Abstract
MicroRNAs are important regulators in numerous cellular processes, including cell differentiation, proliferation, and apoptosis. Recently, miR-143 was identified as a tumor suppressor in prostate cancer (PCa). To explore the mechanism of dysregulation and anti-tumor function of miR-143 in PCa, we first found a single-nucleotide polymorphism rs4705342T>C in the promoter region of miR-143 through bioinformatics tools and then performed a case–control study including 608 PCa patients and 709 controls. Results suggested that subjects with TC/CC genotypes had significantly decreased risk of PCa compared with those with TT genotype (adjusted OR 0.68, 95 % CI 0.55–0.85). Further functional assays showed that the risk-associated T allele increased the protein-binding affinity and reduced the activity of the promoter compared with C allele. In addition, restoration of miR-143 by mimics in PCa cells significantly inhibited cell proliferation and migration and down-regulated the expression level of kallikrein-related peptidase 2 (KLK2) mRNA and protein. The miR-143-KLK2 axis was also confirmed by luciferase reporter assay in vitro. In conclusion, our findings demonstrate that there is the significant association between the functional promoter variant rs4705342T>C in miR-143 and PCa risk and newly describe the miR-143-KLK2 interaction which provided another potential mechanism for miR-143 anti-tumor function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) is the commonest diagnosed malignant tumor and the third leading cause of cancer deaths among men in Western developed countries (Jemal et al. 2011). There were an estimated 241,740 new cases and 28,170 deaths in the USA in 2012 (Siegel et al. 2012). The prevalence of early prostate-specific antigen (PSA) testing has contributed to decrease the incidence rates in recent years (Baade et al. 2009). However, the efficacy of PSA testing in reducing mortality from PCa remains disputed, and the PSA-based screening programs have led to non-negligible overdiagnosis and over-treatment (Andriole et al. 2009; Schroder et al. 2009). Therefore, novel and valid biomarkers able to drive the diagnosis, treatment, and prognosis are urgently needed.
MicroRNAs (miRNAs) are a class of small, endogenous, evolutionarily conserved non-coding RNA molecules encoded in the genomes of animals (Ambros 2004). Growing evidence indicates that miRNAs regulate almost all basic cellular functions including cell differentiation, proliferation, mobility, and apoptosis in diverse cancer-related biologic processes (Ambros 2004). miRNAs play their important biologic functions normally through negative-regulating protein expression of oncogenes or tumor suppressor genes at the posttranscriptional level (Ambros 2004; Bartel 2004). Thus, miRNAs could participate in human carcinogenesis as either tumor suppressors or oncogenes (Croce 2009). Over-expression of oncogenic miRNAs or down-expression of tumor suppressor miRNAs plays pivotal roles in tumorigenesis.
One major tumor suppressor miRNA, miR-143, which is down-regulated in such neoplasms as liposarcoma, gastric, bladder, and colorectal tumor (Chen et al. 2009; Lin et al. 2009; Takagi et al. 2009; Ugras et al. 2011), is also discovered to be down-regulated in miRNA expression profiles of PCa (Ambs et al. 2008; Tong et al. 2009; Wach et al. 2012). Functional studies have identified that miR-143 inhibited PCa cells proliferation and migration and enhanced their sensitivity to docetaxel through the suppression of V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) in vitro (Clape et al. 2009; Xu et al. 2011a). Moreover, restoration of miR-143 expression could abrogate PCa progression in mice which was probably mediated by the inhibition of extracellular signal-regulated kinase-5 (ERK5) activity (Clape et al. 2009).
However, why miR-143 level is reduced in patients with PCa remains unresolved. Zhou et al. (2007) showed that most known miRNA genes have the same type of promoters as protein-coding genes and could influence transcription initiation and regulation of miRNA genes. Several later studies also demonstrated that mutations in the promoter region of miRNAs could alter the expression level of mature miRNA and cancer risk. Xu et al. (2011b) exhibited a potentially functional polymorphism in the promoter region of miR-34b/c which was associated with an increased risk of primary hepatocellular carcinoma. Luo et al. (2011) discovered that the expression level of miR-146a was modulated by a genetic variant of its promoter by altering its binding affinity for v-ets avian erythroblastosis virus E26 oncogene homolog 1 (Ets-1) and the variant conferred disease risk of systemic lupus erythematosus. In the present study, we explored the association between genetic variants in miR-143 promoter region and PCa risk. Through our subsequent case–control study of 608 PCa patients and 709 cancer-free controls in a Chinese population and further functional studies, we identified the single-nucleotide polymorphism (SNP) rs4705342T>C in the miR-143 promoter which affected its expression activity and contributed to PCa risk.
Materials and methods
Study subjects
From September 2003 to the beginning date of our present study, 620 cases and 715 cancer-free controls were recruited from the First Affiliated Hospital of Nanjing Medical University. All subjects were genetically unrelated Han Chinese, and the recruitment has been previously described (Xue et al. 2013). Briefly, the cases were all histologically confirmed to have PCa for the first time, while the controls were healthy people who were seeking health care at the hospital. Based on the international tumor–node–metastasis staging system, the tumor stage was divided into localized and advanced cancer (localized: T1–2N0M0; advanced: T3–4N x M x or T x N1M x or T x N x M1). Gleason score represented the pathologic grade which was estimated by pathologists of the hospital using the Gleason scoring system. Controls were cancer-free subjects seeking health care in the outpatient departments at the hospital and were excluded if they had abnormal PSA level, or abnormal digital rectal examination. Moreover, controls were frequency-matched to the cases on age (±5 years). Informed consent was obtained from all the eligible subjects. After that, detailed information of subjects such as age, race, tobacco use, and alcohol use was collected through face-to-face interview by trained interviewers. Then, each subject donated 5 ml of blood for genomic DNA extraction. The study was approved by the institutional review board of Nanjing Medical University.
Tissue samples, cell lines, and cell culture
A total of 22 pairs of primary PCa and their matched adjacent normal prostate epithelial tissues were collected from surgically removed specimens of individual cases, eight pairs of which were frozen in liquid nitrogen upon collection from the hospital and the others were paraffin-embedded tissues from the hospital. Tissue samples were at least 80 % composed of viable-appearing tumor or normal cells on histological assessment.
Two metastatic PCa cell lines DU145 and PC3, a human embryonic kidney cell line HEK293T, and a human gastric cancer cell line SGC7901 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI 1640 for DU145 and PC3 and DMEM for HEK293T and SGC7901 (GIBCO-BRL) medium supplemented with 10 % fetal bovine serum (10 % FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Carlsbad, USA) in humidified atmosphere of 5 % CO2 at 37 °C.
RNA extraction and RT-PCR analysis
Total RNA was isolated from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol. The stem-loop reverse transcription-polymerase chain reaction (RT-PCR) technique was used to examine mature miR-143 (Chen et al. 2005). The primers used to amplify mature miR-143 were designed according to Chen et al. (2005). U6 was used as an internal control. For the detection of common protein-coding genes mRNA, the total RNA was reverse-transcribed with a 5× PrimeScript RT Master Mix (TaKaRa Biotechnology, Dalian, China) and the transcription level of GAPDH was validated as the normalizer. RT-PCR was performed with SYBR Green PCR Master Mix on an ABI 7900HT Real-Time PCR detection system (Applied Biosystems, CA, USA). ∆C t was calculated by subtracting the C t of U6 or GAPDH mRNA from the C t of the mRNA of interest. The miRNA or mRNA level was calculated according to the equation \(2^{{ -\Delta C_{\text{t}} }}\). All reactions were done in triplicate.
SNP selection and genotyping
We selected the SNPs in the region of 1,000-bp upstream from the transcription start site of miR-143 precursor from the dbSNP database. With the criteria that the minor allele frequency (MAF) >0.05 in a Chinese population, we chose a potentially functional common variant rs4705342T>C (510-bp upstream) and the other 19 SNPs were excluded. The sites of the 20 SNPs are showed in Figure S1.
The rs4705342T>C SNP was determined using the TaqMan SNP Genotyping Assay with the ABI 7900HT Real Time PCR System (Applied Biosystems, CA, USA). Before this, we conducted DNA quality control and excluded 16 degradated DNA samples, with 610 cases and 709 controls remaining. The sequences of primers and probes for the SNP are listed in Table S1. The polymorphism analysis was performed by two people independently in a blinded manner. Ten percent of random samples were selected for validation, and the results were 100 % concordant. Two DNA samples of the cases failed to generate reliable results, and only 608 cases were included in the final analysis.
Construction of reporter plasmids and cell transfection
To compare the activities of miR-143 promoters containing the different rs4705342 alleles, the miR-143 promoter–luciferase reporter constructs were created. The constructs consisted of a 700-bp DNA fragment corresponding to the upstream region of the transcription start site of pre-miR-143, which were amplified from individual homozygous templates and cloned into the pGL3-basic luciferase vector (Promega, WI, USA; Fig. 1a). The amplified fragments were then sequenced to confirm that there were no nucleotide errors.
Allelic difference of rs4705342 in miR-143 promoter activity and in binding affinity to nuclear proteins. a Schematic representation of reporter plasmids containing the rs4705342 T or C allele, which was inserted upstream of the luciferase reporter gene in the pGL3 basic plasmid. b The relative luciferase activity of the two constructs in DU145 and PC3 cells was normalized against the internal control of Renilla luciferase. The data are mean ± SEM and are representative of three independent experiments performed in triplicate. **P < 0.01 compared with the construct counterpart. c EMSA with biotin-labeled probes (T or C) and nuclear extracts of DU145, PC3, HEK293T, or SGC7901 cell lines. The rs4705342 T-allelic probe had stronger binding affinity with nuclear proteins than the C allelic probe (lane 3 vs. lane 4, lane 5 vs. lane 6, lane 7 vs. lane 8, lane 9 vs. lane 10). Lanes 1, 2 are negative and positive control, respectively. d EMSA with biotin-labeled probes (T or C) and nuclear extracts from PC3 cell and a special nuclear protein binding can be abolished by 100-fold unlabeled T-allelic probe
DU145 and PC3 cells were transfected with vector DNA containing either the T or C allele of rs4705342, and using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Transfection efficiency was verified by fluorescence microscopy.
Electrophoretic mobility shift assay (EMSA)
To further examine whether rs4705342T>C had an effect on nuclear factor-binding affinity, we performed EMSA with the LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, USA). Synthetic double-stranded oligonucleotides 5′-AAGTACCATGAATGATGCTGTTAGG-3′ and 5′-AAGTACCACGAATGATGCTGTTAGG-3′ (i.e., bold base was the variant site) corresponding to the rs4705342T>C sequence from the miR-143 promoter region were labeled with biotin. For each gel-shift reaction (20 μl), a total of 100-fmol-labeled probe were combined with 4 μg nuclear protein extracted from DU145, PC3, HEK293T or SGC7901 cells, 1 μg poly (deoxyinosinic-deoxycytidylic acid), and 10× binding buffer. The reaction process took up 30 min at room temperature. The competition assay was performed by adding cognate unlabeled oligonucleotides and preincubating for 40 min at room temperature with nuclear extracts before the addition of labeled probe. After incubation, the protein–DNA complexes were separated on a native nondenaturing 4.5 % polyacrylamide gel and then transferred to a nylon membrane. The signals were detected by a chemiluminescent reaction with the stabilized streptavidin-horseradish peroxidase conjugate according to the manufacturer’s instructions and visualized by autoradiography.
Cell proliferation and migration assay
To detect the function of miR-143 on suppression of cell proliferation and migration, transfections with hsa-miR-143 mimics (5′-UGAGAUGAAGCACUGUAGCUC-3′) and stable negative control (NC) (5′-UUCUCCGAACGUGUCACGUTT-3′) (GenePharma, Shanghai, China) were first performed at concentration of 40 nmol/l with Lipofectamine 2000. Cell culture was then continued for 24, 48, and 72 h and assessed using the Cell Counting Kit-8 (CCK-8; Dojindo, Tokyo, Japan). The absorbance was measured at 450 nm using the Infinite M200 spectrophotometer (Tecan, Männedorf, Schweiz).
The anti-proliferative effect was also assessed by colony formation assay. Briefly, cells were collected 48 h after transfection with NC or miR-143, and 2,000 cells were placed into a six-well plate. The cells were then maintained in RPMI 1,640 containing 10 % FBS. After 10 days, cells were fixed with 95 % methanol and stained with 0.1 % crystal violet. Visible colonies were manually counted. Triplicate wells were measured in each treatment group.
For the migration assay, 1 × 105 transfected cells in serum-free media were placed into the upper chamber of a transwell. Media containing 10 % FBS were added to the lower chamber. After 12 h of incubation, the cells remaining on the upper membrane were removed with cotton wool, whereas the cells that had migrated through the membrane were fixed with 95 % methanol and stained with 0.1 % crystal violet. For quantification, the cells were counted by photographing the membrane under a microscope in five predetermined fields at 200× magnification.
Identify the potential target genes of miR-143
Through reviewing the previous articles, we found that kallikrein-related peptidases (KLKs) play important roles in the development and progression of PCa (Clements et al. 2004). Recently, studies showed that miRNAs represent a novel mechanism for posttranscriptional control of KLKs expression which may be involved in cancer pathogenesis (Chow et al. 2008; White et al. 2010a, b, 2012). To predict potential KLKs as miR-143 target, we used 4 computer-aided algorithms, including TargetScan, PicTar, DIANAmT and miRanda software. The combination of these 4 approaches was used to provide a more accurate assessment of real miRNA targets. As a result, KLK2 and KLK13 were predicted by all of these software packages. We then chose KLK2 for further identification based on its potential roles in PCa as reported previously (Mize et al. 2008; Recker et al. 2000; Rehault et al. 2001).
Reporter plasmids construction and luciferase reporter assay
To experimentally validate the predicted miR-143-KLK2 interaction, luciferase reporter plasmids were constructed and fragments of 3′ untranslated region (3′ UTR) of KLK2 (207-bp) with the wild or mutant miR-143 binding site (Fig. 3a) were inserted downstream of the luciferase gene in the pGL3-promoter vector (Promega, WI, USA). Constructs were verified by DNA sequencing.
Forty-eight hours after co-transfection with miR-143 or NC in DU145 cells, luciferase activity in lysates was measured with a Dual-Luciferase Reporter Assay System (Promega, WI, USA) and normalized to Renilla activity.
Western blotting analysis
Western blotting assay was performed to detect the expression of KLK2 protein in DU145 cell. Forty-eight hours after transfection with miR-143 or NC, cells were harvested and then lysed in RIPA buffer with 0.5 % PMSF and stood for 30 min on ice. After centrifugation at 12,000g for 15 min at 4 °C, the supernatants were collected as protein samples. Protein was then separated by 12.5 % SDS-PAGE minigel and transferred onto PVDF membrane (Millipore, MA, USA). After probed with 1:1,000 diluted rabbit monoclonal KLK2 antibody (Epitomics, CA, USA) at 4 °C overnight, membranes were incubated with goat anti-rabbit peroxidase-conjugated secondary antibody (Zhongshan Goldenbridge Biotechnology, Beijing, China) and visualized using enhanced chemiluminescence (Millipore, MA, USA). α-Tubulin was used as an endogenous protein for normalization.
Statistical analysis
The differences in the distribution of selected variables and genotype of rs4705342T>C between PCa cases and controls were assessed using the Student’s t test (for continuous variables) or Chi-square test (for categorical variables). Hardy–Weinberg equilibrium (HWE) was tested by a goodness-of-fit Chi-square test. The associations between rs4705342T>C and PCa risk were estimated with unconditional univariate and multivariate logistic regression analyses by calculating the odds ratios (ORs) (Shahi et al. 2006) and their 95 % confidence intervals (CIs). All the ORs were adjusted for age, smoking, and drinking status. The expression of miR-143 in PCa and paired adjacent normal tissues were compared by paired t test. The Spearman test was used to analyze the correlation between miR-143 expression and KLK2 mRNA expression levels. P < 0.05 was considered statistically significance. All tests were two-sided and were conducted using SAS software (version 9.1.3; SAS Institute, Inc., Cary, NC, USA).
Results
miR-143 expression in PCa tissues and cell lines
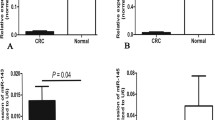
We performed RT-PCR analysis to detect the expression level of mature miR-143 in PCa tissues and cell lines. As shown in Figure S2A, the expression level of miR-143 was decreased in 18 (81.8 %) PCa tissues compared with that in the paired adjacent normal tissues, which had statistical significance (P = 0.014). In addition, the expression levels of miR-143 in DU145 and PC3 cell lines were also lower than that in the normal HEK293T cell (P = 0.003 for DU145 and P < 0.001 for PC3, Figure S2B).
Characteristics of the study population
The distribution of selected characteristics between 608 PCa cases and 709 control subjects in the present study is shown in Table 1. There were no differences in the distribution of age (P = 0.218) and alcohol drinking (P = 0.093) between cases and controls. However, there were more smokers among the cases (55.9 %) than among the controls (50.2 %), and the proportion of subjects who smoked greater than 20 pack-years was larger in the cases than in the controls (P < 0.001). Besides, there was statistically significant difference in the distribution of family history of cancers between the cases and controls (P < 0.001). Of the 608 PCa cases, 62.0 % were in localized stage and the other 38.0 % were in advanced stage.
Association between miR-143 rs4705342T>C and PCa risk
The genotype and allele distributions of rs4705342T>C in the cases and controls are shown in Table 2. The observed genotype frequencies for the SNP among the control subjects were in agreement with HWE (χ 2 = 0.304, P = 0.580). The frequency of rs4705342 C allele was higher in the controls (34 %) than in the cases (28 %) (P < 0.001). There was also statistically significant difference in the distribution of genotypes between cases and controls (P = 0.003). miR-143 rs4705342T>C decreased the risk of PCa in all genetic models (adjusted OR = 0.71, 95 % CI = 0.56–0.89, P = 0.004 for TC vs. TT; 0.58, 0.39–0.85, 0.007 for CC vs. TT; 0.68, 0.55–0.85, 0.001 for TC/CC vs. TT). In addition, we also found that C allele was associated with the decreased risk of PCa in a dose–response manner (P trend = 0.001).
We further evaluated the effect of the SNP on PCa risk with stratifying analyses by age, pack-years smoked, drinking status, and family history of cancers. As presented in Table S2, the decreased risk was more pronounced among younger subjects (adjusted OR = 0.61, 95 % CI = 0.45–0.84, P = 0.002), heavy smokers (pack-years >20) (0.54, 0.36–0.81, 0.002), and the subjects without family history of cancers (0.63, 0.50–0.81, 0.001). Furthermore, we also evaluated the association between rs4705342T>C and clinicopathological characteristics of PCa (Table S3). Results showed that the reduced risk of PCa was statistically significant in localized stage (adjusted OR = 0.72, 95 % CI = 0.56–0.93, P = 0.016), advanced stage (0.62, 0.46–0.83, 0.002), Gleason score ≥7 (0.71, 0.52–0.98, 0.031 for = 7; 0.58, 0.42–0.80, 0.001 for >7), PSA level ≤ 20 (0.72, 0.54–0.97, 0.034), and PSA level >20 (0.66, 0.51–0.85, 0.002).
Effect of rs4705342T>C on miR-143 promoter activity
To investigate the effect of the variant on miR-143 promoter activity, luciferase reporter vectors containing either rs4705342 T or C allele (Fig. 1a) were constructed and transfected into PCa cells. As shown in Fig. 1b, the vectors carrying the C allele had 43 and 36 % increase in the relative luciferase activities, compared with the ones carrying the risk-associated T allele respectively in DU145 and PC3 cells (P < 0.01 for both). This finding was consistent with our previous observation of an association between the C allele and decreased PCa risk.
Effect of rs4705342 on nuclear protein-binding activity
To explore the molecular mechanisms of the association between rs4705342 and miR-143 promoter activity, we performed EMSA to examine whether the allelic difference may influence the binding capacities of miR-143 promoter for nuclear factors. Two probes containing either T or C allele were synthesized, and biotin-labeled and unlabeled oligonucleotides were used as competitors. As shown in Fig. 1c, rs4705342T allele probe formed much more DNA–protein complexes with the nuclear extracts from resting DU145, PC3, and HEK293T cells than did C allele probe (lane 3 vs. lane 4, lane 5 vs. lane 6, lane 7 vs. lane 8). Moreover, the addition of excessive unlabeled T allele probe caused much more obvious reduction in the DNA–protein (extracted from PC3 cells) complexes than the unlabeled C allele probe (lane 3 vs. lane 5, Fig. 1d) and almost abolishment of the C allele probe–protein complexes. The results indicated that the promoter carrying the rs4705342T allele binds more robustly to nuclear factors, which was not in agreement with our previous results of case–control study and miR-143 promoter activity analyses. We deduced that the special nuclear factors were transcription inhibitors although we failed to reveal the reality.
Effect of miR-143 on cell proliferation and cell migration
In an attempt to determine the functional role of miR-143 in PCa carcinogenesis, we assessed its impact on PCa cells growth and migration by transfecting miR-143 mimics or stable negative control. The level of miR-143 was significantly increased in miR-143-transfected DU145 and PC3 cells (Fig. 2a). The CCK-8 results showed that the increased expression of miR-143 induced significant inhibition on cell growth. Forty-eight hours after transfected with miR-143, cells proliferation was significantly decreased compared with cells transfected with NC (P = 0.001 for DU145; P = 0.003 for PC3). The inhibition effect decreased after 48 h possibly because cell growth could be slow when there were a large number of cells (Fig. 2b). Similarly, the results of colony formation assays revealed that clonogenic survival was decreased following over-expression of miR-143 in PCa cells. The rate of colony formation decreased 28 % (P = 0.039) and 39 % (P = 0.022), respectively, for DU145 and PC3 cells (Fig. 2c). Furthermore, miR-143 affected not only cell growth but also cell migration. Cells transfected with miR-143 mimics exhibited significant impairment of migratory ability when comparing with NC-transfected cells. The number of transferred cells was reduced to 48 % (DU145) and 45 % (PC3) (both P < 0.001) (Fig. 2d).
Effects of miR-143 on proliferation, colony formation, and migration of DU145 and PC3 cell lines. a Transfecting miR-143 mimics significantly increased the expression level of miR-143 in PCa cell lines compared with transfecting negative control. **P < 0.01. b Over-expression of miR-143 inhibited cell growth using CCK-8 assay. c Colony-forming growth assay showed that miR-143 significantly reduced colony formation of PCa cell lines. *P < 0.05. d Inhibition of migration ability of PCa cell lines transfected with miR-143, compared with the negative control. ***P < 0.001. The assays were performed in triplicate independent experiments
Identification of miR-143 targeting on KLK2
To confirm the potential relationship between miR-143 and KLK2, we first detected the KLK2 mRNA expression level and miR-143 in seven paired prostate tissues (RNA extracted from the paraffin-embedded sections and one paired new tissues was degradated, and the KLK2 mRNA level could not be detected). RT-PCR results showed that KLK2 mRNA levels in PCa tissues were higher than those in paired adjacent normal tissues (P = 0.028) (Fig. 3b), and there was an inverse correlation of expression between miR-143 and KLK2 mRNA (r = −0.741, P = 0.002) (Fig. 3c). We then detected the effect of miR-143 on KLK2 protein expression by transfecting DU145 cell with miR-143 or NC as it had moderate expression of KLK2. As shown in Fig. 3d, the restoration of miR-143 in DU145 cell significantly repressed KLK2 protein level compared with the negative control. To further investigate whether the predicted binding site of miR-143 to KLK2 3′-UTR is responsible for this regulation, we constructed luciferase reporter vectors by using pGL3-promoter vector containing fragments of KLK2 3′-UTR with the wild or mutant miR-143 binding site and then co-transfected them with miR-143 or NC into DU145 cells. As shown in Fig. 3e, wild vector co-transfected with miR-143 had a 54 % decrease in relative luciferase activity compared with that co-transfected with NC (P = 0.014). Meanwhile, miR-143 could not affect the luciferase reporter activity of the mutant plasmid (Fig. 3e).
miR-143 directly targets KLK2 by binding to its 3′-UTR. a Predicted miR-143 binding site within KLK2 3′-UTR and its mutated version. b The expression level of KLK2 mRNA was higher in PCa tissues than that in the adjacent normal tissues (P = 0.028). c An inverse correlation between KLK2 mRNA and miR-143 level was observed in seven paired PCa tissues (Spearman’s correlation, r = −0.741, P = 0.002). d Western blotting assay showed the expression level of KLK2 protein following treatment of DU145 cells with miR-143 mimics or NC. α-Tubulin was used as control. e The luciferase reporter plasmid containing wild or mutant KLK2 3′-UTR was co-transfected into DU145 cells with miR-143 mimics or NC. The luciferase activity was normalized against the internal control of Renilla luciferase. *P < 0.05
Discussion
In the present study, we first identified the reduction of miR-143 expression in PCa both in vivo and in vitro which was consistent with the previous miRNA profiling studies in PCa (Ambs et al. 2008; Schaefer et al. 2010; Szczyrba et al. 2010). We then discovered that a novel functional variant rs4705342T>C located in the promoter of miR-143 precursor was associated with PCa risk and played a critical role on the down-regulation of miR-143 by affecting the promoter activity in PCa. Moreover, we confirm that miR-143 exerts its anticancer function through the suppression on cell proliferation and migration, at least in part by specifically targeting the KLK2 gene.
The development of tumor is complicated, and accumulating evidence suggests that dysregulation of miRNAs participates in the pathogenesis of most human malignancies, which can be caused by various mechanisms, including deletions, amplifications, or mutations involving miRNA loci, epigenetic silencing, or the dysregulation of transcription factors that target specific miRNAs (Croce 2009; Vrba et al. 2011). Recently, it has been proposed that SNPs in miRNA genes are likely to account for their abnormal expression and increased cancer risk (Ryan et al. 2010). On the bases that the promoter of a gene is a crucial control region for its transcription initiation (Smale and Kadonaga 2003; Weis and Reinberg 1992), SNPs occurring in the promoter of miRNAs would also be predicted to affect miRNA levels. Sevignani et al. (2007) found that most of the sequence differences in miRNA genes in tumor-susceptible mice rather than tumor-resistant mice occur in the promoters. For human common cancer, there have been less reports on the association between miRNA promoter SNPs and cancer risk (Luo et al. 2011; Xu et al. 2011b).
In the present study, we selected SNPs in the 1,000-bp upstream from the transcription start site of miR-143 precursor in dbSNP with the condition that MAF >0.05 in Chinese population and finally identified that rs4705342T>C was associated with the risk of PCa and the C allele had a protective effect. In agreement with the case–control study, functional studies showed that the miR-143 promoter with the C allele had an increased transcription activity in vitro. However, the result of EMSA displayed that the C allele decreased the binding affinity with transcriptional factors. We deduced that the SNP rs4705342 altered the binding affinity with some special transcriptional inhibitors rather than activators although we failed to find out the matter. For lack of tissue samples of the subjects with the genotyping of rs4705342, we could not confirm the effect of the SNP on miR-143 expression in vivo. The association between the potentially functional variant rs4705342 and PCa risk should be investigated in other ethnic groups, and the molecular mechanism is worthy for further study.
Dysregulation of miRNAs may lead to alterations in a variety of biologic processes, including organ development, cell proliferation, cell differentiation, and apoptosis, which play critical roles in carcinogenesis (Bartel 2004; Chitwood and Timmermans 2010; Kosik 2010). Indeed, several key functions on antitumor have been attributed to miR-143 in the context of tumorigenesis. For instance, down-regulated expression of miR-143 and miR-145 was observed in bladder cancer cells, whereas enhanced miR-143 and miR-145 induced growth suppression through repressing the mitogen-activated protein kinase (MAPK) and phosphoinositide-3 kinase/activation of protein kinase B (PI3K/Akt) signaling pathways (Noguchi et al. 2013). Zhang et al. (2012) also reported that miR-143 inhibited cell invasion and migration in colorectal cancer through the regulation of metastasis-associated in colon cancer-1 (MACC1). For PCa, Xu et al. (2011a) suggested that miR-143 displayed antitumor function through the suppression of KRAS. Clape et al. showed that miR-143 could down-regulate the expression of ERK5 and abolish PCa progression in mice (Clape et al. 2009). In the present study, we identified that miR-143 inhibited PCa cell proliferation and migration by inhibition of KLK2.
KLK2 is a member of the tissue kallikrein family of genes, almost exclusively produced by the epithelial cells of the prostate, and is considered prostate-specific (Clements et al. 2004; Obiezu and Diamandis 2005). KLK2 may mediate cell proliferation in both the normal and malignant prostate by interaction with the insulin-like growth factor (IGF) axis through cleaving several insulin-like growth factor-binding proteins (IGFBPs), a process that enhances IGF-1 bioavailability (Rehault et al. 2001). As a member of kallikreins, KLK2 is also involved in direct or indirect extracellular matrix degradation to facilitate tissue remodeling and thus may regulate (positively or negatively) re-epithelialization and cell migration and tumor invasion (Clements et al. 2004; Deperthes et al. 1996; Lovgren et al. 1999; Mikolajczyk et al. 1999). The expression of KLKs was considered to be commonly regulated by hormone (Riegman et al. 1991; Sun et al. 1997; Young et al. 1991, 1992). miRNAs were explored to participate in regulation of KLKs expression, and miRNA-KLKs axis of interaction was suggested to a new dimension in the pathogenesis of some neoplasms. Chow et al. (2008) first revealed that about 60 % of known miRNAs were predicted to target KLKs by silico analyses. In this study, we used four computer-aided algorithms including TargetScan, PicTar, DIANAmT and miRanda software to predict potential KLKs as miR-143 target. As a result, KLK2 and KLK13 were predicted by all of these software packages. Previous studies had demonstrated that KLK2 contributed to an enhanced diagnosis of PCa (Becker et al. 2000), which shared an 80 % sequence homology with PSA (Kumar et al. 1997). miRNA-kallikrein network was involved in the development of PCa, and KLK2 was predicted to be targeted by miRNAs in PCa (Samaan et al. 2014). Avgeris et al. also found that miR-378 regulated the expression levels of KLK2 and KLK4 to affect the risk of PCa progression and relapse. However, few studies reported the effect of KLK13 in PCa risk. Change and his colleagues had demonstrated that KLK13 contributed to an independent favorable prognostic marker for breast cancer (Chang et al. 2002). Additionally, a PCa case and control study revealed that KLK13 SNPs were not associated with PCa risk (Lose et al. 2013). Therefore, we chose KLK2 for further identification based on its potential roles in PCa as reported previously. At present study, we observed that miR-143 reduced KLK2 protein expression level in PCa cell which was consistent with the results of luciferase assay. Besides, KLK2 mRNA level in prostate tissues was inversely correlated with the expression of miR-143. Thus, miR-143-KLK2 interaction may project a new element in the mechanism of development and progression of PCa.
In conclusion, we firstly provide evidence that the functional variant rs4705342 in the promoter region of miR-143 contributes to the risk of PCa in a Chinese population. Moreover, miR-143 can suppress PCa through regulating KLK2. Further prospective studies and functional studies are needed to confirm our findings.
References
Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355. doi:10.1038/nature02871nature02871
Ambs S, Prueitt RL, Yi M et al (2008) Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 68(15):6162–6170. doi:10.1158/0008-5472.CAN-08-0144
Andriole GL, Crawford ED, Grubb RL 3rd et al (2009) Mortality results from a randomized prostate-cancer screening trial. N Engl J Med 360(13):1310–1319. doi:10.1056/NEJMoa0810696
Baade PD, Youlden DR, Krnjacki LJ (2009) International epidemiology of prostate cancer: geographical distribution and secular trends. Mol Nutr Food Res 53(2):171–184. doi:10.1002/mnfr.200700511
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Becker C, Piironen T, Pettersson K et al (2000) Discrimination of men with prostate cancer from those with benign disease by measurements of human glandular kallikrein 2 (HK2) in serum. The Journal of urology 163(1):311–316
Chang A, Yousef GM, Scorilas A et al (2002) Human kallikrein gene 13 (KLK13) expression by quantitative RT-PCR: an independent indicator of favourable prognosis in breast cancer. Br J Cancer 86(9):1457–1464
Chen C, Ridzon DA, Broomer AJ et al (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20):e179. doi:10.1093/nar/gni178
Chen X, Guo X, Zhang H et al (2009) Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28(10):1385–1392. doi:10.1038/onc.2008.474
Chitwood DH, Timmermans MC (2010) Small RNAs are on the move. Nature 467(7314):415–419. doi:10.1038/nature09351
Chow TF, Crow M, Earle T, El-Said H, Diamandis EP, Yousef GM (2008) Kallikreins as microRNA targets: an in silico and experimental-based analysis. Biol Chem 389(6):731–738. doi:10.1515/BC.2008.07110.1515/BC.2008.071
Clape C, Fritz V, Henriquet C et al (2009) miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS ONE 4(10):e7542. doi:10.1371/journal.pone.0007542
Clements JA, Willemsen NM, Myers SA, Dong Y (2004) The tissue kallikrein family of serine proteases: functional roles in human disease and potential as clinical biomarkers. Crit Rev Clin Lab Sci 41(3):265–312. doi:10.1080/10408360490471931
Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10(10):704–714. doi:10.1038/nrg2634
Deperthes D, Frenette G, Brillard-Bourdet M et al (1996) Potential involvement of kallikrein hK2 in the hydrolysis of the human seminal vesicle proteins after ejaculation. J Androl 17(6):659–665
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90. doi:10.3322/caac.20107
Kosik KS (2010) MicroRNAs and cellular phenotypy. Cell 143(1):21–26. doi:10.1016/j.cell.2010.09.008
Kumar A, Mikolajczyk SD, Goel AS, Millar LS, Saedi MS (1997) Expression of pro form of prostate-specific antigen by mammalian cells and its conversion to mature, active form by human kallikrein 2. Cancer Res 57(15):3111–3114
Lin T, Dong W, Huang J et al (2009) MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol 181(3):1372–1380. doi:10.1016/j.juro.2008.10.149
Lose F, Batra J, O’Mara T et al (2013) Common variation in Kallikrein genes KLK5, KLK6, KLK12, and KLK13 and risk of prostate cancer and tumor aggressiveness. Urologic oncology 31(5):635–643
Lovgren J, Airas K, Lilja H (1999) Enzymatic action of human glandular kallikrein 2 (hK2). Substrate specificity and regulation by Zn2+ and extracellular protease inhibitors. Eur J Biochem 262(3):781–789
Luo X, Yang W, Ye DQ et al (2011) A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet 7(6):e1002128. doi:10.1371/journal.pgen.1002128PGENETICS-D-10-00139
Mikolajczyk SD, Millar LS, Kumar A, Saedi MS (1999) Prostatic human kallikrein 2 inactivates and complexes with plasminogen activator inhibitor-1. Int J Cancer 81(3):438–442. doi:10.1002/(SICI)1097-0215(19990505)81:3<438:AID-IJC18>3.0.CO;2-U
Mize GJ, Wang W, Takayama TK (2008) Prostate-specific kallikreins-2 and -4 enhance the proliferation of DU-145 prostate cancer cells through protease-activated receptors-1 and -2. Mol Cancer Res 6(6):1043–1051. doi:10.1158/1541-7786.MCR-08-0096
Noguchi S, Yasui Y, Iwasaki J et al (2013) Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett 328(2):353–361. doi:10.1016/j.canlet.2012.10.017
Obiezu CV, Diamandis EP (2005) Human tissue kallikrein gene family: applications in cancer. Cancer Lett 224(1):1–22. doi:10.1016/j.canlet.2004.09.024
Recker F, Kwiatkowski MK, Piironen T et al (2000) Human glandular kallikrein as a tool to improve discrimination of poorly differentiated and non-organ-confined prostate cancer compared with prostate-specific antigen. Urology 55(4):481–485
Rehault S, Monget P, Mazerbourg S et al (2001) Insulin-like growth factor binding proteins (IGFBPs) as potential physiological substrates for human kallikreins hK2 and hK3. Eur J Biochem 268(10):2960–2968
Riegman PH, Vlietstra RJ, van der Korput HA, Romijn JC, Trapman J (1991) Identification and androgen-regulated expression of two major human glandular kallikrein-1 (hGK-1) mRNA species. Mol Cell Endocrinol 76(1–3):181–190
Ryan BM, Robles AI, Harris CC (2010) Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 10(6):389–402. doi:10.1038/nrc2867
Samaan S, Lichner Z, Ding Q et al (2014) Kallikreins are involved in an miRNA network that contributes to prostate cancer progression. Biol Chem 395(9):991–1001
Schaefer A, Jung M, Mollenkopf HJ et al (2010) Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer 126(5):1166–1176. doi:10.1002/ijc.24827
Schroder FH, Hugosson J, Roobol MJ et al (2009) Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 360(13):1320–1328. doi:10.1056/NEJMoa0810084
Sevignani C, Calin GA, Nnadi SC et al (2007) MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA 104(19):8017–8022. doi:10.1073/pnas.0702177104
Shahi P, Loukianiouk S, Bohne-Lang A et al (2006) Argonaute–a database for gene regulation by mammalian microRNAs. Nucleic acids res 34(database issue):D115-8. doi:10.1093/nar/gkj093
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62(1):10–29. doi:10.3322/caac.20138
Smale ST, Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72:449–479. doi:10.1146/annurev.biochem.72.121801.161520121801.161520
Sun Z, Pan J, Balk SP (1997) Androgen receptor-associated protein complex binds upstream of the androgen-responsive elements in the promoters of human prostate-specific antigen and kallikrein 2 genes. Nucleic Acids Res 25(16):3318–3325
Szczyrba J, Loprich E, Wach S et al (2010) The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res 8(4):529–538. doi:10.1158/1541-7786.MCR-09-0443
Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y (2009) Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 77(1):12–21. doi:10.1159/000218166
Tong AW, Fulgham P, Jay C et al (2009) MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther 16(3):206–216. doi:10.1038/cgt.2008.77
Ugras S, Brill E, Jacobsen A et al (2011) Small RNA sequencing and functional characterization reveals MicroRNA-143 tumor suppressor activity in liposarcoma. Cancer Res 71(17):5659–5669. doi:10.1158/0008-5472.CAN-11-0890
Vrba L, Garbe JC, Stampfer MR, Futscher BW (2011) Epigenetic regulation of normal human mammary cell type-specific miRNAs. Genome Res 21(12):2026–2037. doi:10.1101/gr.123935.111
Wach S, Nolte E, Szczyrba J et al (2012) MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer 130(3):611–621. doi:10.1002/ijc.26064
Weis L, Reinberg D (1992) Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J 6(14):3300–3309
White NM, Bui A, Mejia-Guerrero S et al (2010a) Dysregulation of kallikrein-related peptidases in renal cell carcinoma: potential targets of miRNAs. Biol Chem 391(4):411–423. doi:10.1515/BC.2010.041
White NM, Chow TF, Mejia-Guerrero S et al (2010b) Three dysregulated miRNAs control kallikrein 10 expression and cell proliferation in ovarian cancer. Br J Cancer 102(8):1244–1253. doi:10.1038/sj.bjc.6605634
White NM, Youssef YM, Fendler A, Stephan C, Jung K, Yousef GM (2012) The miRNA-kallikrein axis of interaction: a new dimension in the pathogenesis of prostate cancer. Biol Chem 393(5):379–389. doi:10.1515/hsz-2011-0246/j/bchm.2012.393.issue-5/hsz-2011-0246/hsz-2011-0246.xml
Xu B, Niu X, Zhang X et al (2011a) miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem 350(1–2):207–213. doi:10.1007/s11010-010-0700-6
Xu Y, Liu L, Liu J et al (2011b) A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer 128(2):412–417. doi:10.1002/ijc.25342
Xue Y, Wang M, Kang M et al (2013) Association between lncrna PCGEM1 polymorphisms and prostate cancer risk. Prostate cancer and prostatic diseases 16(2):139–144. doi:10.1038/pcan.2013.6
Young CY, Montgomery BT, Andrews PE, Qui SD, Bilhartz DL, Tindall DJ (1991) Hormonal regulation of prostate-specific antigen messenger RNA in human prostatic adenocarcinoma cell line LNCaP. Cancer Res 51(14):3748–3752
Young CY, Andrews PE, Montgomery BT, Tindall DJ (1992) Tissue-specific and hormonal regulation of human prostate-specific glandular kallikrein. Biochemistry 31(3):818–824
Zhang Y, Wang Z, Chen M et al (2012) MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol Cancer 11:23. doi:10.1186/1476-4598-11-23
Zhou X, Ruan J, Wang G, Zhang W (2007) Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 3(3):e37. doi:10.1371/journal.pcbi.0030037
Acknowledgments
This study was partly supported by National Natural Science Foundation of China (81230068, and 81102089), Natural Science Foundation of Jiangsu Province (BK2011773), the Key Program for Basic Research of Jiangsu Provincial Department of Education (11KJB330002 and 12KJA330002), Jiangsu Provincial Six Talent Peaks Project (2012-SWYY-028), Specialized Research Fund for the Doctoral Program of Higher Education (20123234110001), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Haiyan Chu, Dongyan Zhong, and Jialin Tang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chu, H., Zhong, D., Tang, J. et al. A functional variant in miR-143 promoter contributes to prostate cancer risk. Arch Toxicol 90, 403–414 (2016). https://doi.org/10.1007/s00204-014-1396-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-014-1396-2