Abstract
Protosappanin A as one major and effective ingredient from Caesalpinia sappan L. exhibited antirejection activity obviously in heart-transplanted rat. The present study was designed to screen out the potential target genes of protosappanin A with microarray technology and reveal some molecular mechanism of immunosuppressive effect. Rats performed with ectopic peritoneal heart transplantation were randomized into three groups receiving different treatments for 7 days: protosappanin A group (25 mg kg−1), cyclosporine A group (10 mg kg−1), and control group. The differentially expressed genes responding to protosappanin A were analyzed with microarrays. Among common differentially expressed genes, the ones of interest were selected for further evaluation by real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), Western blot, immunochemistry, immunofluorescence, and ELISA. Among the 146 common differentially expressed genes, NF-κB and related genes like IκBa, IFN-r, and IP10 were selected for verification. The results of qRT-PCR, Western blot, immunochemistry, and ELISA showed that protosappanin A significantly reduced the expression of NF-κB, IFN-r, and IP10 (p < 0.05) and increased IκBa expression (p < 0.05) in graft. Moreover, the immunochemistry staining of NF-κB and IκBa was mainly observed in infiltrating mononuclear cells. Strikingly, immunofluorescent staining localized NF-κB to the TCR-positive T cells in graft. Furthermore, protosappanin A exhibited inhibitory effect on T cell proliferation in recipients after 7-day treatment. In conclusion, protosappanin A might act on T cells through inhibiting NF-κB activation and downstream gene expressions of IFN-r and IP10, meanwhile reducing T cell proliferation responding to alloantigen, so as to induce immunosuppressive effect. The results encourage a potential therapeutic evaluation of protosappanin A for clinical organ transplantation or other T cell-mediated immune disorders. Additionally, our study also verified the feasibility of microarray utilization in Chinese herb research to explore molecular mechanism and promote development of scientific theories.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chinese herb Caesalpinia sappan L., which is a member of the leguminous plant family, exhibits multiple biological activities and offers therapeutic potential for multiple diseases (Badami et al. 2003; Baek et al. 2000; Hikino et al. 1977; Kim et al. 2005; Oh et al. 1998; Ye et al. 2006). Its ethanol extract has been proposed and proven for its potential immunosuppressive effects on transplantation rejection. Some studies demonstrated that it prolonged rat heterotopic cardiac allograft survival. Furthermore, an active ingredient of it, protosappanin A (PrA), has been isolated and identified (Jian et al. 2008). In our previous work, PrA exhibited obvious antirejection activity, which indeed prolonged graft heart survival and alleviated pathologic damages (Jian et al. 2008). Moreover, we also proved that PrA suppressed peripheral T cell CD4+/CD8+ ratio, perforin, and granzyme B expression in graft, which are secreted by T cells to kill donor cells; it seems that PrA has some effects on T cells (Jian et al. 2008). However, the mechanism of PrA-inducing immunosuppression remains to be elucidated.
Nuclear factor kappa B (NF-κB) is an ubiquitous transcription factor associated with the regulation of numerous immune and inflammatory responses, which mediate subsequent transcriptional activation of genes encoding proinflammatory cytokines and chemokines that play important roles in the onset of alloimmunity (Baldwin 1996; Mason et al. 2004). Moreover, NF-κB has been implied in lymphocyte activation, proliferation, and survival of transplantation. Inhibition of NF-κB prolonged allograft survival and can induce potent immunosuppression, which may become a new modality in controlling allograft rejection (Cooper et al. 1998; Suzuki et al. 2000). Additionally, it was reported that normal T cell-intrinsic NF-κB activation is necessary for cardiac allograft rejection and inhibited NF-κB activation in T cell-induced acceptance of allogeneic cardiac transplants (Finn et al. 2001; Zhoua et al. 2003).
DNA microarray provides a powerful tool for high throughput, analyzing thousands of genes, which is today frequently used to approach the mechanism of drug action (Erickson et al. 2003; Mizuarai et al. 2008; Zarkhin and Sarwal 2008). Moreover, in the recent years, microarrays have been applied in the field of Chinese medications for its superiority of high throughput and parallel and high density (Chen and Leung 2007; Cheng et al. 2008; Gramowski et al. 2006; Iizuka et al. 2003). Screening potential target genes of Chinese medication action by analyzing differentially expressed genes before and after administrations with microarray may further reveal molecular mechanism on genetic level and promote development of scientific theories of Chinese medications.
In the study, we utilized microarray to analyze the significant differentially expressed genes of PrA action of immunosuppression in alloimmunity and further verified the contribution of these genes to the immunosuppressive effect of PrA. Most notably, NF-κB and its related genes were screened out by microarray, and further studies confirmed that PrA depressed NF-κB activation and subsequent cytokines, such as interferon-gamma (IFN-r) and interferon-gamma-inducible protein 10 (IP10), which may partially explain the beneficial immunosuppression effects of PrA. Moreover, T cells were identified responsible for NF-κB activity, and PrA also reduced T cell proliferation response. All together, PrA might act on T cells through inhibiting NF-κB activation and downstream gene expressions of IFN-r and IP10, meanwhile inhibiting T cell proliferation response to alloantigen, so as to induce immunosuppressive effect.
Materials and methods
Drug preparation
The heartwood of C. sappan L. was supplied by San Keshu Chinese medical market (Harbin, China), and it was identified by the pharmacy faculty of Heilongjiang University of Chinese medicine.
PrA were obtained as described in our previous work (Jian et al. 2008). In brief, the shade-dried heartwood of C. sappan L. was sequentially extracted by 95% ethanol for three times and acetic ether, finally getting the ingredient with silica gel column chromatography and meanwhile identifying with wave spectrum. The purity of the PrA was more than 98%. Its chemical structure has been reported in our previous work (Jian et al. 2008). Then, it was dissolved in sterile distilled water. Cyclosporine A (CsA) was dissolved in olive oil.
Animals and grouping
All animal care and procedures were in compliance with the “Principles of Animal Care” (National Society for Medical Research) and the “Guide for the Care and Use of Laboratory Animals” (Institute of Laboratory Animal Resources/NIH).
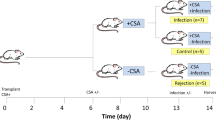
Male Sprague Dawley [SD; recipient, grade specified pathogen-free (SPF)] rats (200–250 g) and male Wistar (donor, grade SPF) rats (180–220 g) were obtained from the experimental animal center of Beijing. After heart transplantation, animals were divided randomly into three groups as follows and given different treatments from days 2 to 7 after operation: PrA group, recipients were given PrA 25 mg kg−1 per os every day; CsA group, recipients were given CsA 10 mg kg−1 per os; and control group, recipients were given olive oil every day.
Heart transplantation
Ectopic peritoneal heart transplantation was performed from Wistar to SD rats with the method reported by Ono and Lindsey (1969). In brief, the ascending aorta and pulmonary artery of donor heart were side-anastomosed, respectively, to the recipient abdominal aorta and inferior vena cava. Graft function was monitored by daily abdominal palpation and scored in a range from 0 (no contractions) to 4 (vigorous contractions). Hearts were considered acutely rejected and excepted from experimental group when palpation scores were <1.
Sample preparation
Blood samples (2 ml) and graft hearts were taken on the seventh postoperative day as the measuring point according to our previous work (Jian et al. 2008). The SD (recipient) rats were anesthetized by 4% pentobarbital sodium and abdomens opened. Blood samples were drawn from inferior vena cava. We separated transplanted grafts and cut half through the left ventricle long axis. One part of grafts were fixed in 10% buffered formalin overnight and embedded in paraffin. The other parts of grafts were frozen by liquid nitrogen.
RNA isolation
Total RNA was obtained from grafts by Tri-Reagent (Invitrogen) and concentrated by precipitation with isopropanol. Its quantity was detected by spectrophotometer, and integrity was verified by formaldehyde-agarose gel electrophoresis.
Microarray analysis
We explore the genes that are differentially expressed with the PrA treatment, using dual colors (Cy3 and Cy5) oligonucleotide microarray to compare the treatment group with PrA and control group. In our microarray experiment, on day 7 posttransplantation, rats were killed, and the hearts were collected for RNA extraction. In each group, every four from 12 RNA samples were pooled for one microarray; thus, three microarrays were performed, which was recommended by Kendziorski et al. (2005) in this condition.
Briefly, the total RNA samples were reversely transcribed to complementary DNA (cDNA) in the presence of fluorescent Cy3 or Cy5 dye. Usually, the treatment group is labeled with Cy3, while the control group is labeled with Cy5. Probes were then hybridized onto the rat genome 70-mer oligonucleotide microarray (Rat Genome Version 3.0.5), which was obtained from CapitalBio Corporation (Beijing, China). After two subsequent washings, all the hybridized microarrays were scanned using LuxScan Scanner (CapitalBio Company), and images were further analyzed by the GenePix Pro 4.0 software (Axon Instruments Company) to export data. The data were statistically analyzed in locally weighted scatter plot smoothing way. After normalization and correction, the Cy-5/Cy-3 value was looked as standard ratio, and ratio >2 or <0.5 was the statistical standard to judge differentially expressed genes. PrA and control groups were tested in three individual hybridization experiments, and only the genes differentially expressed in three chips can be judged as common differentially expressed genes. The pathway classification and function of these genes were investigated using PathWay Miner, which includes information of Kyoto Encyclopedia of Genes and Genomes, BioCarta, and CenMAPP database (Kanehisa and Goto 2000).
Real-time RT-PCR analysis
For quantitative measurement of messenger RNA (mRNA), 2 µg of total RNA was used for cDNA synthesis with Reverse Transcription System (Promega, USA). Real-time PCR was performed using SYBR Green I Real-Time Kit (Takara, Japan) on a LightCycler system (Roche LightCycler, USA). Specific primer pairs were used. Primer sequences and reactive condition are shown in Table 1. Reaction specificity was controlled by postamplification melting curve analyses as well as by gel electrophoresis of the obtained products. Results were expressed relative to the number of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts, which were used as internal controls. The specific primers sequences were as follows: NF-κB, forward 5′-CACTCTCTT TTTGGAGGT-3′, reverse 5′-TGGATATAAGGCTTTACG-3′; IκBa, forward 5′-GAAATACCCCTCTCCATC-3′, reverse 5′-ATCAGCACCCAAAGTCAC-3′; IFN-r, forward 5′-AGGCCATCAGCAACAACATAAGTG-3′, reverse 5′-GACAGCTTT GTGCTGGATCTGTG-3′; IP10, forward 5′-TTATTGAAAGCGGTGAGCCA AAG-3′, reverse 5′-GGACAGTTAGGACTAGCCGCAC-3′; GAPDH, forward 5′-TTCA TTGACCTCAACTAC-3′, reverse, 5′-AGACTCCACGACATACTC-3′. The following protocol was used for each reaction: initial denaturation at 94°C for 5 min, three-step cycling with 30 cycles consisting of denaturation at 94°C for 30 s, annealing at 50°C for 30 s and extension at 68°C for 30 s, and a final extension at 68°C for 10 min.
Western blot analysis
A previously reported semiquantitative Western blot approach was used to assess NF-κB and IκBa expressions in the grafts. Briefly, total proteins were extracted from the grafts, and then protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were incubated with first antibody and then followed by peroxidase-labeled secondary antibodies. Signals were visualized using gel imaging system (Tanon GIS-2010 Shanghai Pharmacia Biotech, China). Antibodies to NF-κB (Newmark, USA), IκBa (Santa Cruz, USA), and β-actin (Sigma) were used. Protein expression levels were expressed as their ratio to β-actin.
Immunohistochemistry analysis
Immunohistochemical studies for the expression of NF-κB and IκBa in graft utilized an avidin–biotin peroxidase method with a 3,3′-diaminobenzidine chromatogen. After antigen retrieval (microwave oven for 30 min), immunohistochemistry was carried out following the manufacturer’s instructions. The following primary antibodies were used: anti-NF-κB (Newmark, USA) and anti-IκBa (Santa Cruz, USA).
From three samples of each group, ones in six series of section were selected randomly. In each section, ten microscopic fields were examined under a microscope at a magnification of ×200. The intensity of the reaction product of NF-κB and IκBa immunohistochemistry was measured quantitatively using a Nikon Eclipse E-600 microscope and Image Pro Plus Software 4.1. The expressions were evaluated by density mean (density mean = integrated optical density/area sum; Kira et al. 2008; Xavier et al. 2005). Sections were processed in parallel under identical conditions to minimize variations in staining density.
ELISA analysis
Cytokine levels in serum were detected by two-site sandwich ELISA using Jingmei Biotech Company antibody pairs (IFN-r). Samples were assayed in duplicate and were quantitated by comparison with standard curves obtained with purified recombinant cytokines. Results are presented as the means of duplicates.
Immunofluorescence
To investigate whether NF-κB activation was interfered by PrA located in T cell receptor-positive T cells, NF-κB staining was combined on a triple staining with TCR and 4′-6-diamidino-2-phenylindole (DAPI) to determine overlap in the staining pattern. On day 7 posttransplantation, the grafts were dissected and embedded in optimum cutting temperature compound, snap-frozen in liquid nitrogen, and cut into sections and then blocked with goat serum and incubated in solution of primary antibodies complex and 1% bovine serum albumin overnight at 4°C. Primary antibodies included rabbit anti-NF-κB p65 (1:50, Cell Signaling) and mouse anti-TCR (1:50, Santa Cruz) antibodies. After washing, sections were incubated with the mixture of fluorescein isothiocyannate-conjugated goat antirabbit antibody (1:200, Santa Cruz) and tetramethylrhodamine isothiocyanate-conjugated goat antimouse antibody (1:200, Santa Cruz) for 1 h at 37°C. Nuclei were counterstained with DAPI (Sigma). Fluorescent images were acquired with a confocal laser-scanning microscope (Olympus FluoView V5.0 FV300).
BrdU cell proliferation assay
T cells were used at different numbers (0–15 × 103) as stimulators isolated from Wistar rat spleen by nonadherence to nylon wool. Responder allogenic T cells from cardiac recipients were also isolated from spleen on day 7 posttransplantation. After pretreatment of stimulator T cells with mitomycin C, stimulator and responder T cells were cultured (4 days) in 96-well round-bottom microplates at different ratios. Cells were incubated with bromodeoxyuridine (BrdU) (18 h), and BrdU incorporation by proliferating responder cells was quantified using a BrdU cell proliferation assay according to the manufacturer’s instructions (Chemicon, USA). All proliferation assays were performed in quadruplicate.
Statistical analysis
Data were expressed as means ± standard errors of the means. Differences between nonparametric groups were examined by the Mann–Whitney test. Other data were compared by one-way ANOVA. Differences were considered statistically significant at a value of p < 0.05.
Results
Analyses of differentially expressed genes responding to PrA in microarray data
The microarray analysis of grafts on the seventh day after operation showed that there were approximately 14,768 expressed genes detectable on chip 1, and 609 of them were differentially expressed (290 upregulation and 319 downregulation). Those indices were 11045,673(254,419) on chip 2 and 11123,1010(651,359) on chip 3, respectively. There were 146 common differentially expressed genes in three microarrays, which was shown in Fig. 1 (120 downregulated and 20 upregulated). However, the complete file with common differentially expressed genes is provided as additional file 1.
Differentially expressed genes screened out by microarray respond to PrA. Comparisons were performed to identify genes with significant changes in expression levels in PrA group comparing with control group. After normalization and correction, the Cy-5/Cy-3 value was looked as the standard ratio, and ratio >2 or <0.5 was the statistical standard to judge differentially expressed genes. The black and white columns, respectively, mean the numbers of upregulated and downregulated genes in arrays 1, 2, and 3 and common differential expressed genes in three arrays
These common differentially expressed genes were subjected to pathway miner and then distributed into five major functional categories: mitogen-activated protein kinase (MAPK) signaling pathway, toll-like receptor signaling pathway, apoptosis pathway, complement and coagulation cascades pathway, and genes related with energy and substance metabolism. Among these common differentially expressed genes, NF-κB and related genes attracted more attention because they are involved in MAPK signaling pathway, apoptosis, toll-like receptor signaling pathway, T cell receptor signaling pathway, and B cell receptor signaling pathway; moreover, they regulate the subsequent costimulatory molecules, inflammatory cytokines, and chemokine, which are well known to be involved in alloimmunity. IκBa, a major inhibitory protein of NF-κB, plays a key role in nuclear translocation of NF-κB and activation of downstream proinflammatory cytokines. Not only NF-κB and IκBa but also subsequent downstream genes like IFN-r and IP10 are differentially expressed by PrA. It suggests that NF-κB and related genes might be involved in immunosuppressive action of PrA. Additionally, it has been proven that NF-κB inhibitors can prolong allograft survival and induce potent immunosuppression. Based on the above supports, we assumed that NF-κB might be implied in the immunosuppressive action of PrA. Therefore, NF-κB, IκBa, IFN-r, and IP10 from common differentially expressed genes were selected for further evaluation to verify differential expression.
PrA inhibited NF-κB activation and increased IκBa expression in grafts
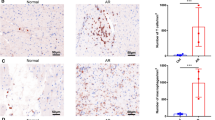
We examined the effect of PrA on NF-κB and IκBa mRNA expression. As shown in Fig. 2a, the mRNA level of NF-κB decreased in the PrA group as well as in the CsA group compared with the control group (p < 0.01), as measured by qRT-PCR. However, the mRNA level of IκBa (Fig. 2a) was significantly increased by PrA and CsA (p < 0.05). Consistent with qRT-PCR data, PrA significantly inhibited NF-κB protein expression (p < 0.05) and increased protein level of IκBa (p < 0.01) measured by Western blot (Fig. 2b).The cellular distribution of NF-κB and IκBa was also examined by immunohistochemistry, and staining density was quantitated as shown in Fig. 3. The staining density mean of NF-κB was significantly lower (p < 0.05), but IκBa was higher (p < 0.05) in the PrA and CsA groups, compared with the control group (Fig. 3). After PrA or CsA treatments, infiltrations in grafts were obviously reduced compared with the control group. Strikingly, the staining of NF-κB and IκBa was mainly observed in infiltrating mononuclear cells. These results confirmed that PrA inhibited NF-κB activation and increased IκBa expression; moreover, immunostaining implies that infiltrating mononuclear cells might be responsible for NF-κB activity interfered by PrA.
Effect of PrA on NF-κB and IκBa expression in graft. a qRT-PCR analysis. The mRNA of NF-κB was significantly decreased in PrA group (p < 0.01, n = 7) and CsA group (p < 0.01, n = 8) compared with control group (n = 6). However, IκBa expression was increased in PrA group (p < 0.05, n = 7) and CsA group (p < 0.01, n = 8) compared with control group (n = 8). Gene expression levels were relative to the reference gene GAPDH. b Protein expressions by Western blot. PrA and CsA exerted similar effects on NF-κB and IκBa protein expression. Protein expression was relative to the reference gene s β-actin. Each value represents the mean ± SEM of independent experiments vs control group. *p < 0.05; **p < 0.01 (ANOVA)
Immunohistochemistry analysis for NF-κB and IκBa. Immunostaining for NF-κB and IκBa in grafts of control group (a, b), PrA group (c, d), and CsA group (e, f) were quantitated, respectively. The staining density mean of NF-κB was significantly lower (p < 0.05), but IκBa was higher (p < 0.05) in PrA group and CsA group, compared with control group. Moreover, the stainings of NF-κB and IκBa were mainly observed in infiltrating mononuclear cells. Each value represents the mean ± SEM of independent experiments vs control group. *p < 0.05; **p < 0.01 (ANOVA), ×200
PrA exerted marked depression on subsequent downstream genes IFN-r and IP10
As for the subsequent downstream genes of NF-κB, PrA also exerted marked depression on IFN-r and IP10 mRNA expression in graft (shown in Fig. 4). The results confirmed microarray data. We continuously investigated the productions of subsequent downstream genes IFN-r, a secreted cytokine in peripheral blood, with ELISA (Fig. 4). Depressive effect on IFN-r by PrA or CsA was confirmed again (p < 0.05). These results further verified our postulation.
PrA exerted marked depression on subsequent downstream genes IFN-r and IP10. a The expressions of IFN-r and IP10 were significantly decreased in PrA group (p < 0.05, n = 6; p < 0.01, n = 7, respectively) and CsA group (p < 0.01, n = 7; p < 0.01, n = 6, respectively) compared with control group (n = 6). Gene expression levels were relative to the reference gene GAPDH. b PrA and CsA inhibited the serum production of IFN-r (p < 0.05, n = 6; p < 0.05, n = 5, respectively). Each value represents the mean ± SEM of independent experiments vs control group. *p < 0.05; **p < 0.01 (ANOVA)
PrA interfered with NF-κB expression in TCR-positive T cells of graft and exhibited inhibitory effect on T cell proliferation
Previous work has proven that PrA suppressed peripheral T cell CD4+/CD8+ ratio and perforin and granzyme B expression in graft, which secreted by T cells to kill donor cells. Therefore, among these infiltrating mononuclear cells, we presumed that T cell might be the main cell type, which the ingredient is acting on. We performed triple staining for NF-κB (green) and T cell marker TCR (red) in the allografts by immunofluorescence; meanwhile, DAPI staining (blue) for nucleus was also used. The colocalization (yellow color) of NF-κB and TCR combined with blue staining nucleus of DAPI is shown in Fig. 4. Consistent with immunochemistry results, after PrA treatment, mononuclear cell infiltration in grafts was significantly reduced, and NF-κB intensity in TCR-positive cells also decreased compared with untreated recipients shown in Fig. 5. It proved the hypothesis that T cells should be target cells of PrA action by interfering with NF-κB pathway.
Immunofluorescence analysis for NF-κB and IκBa. Immunofluorescence triple staining for NF-κB (green), T cell marker TCR (red), and DAPI staining (blue) for nucleus in the allografts were performed. The colocalization (yellow) of NF-κB and TCR combined with blue staining nucleus of DAPI was observed. In PrA group, mononuclear cell infiltrations in grafts were significantly reduced, and NF-κB intensity in TCR-positive cells was also decreased compared with untreated recipients. The results are representative of three independent experiments
To further investigate the effect of PrA on T cells, T cell proliferation responses to alloantigen and mixed lymphocyte cultures were performed after 7 days administration of PrA. On day 7 after heart transplantation, recipients were killed, and T cells isolated from spleen were incubated with mitomycin-C-pretreated Wistar splenocytes as stimulators. T cell proliferation was measured through BrdU incorporation by BrdU–ELISA. As we expected, PrA inhibited the proliferative response of T cells to alloantigen after 7-day treatments as shown in Fig. 6.
Inhibitory effect of PrA on T cell proliferation. T cell proliferation responses to alloantigen were performed after 7 days administration of PrA treatment. Recipient T cells isolated from spleen were incubated with allogeneic mitomycin C-pretreated T cells as stimulators, and T cell proliferations were measured through BrdU incorporation by BrdU cell proliferation assay. The proliferative responses of T cells were significantly inhibited in PrA group comparing with control group (p < 0.05)
Discussion
Our previous study has proven that PrA, an active ingredient derived from C. sappan L. can induce immunosuppression in rat heart transplantation (Jian et al. 2008), but the mechanism is still unclear. The present study represents a first step in the study of PrA for potential mechanisms of immunosuppressive action.
In this study, we utilized microarray to screen out the potential target genes by analyzing the differentially expressed genes between the group treated with PrA and control group on day 7 postoperation. Among these common differentially expressed genes, NF-κB and related genes, like inhibitor IκBa, downstream genes IFN-r, and IP10 were attracting more attention for their substantial roles in allograft immunity. Further analysis confirmed that PrA inhibited NF-κB activation and increased IκBa expression measured by qRT-PCR, Western blot, and immunohistochemistry, respectively. These results supported our presumption that NF-κB pathway might imply in the immunosuppressive action of PrA. Our results are in line with the study of Cooper et al. (1998). As we have known, NF-κB activation is directly involved in the rejection of transplanted organs; moreover, it has been implicated as a key-signaling mediator for alloimmunity. The importance of NF-κB in alloimmunity was underscored by the fact that inhibition of NF-κB activation or translocation prolonged survival of graft (Cooper et al. 1998; Green 2003; Ping Zhou et al. 2005). It has been proposed that NF-κB could be an effective target of treatment for transplant rejection. Moreover, it is well known that NF-κB is usually located in the cytoplasm in an inactive form complexed with IκBa, a major inhibitor of NF-κB. When cells are induced by various stimuli, IκBa is phosphorylated and then degraded, which causes NF-κB to be released from the complex and translocated into nucleus. The increased cytosolic IκBa more frequently combined with NF-κB can prevent the translocation of NF-κB to the nucleus and the subsequent transcriptional activation of genes encoding inflammatory cytokines and chemokines. Thus, we can indicate that the upregulation of IκBa expression might be involved in depressing NF-κB activation, but it still needs further study.
We continuously analyzed the subsequent transcriptional activation of genes, which is mediated by NF-κB: IFN-r and IP10. The depressive effects of PrA on these genes were confirmed by qRT-PCR and ELISA. IFN-r and IP10 play important roles in organ transplantation. The expressions and roles of IFN-r and IP10 during cardiac rejection have been investigated previously (Colvin and Thomson 2002; Kapoor et al. 2000; Miura et al. 2001). Several studies showed that IFN-r primes Th1 development and negatively regulates the growth of Th2 cells (Lu et al. 1998; Wenner et al. 1996). In our study, mRNA expressions of IFN-r and IP10 were greatly depressed by PrA or CsA. It may explain the reduction of recruitment of effector cells and immunologic reaction during allograft rejection. The results can be supported by the previous studies that the presence of CXCR3(+) T cells and the CXCR3 ligand IP10 within endomyocardial biopsies is strongly associated with acute rejection. Moreover, the treatment with anti-CXCR3 peptide nucleic acid to recipient mouse after skin transplant prolonged the survival of graft (Melter et al. 2001; Jiankuo et al. 2003; Zhao et al. 2002). Therefore, inhibition of subsequent downstream genes IFN-r and IP10 also contribute to immunosuppressive action of PrA, which might be associated with reduction of CXCR3(+) T cells.
Furthermore, we try to explore which types of cell in grafts PrA is acting on and should be responsible for NF-κB activity. Immunochemistry results showed that stainings of NF-κB and IκBa in graft were mainly located in infiltrating mononuclear cells. Among the infiltrating mononuclear cells, T cells play key roles in allograft immunity (Jones et al. 2000). Consistent with our hypotheses, immunofluorescence triple staining for NF-κB-, TCR-, and DAPI-identified T cells should be responsible for NF-κB activity. It was supported by the reports that T cell functions were affected by NF-κB activation in vitro and recipients with impaired NF-κB activation in T cells that had accepted a cardiac allograft (Finn et al. 2001; Ping Zhou et al. 2005). Meanwhile, our previous work has proven that this ingredient suppressed peripheral T cell CD4+/CD8+ ratio and perforin and granzyme B expression in graft, which is secreted by T cells to kill donor cells. Therefore, it further proved that T cells should be the target cells of PrA. The effect of PrA on T cells was further studied by mixed lymphocyte cultures, and PrA also exhibited inhibitory effect on T cell proliferation to alloantigen, which can be explained by incomplete T cell-intrinsic NF-κB activation. The results were in line with the report of Zhoua et al. (2003). Normal T cell-intrinsic NF-κB activation is necessary for cardiac allograft rejection, and reduced NF-κB activation in T cells can induce immunosuppression of allografts (Finn et al. 2001; Zhoua et al. 2003). Therefore, PrA might act on T cells via interfering with NF-κB pathway, so as to induce immunosuppression. Maybe there are some other cell types like monocytes that were also affected by PrA in grafts, but it still needs more studies to confirm this. According to the above studies, we can conclude that T cells should be one of target cells of PrA at least.
In conclusion, PrA might act on T cells through inhibiting NF-κB activation and downstream gene expressions of IFN-r and IP10, which results in reducing T cell proliferation response to alloantigen. These all contribute to immunosuppression effect of PrA. The results also encourage a potential therapeutic evaluation of PrA for clinical organ transplantation or other T cell-mediated immune disorders. Additionally, our study also verified the feasibility of microarray utilization in Chinese herb research to explore molecular mechanism and promote development of scientific theories.
Abbreviations
- PrA:
-
Protosappanin A
- CsA:
-
Cyclosporin A
- NF-κB:
-
Nuclear factor kappa B
- IκBa:
-
Inhibitor of nuclear factor kappa B alpha
- IFN-r:
-
Interferon-gamma
- IP10:
-
Interferon-gamma-inducible protein 10
- qRT-PCR:
-
Real-time quantitative reverse transcriptase polymerase chain reaction
- FITC:
-
Fluorescein isothiocyannate
- TRITC:
-
Tetramethylrhodamine isothiocyannate
References
Badami S, Moorkoth S, Rai SR, Kannan E, Bhojraj S (2003) Antioxidant activity of Caesalpinia sappan heartwood. Biol Pharm Bull 26(11):1534–1537
Baek NI, Jeon SG, Ahn EM, Hahn JT, Bahn JH, Jang JS, Cho SW, Park JK, Choi SY (2000) Anticonvulsant compounds from the wood of Caesalpinia sappan L. Arch Pharm Res 23(4):344–348
Baldwin AS (1996) The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol 14:649–681
Chen CF, Leung AY (2007) Gene response of human monocytic cells for the detection of antimigraine activity of feverfew extracts. Can J Physiol Pharmacol 85(11):1108–1115
Cheng WY, Wu SL, Hsiang CY, Li CC, Lai TY, Lo HY, Shen WS, Lee CH, Chen JC, Wu HC, Ho TY (2008) Relationship between San-Huang-Xie-Xin-Tang and its herbal components on the gene expression profiles in HepG2 cells. Am J Chin Med 36(4):783–797
Colvin BL, Thomson AW (2002) Chemokines, their receptors, and transplant outcome. Transplantation 74(2):149–155
Cooper M, Lindholm P, Pieper G, Seibel R, Moore G, Nakanishi A, Dembny K, Komorowski R, Johnson C, Adams M, Roza A (1998) Myocardial nuclear factor-kappa B activity and nitric oxide production in rejecting cardiac allografts. Translantation 66(7):838–844
Erickson LM, Pan F, Ebbs A, Kobayashi M, Jiang H (2003) Microarray-based gene expression profiles of allograft rejection and immunosuppression in the rat heart transplantation model. Transplantation 76(3):582–588
Finn PW, Stone JR, Boothby MR, Perkins DL (2001) Inhibition of NF-kappaB-dependent T cell activation abrogates acute allograft rejection. J Immunol 167:5994–6001
Gramowski A, Jugelt K, Stuwe S, Schulze R, McGregor GP, Wartenberg-Demand A, Loock J, Schroder O, Weiss DG (2006) Functional screening of traditional antidepressants with primary cortical neuronal networks grown on multielectrode neurochips. Eur J NeuroSci 24(2):455–465
Green D (2003) Death and NF-kappaB in T cell activation: life at the edge. Mol Cell 11(3):551–552
Hikino H, Taguchi T, Fujimura H, Hiramatsu Y (1977) Antiinflammatory principles of Caesalpinia sappan wood and of Haematoxylon campechianum wood. Planta Med 31(3):214–220
Iizuka N, Oka M, Yamamoto K, Tangoku A, Miyamoto K, Miyamoto T, Uchimura S, Hamamoto Y, Okita K (2003) Identification of common or distinct genes related to antitumor activities of a medicinal herb and its major component by oligonucleotide microarray. Int J Cancer 107(4):666–672
Jian Wu, Jingbo H, Maomao Z, Yongpeng Z, Bo Y (2008) Protosappanin A, a immunosuppressive constituent from Chinese herb, prolongs graft survival and attenuates acute rejection in rat heart allografts. Transplant Proc 40(10):3719–3722
Jiankuo M, Xingbing W, Baojun H (2003) Peptide nucleic acid antisense prolongs skin allograft survival by means of blockade of CXCR3 expression directing T cells into graft. J Immunol 170(3):1556–1565
Jones ND, Van Maurik A, Hara M, Spriewald BM, Witzke O, Morris PJ, Wood KJ (2000) CD40-CD40 ligand-independent activation of CD8+ T cells can trigger allograft rejection. J Immunol 165(2):1111–1118
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28(1):27–30
Kapoor A, Morita K, Engeman TM, Koga S, Vapnek EM, Hobart MG, RL F (2000) Early expression of interferon-gamma inducible protein 10 and monokine induced by interferon-gamma in cardiac allografts is mediated by CD8+ T cells. Transplantation 69:1147–1155
Kendziorski C, Irizarry RA, Chen K-S, Haag JD, Gould NM (2005) On the utility of pooling biological samples in microarray experiments. PNAS 102(12):4252–4257
Kim EC, Hwang YS, Lee HJ, Lee SK, Park MH, Jeon BH, Jeon CD, Yu HH, You YO (2005) Caesalpinia sappan induces cell death by increasing the expression of p53 and p21WAF1/CIP1 in head and neck cancer cells. Am J Chin Med 33(3):405–414
Kira VM, Fagundes DJ, Bandeira COP, Kaufman O, Fagundes ATN, Ortiz V (2008) Effects of repeated extracorporeal shock wave on kidney apoptosis of normal and diabetic Rat. Investig Urol 34(1):91–96
Lu B, Ebensperger C, Dembic Z, Wang Y, Kvatyuk M, Lu T, Coffman RL, Pestka S, Rothman PB (1998) Targeted disruption of the interferon-gamma receptor 2 gene results in severe immune defects in mice. Proc Natl Acad Sci USA 95(14):8233–8238
Mason NJ, Artis D, Hunter CA (2004) New lessons from old pathogens: what parasitic infections have taught us about the role of nuclear factor-kappaB in the regulation of immunity. Immunol Rev 201:48–56
Melter M, Exeni A, Reinders ME, Fang JC, McMahon G, Ganz P, Hancock WW, Briscoe DM (2001) Expression of the chemokine receptor CXCR3 and its ligand IP-10 during human cardiac allograft rejection. Circulation 104(21):2558–2564
Miura M, Morita K, Kobayashi H, Hamilton TA, Burdick MD, Strieter RM, Fairchild RL (2001) Monokine induced by IFN-gamma is a dominant factor directing T cells into murine cardiac allografts during acute rejection. J Immunol 167(6):3494–3504
Mizuarai S, Irie H, Schmatz DM, Kotani H (2008) Integrated genomic and pharmacological approaches to identify synthetic lethal genes as cancer therapeutic targets. Curr Mol Med 8(8):774–783
Oh SR, Kim DS, Lee IS, Jung KY, Lee JJ, Lee HK (1998) Anticomplementary activity of constituents from the heartwood of Caesalpinia sappan. Planta Med 64(5):456–458
Ono K, Lindsey ES (1969) Improved technique of heart transplantation in rats. J Thorac Cardiovasc Surg 57(2):225–229
Ping Zhou J, Balin S, Mashayekhi M (2005) Transplantation tolerance in NF-kappaB-impaired mice is not due to regulation but is prevented by transgenic expression of Bcl-xL. J Immunol 174:3447–3453
Suzuki J, Morishita R, Amano J, Kaneda Y, Isobe M (2000) Decoy against nuclear factor-kappa B attenuates myocardial cell infiltration and arterial neointimal formation in murine cardiac allografts. Gene Ther 7(21):1847–1852
Wenner CA, Guler ML, Macatonia SE, O’Garra A, Murphy KM (1996) Roles of IFN-r and IFN-r in IL-12-induced T helper cell-1 development. J Immunol 156:1442
Xavier LL, Viola GG, Ferraz AC, Da Cunha C, Deonizio JM, Netto CA, Achaval M (2005) A simple and fast densitometric method for the analysis of tyrosine hydroxylase immunoreactivity in the substantia nigra pars compacta and in the ventral tegmental area. Brain Res Brain Res Protoc 16(1–3):58–64
Ye M, Xie WD, Lei F, Meng Z, Zhao YN, Su H, Du LJ (2006) Brazilein, an important immunosuppressive component from Caesalpinia sappan L. Int Immunopharmacol 6(3):426–432
Zarkhin V, Sarwal MM (2008) Microarrays: monitoring for transplant tolerance and mechanistic insights. Clin Lab Med 28(3):385–410
Zhao DX, Hu Y, Miller GG, Luster AD, Mitchell RN, Libby P (2002) Differential expression of the IFN-gamma-inducible CXCR3-binding chemokines, IFN-inducible protein 10, monokine induced by IFN, and IFN-inducible T cell alpha chemoattractant in human cardiac allografts: association with cardiac allograft vasculopathy and acute rejection. J Immunol 169(3):1556–1560
Zhoua P, Hwanga KW, Paluckia DA, Guob Z, Boothbyc M, Newellb KA, Alegrea M-L (2003) Impaired NF-κB activation in T cells permits tolerance to primary heart allografts and to secondary donor skin grafts. Am J Transplant 3:139–147
Acknowledgments
Our projects are supported by the National Nature Science Foundation of China (C03020504), the Heilongjiang Province Nature Science Foundation (D200660), the Heilongjiang Province Nature Foundation of Great Subject (ZJY03-7), and Heilongjiang Province Scientific Technique Project (GC02C122).
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Wu and M. Zhang contributed equally in the study.
Rights and permissions
About this article
Cite this article
Wu, J., Zhang, M., Jia, H. et al. Protosappanin A induces immunosuppression of rats heart transplantation targeting T cells in grafts via NF-κB pathway. Naunyn-Schmied Arch Pharmacol 381, 83–92 (2010). https://doi.org/10.1007/s00210-009-0461-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-009-0461-5