]. For coarse-graining a sizable molecular system such as a protein, a useful starting point is the heat current operator in harmonic approximation [69]
where α and β are two modes of the protein, and a † and a are, respectively, raising and lowering operators. The coefficient, S αβ , corresponding to the protein as a whole can be expressed in terms of the Hessian matrix, H, and eigenmodes, e, of the object. We break the coefficient up into contributions from various regions. The contribution to the energy flux between regions A and A′ to the matrix elements of S is [43]
where R l is the position of atom l and r is a coordinate (x, y or z). When the regions A and A′ span the protein, Eq. (2) expresses the matrix elements of the heat current operator for the whole system in harmonic approximation given in Ref. [69]. Similarly, we can sum over all regions to obtain S ∝β for the whole molecule. Considering only energy flow between the local regions A and A′, we write the local energy diffusivity in mode α in harmonic approximation as
When A and A′ span the molecule, Eq. (3) gives the mode diffusivity, from which the coefficient of thermal conductivity, κ, can be expressed for the whole molecule, κ = Σ α C α D α , where C α is the heat capacity per unit volume of the molecule for mode α, given by \( C_{\alpha } = k_{B} \left( {\beta \hbar \omega_{\alpha } } \right)^{2} \frac{{e^{{\beta \hbar \omega_{\alpha } }} }}{{\left( {e^{{\beta \hbar \omega_{\alpha } }} - 1} \right)^{2} }} \). Otherwise \( D_{\alpha }^{{\{ AA'\} }} \) is a local mode diffusivity for energy flow between A and A′. The coefficient of thermal conductivity has been calculated for proteins [45, 59, 60], and here, we identify the regions of the protein that primarily contribute to thermal transport through the molecule. For a practical calculation on a finite-sized system, we substitute a rectangular window of width η for the delta function, which should be large enough to envelop several vibrational modes. For the results presented here, we calculated the mode diffusivity using η = 15 cm−1, which is large enough to include many modes in the averaging; results for D α did not change significantly with larger η. Communication maps can then be constructed by plotting \( D_{\alpha }^{{\{ AA'\} }} \) for all A and A′ at frequencies, ω α . We note that we have considered the extent to which anharmonicity might affect the pathways we calculate in the communication maps in a previous study. In a recent calculation of communication maps for PYP [70], we identified the same energy transport pathways from the chromophore as Ishikura and Yamato [71] did using an anharmonic treatment.
In Sect. 3, we use communication maps to locate energy transport channels that include the heme. We present results for the thermal average over the frequency-resolved communication maps for the structures at 300 K, using for averaging the communication maps constructed at 50, 100 cm−1, and continuing in 50 cm−1 intervals until 400 cm−1, above which the communication maps contribute very little to the thermal average due to the small values of \( D_{\alpha }^{{\{ AA'\} }} \) at higher frequency. Each of these frequency-resolved maps is actually an average over maps for 4 modes closest in frequency to the designated frequency. The thermally averaged communication map is obtained assigning a Boltzmann weight to the frequency-resolved communication maps and is specifically calculated as \( D^{{\{ AA'\} }} = \int {d\omega^{{}} } \rho (\omega )_{{}} D^{{\{ AA'\} }} (\omega )e^{{ - \hbar \omega /k_{B} T}} \). Using the thermally averaged frequency-resolved communication map, we calculate, in addition to the local thermal diffusivity, a response rate of one residue to vibrational excitation in another, corresponding to the inverse of the first mean passage time, which we define as \( D^{{\{ AA'\} }} /R^{2} \) [14], where R is the distance from the center of mass of A to the center of mass of A′.
To complement the analysis of energy transport using communication maps, we also carried out classical non-equilibrium simulations of energy transport from the heme. We in effect heated the Fe and neighboring atoms of the porphyrin to 300 K and followed the transfer of energy over the next few picoseconds to the rest of the molecule, which was initially at 0 K. Similar starting conditions have been applied in earlier molecular simulations of proteins to elucidate energy transport pathways [72] and by us for a homodimeric hemoglobin [14]. We carried out this analysis in harmonic approximation to provide a realistic representation of the thermal population of the vibrational modes of the protein. On the short times of this analysis, there is no time for configurational changes of the protein or confined water molecules [73]. Details of our non-equilibrium simulation approach, including other applications to heme proteins, are given in Refs. [60, 74, 75].
2.2 Cytochrome c–water thermal boundary conductance
The thermal boundary conductance, h Bd , between two subsystems, 1 and 2, the inverse of the thermal boundary resistance or Kapitza resistance [65, 76], is expressed in terms of the heat flow, \( \dot{Q} \), driven by the difference in temperature, ΔT, between the two sides of the interface and the area of the interface, A, \( h_{Bd} = \frac{{\dot{Q}}}{A \Updelta T} \). Heat flow across the interface can be expressed in harmonic approximation in terms of the vibrational excitations, each with energy \( \hbar \omega \), that pass through it. The vibrational mode density per unit volume on side j is \( \bar{\rho }_{j} (\omega ) \) and the mode occupation number is \( n(\omega ,T) = \left( {\exp (\beta \hbar \omega ) - 1} \right)^{ - 1} \), where \( \beta = 1/k_{B} T \) and the speed of propagation on side j is \( v_{j} (\omega ) \). The thermal boundary conductance is then expressed as [76] \( h_{Bd} = \frac{1}{4}\frac{\text{d}}{{{\text{d}}T}}\int {d\omega_{{}} } \hbar \omega_{{}} v_{1} (\omega )\bar{\rho }_{1} (\omega )n(\omega ,T)\alpha (\omega ) \), where α(ω) is the transmission probability. In a convenient approach to eliminating the transmission probability, α(ω), the diffuse mismatch model assumes that after crossing the boundary, a vibrational excitation has no memory of which side of the interface it emerged from, i.e., there is diffuse scattering of vibrational energy at the boundary between the protein and water, giving [76]
We apply Eq. (4) to estimate the boundary conductance at the interface between cytochrome c and water. For the calculations discussed in the following section, the volume of the protein is estimated assuming a sphere with radius corresponding to the protein’s radius of gyration, which for cytochrome c is 14 Å. We have used the vibrational mode density of cytochrome c reported in Ref. [74] and for water in Ref. [77], both calculated in harmonic approximation. For the speed of sound in water and cytochrome c, we have used the values 15 Å ps−1 [77] and 22 Å ps−1 [41, 74], respectively.
3 Results and discussion
3.1 Communication maps of vibrational energy flow from heme into protein
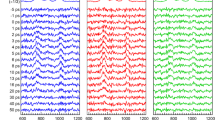
Before discussing the communication maps, we first examine results of non-equilibrium simulations of vibrational energy flow from the center of the heme to the rest of the protein. In Fig. 1, we plot the time evolution to 3 ps of vibrational energy in cytochrome c, which is initially cold everywhere except the Fe atom and the atoms of the porphyrin directly bonded to it.
The non-equilibrium simulations were carried out for the native structure of cytochrome c and can be directly compared with earlier MD simulations of vibrational energy flow in cytochrome c carried out by Bu and Straub [29]. In that study, the entire heme was heated initially, whereas in the simulations we present here, we have considered energy flow from a “hot” region consisting only the Fe and a few atoms of the porphyrin. By 1 ps, we observe that more than 10 % of the total energy in the protein is contained in the Cys14, Cys17, His18, and Met80, i.e., those residues covalently bonded to the heme. However, there is little energy flow into the residues that hydrogen bond to the heme, residues that Bu and Straub [29] found to open up additional energy transport pathways from the heme. The difference is likely due to the different initial conditions in these two studies. By 2–3 ps, we observe that excess energy originally in the heme is found mainly in the two cysteines and Met80.
We calculated communication maps for cytochrome c, not only in its native state with Met80 bonded to the heme, but also for two structures where Met80 is dissociated, which we refer to as upward and downward due to the orientation of Met80 with respect to the heme. The response of the protein to excess energy in the heme, and to excess energy in different parts of the heme, obtained from the communication maps, is plotted in Figs. 2 and 3. We define the response rate of one residue to vibrational excitation in another as \( D^{{\{ AA'\} }} /R^{2} \), where R is the distance from the center of mass of A to the center of mass of A′, and \( D^{{\{ AA'\} }} \) is the local energy diffusivity between A and A′. The largest response rates from the heme that we observed were of order 0.1 fs−1.
Largest values of D/R 2 between the heme, or parts of the heme, and protein residues obtained from communication maps of cytochrome c at 300 K. In a, the heme is complete, whereas in b and c the largest D/R 2 values are plotted for interactions between the residues and the porphyrin ring and heme side chains, respectively
Communication map predictions of the hot residues following excitation of the ring portion of the heme in the bonded (a) and (c) and the dissociated (b) and (d) systems. As in Fig. 2, the largest heme residue D/R 2 values are plotted for the full heme (a) and (b) and for the porphyrin region only (c) and (d). The bonded structure exhibits much faster energy flow into Met80 than the dissociated system, though communication with Met80 remains quite rapid even without the chemical bond between the heme and Met80
Figure 2a shows the residues that respond fastest to excess vibrational energy in the heme of the bonded form of cytochrome c based on the communication maps, which are seen to include Cys14, Cys17, and Met80, all covalently bonded to the heme. Other residues that respond particularly rapidly include Thr49 and Asp50, which are located on the periphery of the heme and hydrogen bond to the side chains, as well as Trp59, which has been probed by Mizutani coworkers by picosecond time-resolved anti-Stokes ultraviolet resonance Raman (UVRR) measurements [26]. In the UVRR experiments, an energy transfer time of 1–3 ps from the heme to Trp59 was found. To examine whether or not the response of these residues is due to excess energy in the Fe-porphyrin portion of the heme or the heme side chains, we computed communication maps in which we separated these two parts of the heme. \( D^{{\{ AA'\} }} /R^{2} \) for the response to excess energy in the Fe-porphyrin is plotted in Fig. 2b, and for the response to excess energy in the heme side chains in Fig. 2c. We observe that the fast response of Met80 is due to interaction with Fe-porphyrin, whereas the fast response of the cysteine residues is due to interaction with both the porphyrin and the side chains. The fast response of Thr49, Asp50, Trp59, as well as some other residues that are peripheral to the heme is due to the interaction with the heme side chains. Similarly, through-space interactions with the heme appear to originate from the heme side chains, e.g., Arg38, which was also found by Bu and Straub [29] to funnel energy from the heme. The results of the communication maps, in which the response to excess energy in the heme core and the heme side chains were separately examined, are consistent with the flow of energy found in the non-equilibrium simulations plotted in Fig. 1. In the latter, only the center of the porphyrin was initially hot, revealing an energy transport pathway through the two cysteines and Met80 but none through Thr49 or Asp50.
We consider now the effect of Met80 dissociation on the response of the protein to excess energy in the heme. Figure 3 compares the response rates for excess energy in the heme for the bonded (Fig. 3a) and upward (Fig. 3b) forms and for excess energy in only the porphyrin region of the bonded (Fig. 3c) and upward orientations of Met80 with respect to the heme (Fig. 3d) (The results for the upward and downward orientations are very similar and only the former are shown.). The most striking difference between the bonded and the unbonded forms is the reduction in the response time of Met80 to excess vibrational energy in the heme when Met80 is not bonded. Indeed, Met80 goes from the residue that responds fastest to excitation in the Fe-porphyrin portion of the heme to a residue that responds more slowly than either of the cysteines or His18 when Met80 is dissociated from the heme. Nevertheless, we still observe a significant through-space response of Met80 to excess energy in the heme, comparable to the response rate of other nearby residues, some of which hydrogen bond to the heme. The rapid response of Met80 to excess energy in the heme indicates that the rate constant for rebinding of Met80 to the heme is not regulated by bottlenecks to vibrational energy flow between them at 300 K.
When the temperature is low, however, bottlenecks to vibrational energy flow within the heme itself could emerge. At low temperatures, anharmonic coupling among the vibrational modes of the heme might not be sufficiently large to ensure rapid energy flow. Straub and coworkers carried out quantum mechanical time-dependent perturbation calculations of vibrational relaxation in an imidazole-ligated Fe-porphyrin system (Fig. 4), examining the anisotropic relaxation from specific vibrational modes of the complex. Using the vibrational mode frequencies that they report, we can estimate the internal energy in this complex that is needed for ergodic flow of vibrational energy in this molecule using local random matrix theory (LRMT) [33, 34]. LRMT predicts that the ergodicity threshold depends on a local density of states, not the total vibrational density of states of the molecule. The ergodicity threshold occurs where the product of the local density of states and the size of the anharmonic matrix elements coupling states on the energy shell is of order 1, or when [33, 34]
where Q is the distance in terms of quantum numbers between two states coupled by the set of matrix elements, V Q , and ρ Q is the local density of states. Cubic anharmonic terms couple states at most a distance Q = 3 in vibrational quantum number space, as a cubic term can transfer one quantum of energy from one mode to two quanta in other modes.
To estimate the ergodicity threshold in terms of the vibrational frequencies of the system, we make use of a scaling relation to estimate average anharmonic matrix elements, V ij, coupling states \( \left| {i\rangle } \right. \) and \( \left| {j\rangle } \right. \). The assumption underlying the scaling is the expectation and observation that coupling between states that lie a “distance” Q from one another in quantum number space, where \( Q = \sum\nolimits_{\alpha } {v_{\alpha } } \), and v α is the occupation number difference in mode α between the two vibrational states, decrease with Q roughly as V ij ~C −Q, where C > 1 [78]. Gruebele and coworkers found empirically that for modest-sized organic molecules [78]
which yields the expected exponential decrease in the matrix elements with Q while accounting for the occupation of individual modes. In Eq. 6, n α is the number of quanta in mode α, ν α is the occupation number difference in mode α between two vibrational states, and a and b are constants. For V ij expressed in cm−1, a and b are chosen to be 3,050 and 270, respectively. We have compared estimates using Eq. (6) for the anharmonic constants with ab initio calculations of the anharmonic constants in peptide–water complexes and found that, on average, the results are quite similar [79].
The value of T(E) calculated with Eq. (5) depends on the energy, E, of the molecule. Since we consider the heme at a given temperature, we take the thermal energy to be \( \left\langle E \right\rangle = \sum\nolimits_{\alpha } {\left\langle {n_{\alpha } } \right\rangle \hbar \omega_{\alpha } } \), where the temperature-dependent mode occupation, \( \left\langle {n_{\alpha } } \right\rangle \), is given by \( \left\langle {n_{\alpha } } \right\rangle = \left( {e^{{\hbar \omega /k_{B} T}} - 1} \right)^{ - 1} \).
The result we obtain for T(E) for the imidazole-ligated Fe-porphyrin complex is plotted in Fig. 5. We find the ergodicity threshold for this system at energy of about 1,040 cm−1. The corresponding temperature dependence of T is plotted in the inset to Fig. 5, where we estimate the temperature of the ergodicity threshold to be around 140 K. That temperature lies below the protein dynamical transition of around 180 K, so that at all temperatures where conformational transitions are relatively facile we expect vibrational energy flow within the heme to be facile as well. The rate of energy flow from an initially excited state is dictated by the local density of states to which it is coupled [33, 34], which mediates the mode-specific pathways observed in calculations [31].
Transition parameter, T, as a function of total energy in the imidazole-ligated Fe-porphyrin system shown in Fig. 2. The ergodicity transition occurs at the energy where T = 1, indicated by the horizontal line, which in this molecule appears around 1,200 cm−1. Inset shows T as a function of temperature, where again the critical T = 1 is indicated by a horizontal line. The ergodicity threshold is found for this molecule to be around 140 K
3.2 Cytochrome c–water thermal boundary conductance
The coefficient of thermal conductivity for several proteins has been computed in linear response approximation from two different approaches, both yielding values of 0.1–0.3 W m−1 K−1 [40, 60], much smaller than the thermal conductivity of water, which is about 0.6 W m−1 K−1. Here, we examine whether the boundary between cytochrome c and water introduces additional resistance to vibrational energy flow beyond the resistance to thermal flow in the protein.
We calculated the thermal boundary conductance, h Bd , for cytochrome c and water with Eq. (4) from 200 to 320 K and plot the result in Fig. 6. In earlier studies, we calculated the thermal boundary conductance between myoglobin and water [41] and between GFP and water [42]. The former turns out to be about 1 % larger than the values we found for cytochrome c and water, so essentially indistinguishable from the cytochrome c–water results plotted in Fig. 6. However, the results for the GFP–water interface that we calculated in the same way are different, and we plot them in Fig. 6 with our calculations for cytochrome c and water. For the cytochrome c–water interface, we find a modest increase in boundary conductance over the plotted range of temperature, from about 240 MW K−1 m−2 at 200 K to about 300 MW K−1 m−2 at 320 K, with a value of 299 MW K−1 m−2 at 300 K. This compares with about 260 MW K−1 m−2 at 200 K to about 340 MW K−1 m−2 at 320 K for the GFP–water interface, with a value at 300 K of 329 MW K−1 m−2, which is about 10 % larger than the thermal boundary conductance we computed for the cytochrome c–water interface. While we are not aware of molecular simulation studies of the thermal boundary conductance between cytochrome c and water with which to compare, we note the values that we compute at 300 K are reasonably consistent with those computed for four different proteins (including GFP but not cytochrome c) by molecular simulations at 300 K and reported in Ref. [40]. We plot the range of those boundary conductance values in Fig. 6 for comparison.
Boundary thermal conductance, h Bd , calculated for cytochrome c and water (solid curve), with the result for GFP and water (dotted curve) plotted for comparison. The vertical line at 300 K represents the range of values for h Bd obtained by molecular simulations for four different proteins reported in Ref. [40]
The thermal boundary conductance between cytochrome c and water is roughly 2–3 times as large as the thermal boundary conductance computed for the interface between water and a monolayer of hydrocarbon chains with –OH head groups [80]. Vibrational energy transport across the cytochrome c–water interface is relatively efficient compared with interfaces involving other organic molecules that hydrogen bond to water.
The relative thermal resistance within a protein to thermal resistance at the boundary between the protein and water is given by the dimensionless Biot number, Bi, defined as Bi = h Bd L/κ, where L is a length scale, often taken as the volume to surface area, and κ is the coefficient of thermal conductivity [40]. At 300 K, we find h Bd = 299 MW K−1 m−2. While we have not computed the coefficient of thermal conductivity for cytochrome c, we use as a representative value κ = 0.25 W K−1 m−1, obtained for myoglobin in an earlier study [60]. We approximate the length, L, as the volume to surface area, R/3, which, using a 14 Å radius of gyration noted earlier, is about 0.5 nm for cytochrome c. These estimates yield a value for the Biot number of Bi ≈ 0.6, so that thermal resistance at the protein–water interface is comparable, perhaps a bit smaller, than thermal resistance in cytochrome c.
4 Concluding remarks
We have examined vibrational energy transfer across the heme–protein and protein–water interfaces of cytochrome c. To characterize energy flow across the heme–protein interface, we calculated frequency-resolved communication maps for cytochrome c in its native structure as well as two forms of the protein with Met80 dissociated from the heme and computed the thermal average at 300 K. We found the response of cytochrome c in its native structure at 300 K to excess energy in the heme to be mediated primarily by covalent and hydrogen bonds to the heme, as well as several specific through-space interactions with the heme side chains. Included in the latter is Trp59, which has been probed by Mizutani coworkers by picosecond time-resolved anti-Stokes UVRR measurements [26]. The communication maps reveal that Trp59 lies along an energy transport pathway that emerges from the heme side chains, consistent with the picosecond response time to excitation of the heme observed in the UVRR experiments [26]. In earlier work, we found vibrational energy transport pathways to be robust with respect to thermal motion of proteins around their native structure [81]. However, the energy transport pathways in cytochrome c, like those for electron transfer [82], could be altered when this protein interacts with its cellular environment.
The communication maps indicted that energy flow from the heme to Met80 remains rapid when it is dissociated from the heme, though not as fast as when it is covalently bonded. The rapid energy flow between heme and dissociated Met80 that we found at 300 K suggests no energy flow bottleneck between them, though bottlenecks may emerge at lower temperature, even within the binding region itself. To examine the latter possibility, we calculated the temperature dependence of the ergodicity threshold in an imidazole-ligated Fe-porphyrin system that constitutes the core of the heme–histidine complex, which we found to correspond to about 140 K, well below the protein dynamical transition of roughly 180 K.
We also addressed vibrational energy flow from cytochrome c into the solvent, adopting a more coarse-grained perspective than our computational study of the response of the protein to excess vibrational energy in the heme. We calculated the thermal boundary conductance between cytochrome c and water over a wide range of temperatures and found that the interface between the protein and water poses no greater resistance to thermal flow than the protein itself.
References
Leitner DM, Straub JE (2009) Proteins: energy, heat and signal flow. Taylor and Francis Press, New York
Smock RG, Gierasch LM (2009) Sending signals dynamically. Science 324:198–203
Fujisaki H, Straub JE (2005) Vibrational energy relaxation in proteins. Proc Natl Acad Sci (USA) 102:7626–7631
Leitner DM (2008) Energy flow in proteins. Ann Rev Phys Chem 59:233–259
Gunasekaran K, Ma B, Nussinov R (2004) Is allostery an intrinsic property of all dynamic proteins? Proteins Struct Func Bioinform 57:433–443
Nussinov R, Tsai C-J (2012) Allostery in disease and in drug discovery. Cell 153:293–305
Agarwal PK (2005) Role of protein dynamics in reaction rate enhancement by enzymes. J Am Chem Soc 127:15248–15256
Fang C, Frontiera RR, Tran R, Mathies RA (2009) Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature 462:200–205
Buchli B, Waldauer SA, Walser R, Donten ML, Pfister R, Bloechliger N, Steiner S, Caflisch A, Zerbe O, Hamm P (2013) Kinetic response of a photoperturbed allosteric protein. Proc Natl Acad Sci USA 110:11725–11730
Nagy AM, Raicu V, Miller RJD (2005) Nonlinear optical studies of heme protein dynamics: implications for proteins as hybrid states of matter. Biochim Biophys Acta 1749:148–172
Sagnella DE, Straub JE, Jackson TA, Lim M, Anfinrud PA (1999) Vibrational population relaxation of carbon monoxide in the heme pocket of carbonmonoxy myoglobin: comparison of time-resolved mid-IR absorbance experiments and molecular dynamics simulations. Proc Natl Acad Sci USA 96:14324–14329
Henry ER, Eaton WA, Hochstrasser RM (1986) Molecular dynamics simulations of cooling in laser-excited heme proteins. Proc Natl Acad Sci USA 83:8982–8986
Lian T, Locke B, Kholodenko Y, Hochstrasser RM (1994) Energy flow from solute to solvent probed by femtosecond ir spectroscopy: malachite green and heme protein solutions. J Phys Chem 98:11648–11656
Gnanasekaran R, Agbo JK, Leitner DM (2011) Communication maps computed for homodimeric hemoglobin: computational study of water-mediated energy transport in proteins. J Chem Phys 135:065103
Champion PM (2005) Following the flow of energy in biomolecules. Science 310:980–982
Sagnella DE, Straub JE (2001) Directed energy “funneling” mechanism for heme cooling following ligand photolysis or direct excitation in solvated carbonmonoxy myoglobin. J Phys Chem B 105:7057–7063
Takayanagi M, Okumura H, Nagaoka M (2007) Anisotropic structural relaxation and its correlation with the excess energy diffusion in the incipient process of photodissociated MbCO: high-resolution analysis via ensemble perturbation method. J Phys Chem B 111:864–869
Nagaoka M, Yu I, Takayanagi M (2009) Energy flow analysis in proteins via ensemble molecular dynamics simulations: time-resolved vibrational analysis and surficial Kirkwood-Buff Theory. In: Leitner DM, Straub JE (eds) Proteins: energy, heat and signal flow. Taylor & Francis Group, CRC Press, Boca Raton, pp 169–196
Mizutani Y, Kitagawa T (1997) Direct observation of cooling of heme upon photodissociation of carbonmonoxy myoglobin. Science 278:443–446
Koyama M, Neya S, Mizutani Y (2006) Role of heme propionates of myoglobin in vibrational energy relaxation. Chem Phys Lett 430:404–408
Sato A, Mizutani Y (2005) Picosecond structural dynamics of myoglobin following photodissociation of carbon monoxide as revealed by ultraviolet time-resolved resonance Raman spectroscopy. Biochem 44:14709–14714
Ye X, Demidov A, Champion PM (2002) Measurements of the photodissociation quantum yields of MbNO and MbO2 and the vibrational relaxation of the six-coordinate heme species. J Am Chem Soc 124:5914–5924
Ow Y-LP, Green DR, Hao Z, Mak TW (2008) Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol 9:532–542
Vos MH (2008) Ultrafast dynamics of ligands within heme proteins. Biochim Biophys Acta 1777:15–31
Zang C, Stevens JA, Link JJ, Guo L, Wang L, Zhong D (2009) Ultrafast proteinquake dynamics in cytochrome c. J Am Chem Soc 131:2846–2852
Fujii N, Mizuno M, Mizutani Y (2011) Direct observation of vibrational energy flow in cytochrome c. J Phys Chem B 115:13057–13064
Zhang P, Malolepsza E, Straub JE (2012) Dynamics of methionine ligand rebinding in cytochrome c. J Phys Chem B 116:6980–6990
Zhang P, Ahn SW, Straub JE (2013) “Strange kinetics” in the temperature dependence of methionine ligand rebinding dynamics in cytochrome c. J Phys Chem B 117:7190–7202
Bu L, Straub JE (2003) Simulating vibrational energy flow in proteins: relaxation rate and mechanism for heme cooling in cytochrome c. J Phys Chem B 107:12339–12345
Zhang Y, Fujisaki H, Straub JE (2009) Mode specific vibrational energy relaxation of amide I and II modes in N-methylacetamide/water clusters: the intra- and inter-molecular energy transfer mechanisms. J Phys Chem A 113:3051–3060
Zhang Y, Fujisaki H, Straub JE (2009) Direct evidence for mode-specific vibrational energy relaxation from quantum time-dependent perturbation theory. I. Five-coordinate ferrous iron porphydin model. J Chem Phys 130:025102
Leitner DM (2001) Vibrational energy transfer in helices. Phys Rev Lett 87:188102
Leitner DM, Wolynes PG (1996) Statistical properties of localized vibrational eigenstates. Chem Phys Lett 258:18–24
Leitner DM, Wolynes PG (1997) Vibrational mixing and energy flow in polyatomic molecules: quantitative prediction using local random matrix theory. J Phys Chem A 101:541–548
Keshavamurthy S (2013) Scaling perspective on intramolecular vibrational energy flow: analogies, insights and challenges. Adv Chem Phys 153:43–110
Semparithi A, Keshavamurthy S (2006) Intramolecular vibrational energy redistributions as diffusion in state space: classical-quantum correspondence. J Chem Phys 125:141101
Leitner DM, Gruebele M (2008) A quantum model of restricted vibrational energy flow on the way to the transition state in unimolecular reactions. Mol Phys 106:433–442
Gruebele M, Bigwood R (1998) Molecular vibrational energy flow: beyond the golden rule. Int Rev Phys Chem 17:91–145
Bu L, Straub JE (2003) Vibrational energy relaxation of ‘tailored’ hemes in myoglobin following ligand photolysis supports energy funneling mechanism of heme ‘cooling’. J Phys Chem B 107:10634–10639
Lervik A, Bresme F, Kjelstrup S, Bedeaux D, Rubi JM (2010) Heat transfer in protein–water interfaces. Phys Chem Chem Phys 12:1610–1617
Leitner DM (2013) Thermal boundary conductance and rectification in molecules. J Phys Chem B 117:12820–12828
Xu Y, Leitner DM (2014) Vibrational energy flow through the green fluorescent protein water interface: communication maps and thermal boundary conductance. J Phys Chem B. doi:10.1021/jp412141z
Leitner DM (2009) Frequency resolved communication maps for proteins and other nanoscale materials. J Chem Phys 130:195101
Berne BJ, Borkovec M, Straub JE (1988) Classical and modern methods in reaction rate theory. J Phys Chem 92:3711–3725
Leitner DM (2005) Heat transport in molecules and reaction kinetics: the role of quantum energy flow and localization. Adv Chem Phys 130B:205–256
Komatsuzaki T, Baba A, Kawai S, Toda M, Straub JE, Berry RS (2011) Ergodic problems for real complex systems in chemical physics. Adv Chem Phys 145:171–220
Leitner DM, Matsunaga Y, Li C-B, Komatsuzaki T, Shojiguchi A, Toda M (2011) Non-brownian phase space dynamics of molecules, the nature of their vibrational states, and non-RRKM kinetics. Adv Chem Phys 145:83–122
Li CB, Matsunaga Y, Toda M, Komatsuzaki T (2005) Phase space reaction network on a multisaddle energy landscape: HCN isomerization. J Chem Phys 123:184301
Shojiguchi A, Li CB, Komatsuzaki T, Toda M (2007) Fractional behavior in multi-dimensional Hamiltonian systems describing reactions. Phys Rev E 76:056205
Toda M (2005) Global aspects of chemical reactions in multidimensional phase space. Adv Chem Phys 130A:337–399
Ezra GS, Martens CC, Fried LE (1987) Semiclassical quantization of polyatomic molecules: some recent developments. J Phys Chem 91:3721–3730
Uzer T (1991) Theories of intramolecular vibrational energy transfer. Phys Rep 199(2):73–146
Keshavamurthy S, Ezra GS (1997) Eigenstate assignments and the quantum-classical correspondence for highly-excited vibrational states of the Baggot H2O Hamiltonian. J Chem Phys 107:156–179
Leitner DM, Wolynes PG (1996) Vibrational relaxation and energy localization in polyatomics: effects of high-order resonances on flow rates and the quantum ergodicity transition. J Chem Phys 105:11226–11236
Logan DE, Wolynes PG (1990) Quantum localization and energy flow in many-dimensional Fermi resonant systems. J Chem Phys 93:4994–5012
Lervik A, Bresme F, Kjelstrup S (2009) Heat transfer in soft nanoscale interfaces: the influence of interface curvature. Soft Matter 5:2407–2414
Nguyen PH, Park SM, Stock G (2010) Nonequilibrium molecular dynamics simulation of the energy transfer through a peptide helix. J Chem Phys 132:025102
Nguyen PH, Hamm P, Stock G (2009) Nonequilibrium molecular dynamics simulation of photoinduced energy flow in peptides: theory meets experiment. In: Leitner DM, Straub JE (eds) Proteins: energy, heat and signal flow. Taylor & Francis Group, CRC Press, Boca Raton, pp 149–168
Yu X, Leitner DM (2003) Vibrational energy transfer and heat conduction in a protein. J Phys Chem B 107:1698–1707
Yu X, Leitner DM (2005) Heat flow in proteins: computation of thermal transport coefficients. J Chem Phys 122:054902
Botan V, Backus EHG, Pfister R, Moretto A, Crisma M, Toniolo C, Nguyen PH, Stock G, Hamm P (2007) Energy transport in peptide helices. Proc Natl Acad Sci USA 104:12749–12754
Kholodenko Y, Volk M, Gooding E, Hochstrasser RM (2000) Energy dissipation and relaxation processes in deoxymyoglobin after photoexcitation in the Soret region. Chem Phys 259:71–87
Helbing J, Devereux M, Nienhaus K, Nienhaus GU, Hamm P, Meuwly M (2012) Temperature dependence of the heat diffusivity of proteins. J Phys Chem A 116:2620–2628
Müller-Werkmeister HM, Bredenbeck J (2014) A donor-acceptor pair for the real time study of vibrational energy transfer in proteins. Phys Chem Chem Phys 16:3261–3266
Hopkins PE (2013) Thermal transport across solid interfaces with nanoscale imperfections: effects of roughness, disorder, dislocations and bonding on thermal boundary conductance (Review Article). ISRN Mech Eng, 2013: 682586
Buldum A, Leitner DM, Ciraci S (1999) Thermal conduction through a molecule. Europhys Lett 47:208–212
Segal D, Nitzan A, Hänggi P (2003) Thermal conductance through molecular wires. J Chem Phys 119:6840–6855
Leitner DM, Wolynes PG (2000) Heat flow through an insulating nanocrystal. Phys Rev E 61:2902–2908
Allen PB, Feldman JL (1993) Thermal conductivity of disordered harmonic solids. Phys Rev B 48:12581–12588
Xu Y, Leitner DM (2014) Communication maps of vibrational energy transport in photoactive yellow protein. J Phys Chem A. doi:10.1021/jp411281y
Ishikura T, Yamato T (2006) Energy transfer pathways relevant for long-range intramolecular signaling of photosensory protein revealed by microscopic energy conductivity analysis. Chem Phys Lett 432:533–537
Ota N, Agard DA (2005) Intramolecular signaling pathways revealed by modeling anisotropic thermal diffusion. J Mol Biol 351:345–354
Gnanasekaran R, Xu Y, Leitner DM (2010) Dynamics of water clusters confined in proteins: a molecular dynamics simulation study of interfacial waters in a dimeric hemoglobin. J Phys Chem B 114:16989–16996
Yu X, Leitner DM (2003) Anomalous diffusion of vibrational energy in proteins. J Chem Phys 119:12673–12679
Yu X, Leitner DM (2006) Thermal conductivity computed for vitreous silica and methyl-doped silica above the plateau. Phys Rev B 74:184305
Swartz ET, Pohl RO (1989) Thermal boundary resistance. Rev Mod Phys 61:605–668
Yu X, Leitner DM (2005) Thermal transport coefficients for liquid and glassy water computer from a harmonic aqueous glass. J Chem Phys 123:104503
Bigwood R, Gruebele M, Leitner DM, Wolynes PG (1998) The vibrational energy flow transition in organic molecules: theory meets experiment. Proc Natl Acad Sci USA 95:5960–5967
Agbo JK, Leitner DM, Myshakin EM, Jordan KD (2007) Quantum energy flow and the kinetics of water shuttling between hydrogen bonding sites on trans-formanilide (TFA). J Chem Phys 127:064315
Acharya H, Mozdzierz NJ, Keblinski P, Garde S (2012) How chemistry, nanoscale roughness, and the direction of heat flow affect thermal conductance of solid–water interfaces. Ind Eng Chem Res 51:1767–1773
Agbo JK, Gnanasekaran R, Leitner DM (2014) Communication maps: exploring energy transport through proteins and water. Isr J Chem. doi:10.1002/ijch.201300139
Volkov AM, vanNuland NAJ (2012) Electron transfer interactome of cytochrome c. PLoS Comput Biol 8:e1002807
Acknowledgments
Support from the National Science Foundation (NSF CHE-0910669 and CHE-1361776 to DML, CHE-1114676 to JES) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Dedicated to Professor Greg Ezra and published as part of the special collection of articles celebrating his 60th birthday.
Rights and permissions
About this article
Cite this article
Agbo, J.K., Xu, Y., Zhang, P. et al. Vibrational energy flow across heme–cytochrome c and cytochrome c–water interfaces. Theor Chem Acc 133, 1504 (2014). https://doi.org/10.1007/s00214-014-1504-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-014-1504-7