Abstract
A novel bacterial strain designated as ADMK78T was isolated from the saline desert soil. The cells were rod-shaped, Gram-stain-negative, and non-motile. The strain ADMK78T grows best at 28 °C. Phylogeny of 16S rRNA gene placed the strain ADMK78T with the members of genera Ciceribacter and Rhizobium, while the highest sequence similarity was with Rhizobium wuzhouense W44T (98.7%) and Rhizobium ipomoeae shin9-1 T (97.9%). Phylogenetic analysis based on 92 core-genes extracted from the genome sequences and average amino acid identity (AAI) revealed that the strain ADMK78T forms a distinct cluster including five species of Rhizobium, which is separate from the cluster of the genera Rhizobium and Ciceribacter. We propose re-classification of Rhizobium ipomoeae, R. wuzhouense, R. rosettiformans and R. rhizophilum into the novel genus Peteryoungia. The average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values of ADMK78T were less than 82 and 81%, respectively, among all type strains included in the genus Peteryoungia. The strain ADMK78T showed differences in physiological, phenotypic, and protein profiles estimated by MALDI-TOF MS to its closest relatives. Based on the phenotypic, chemotaxonomic properties, and phylogenetic analyses, the strain ADMK78T represents a novel species, Peteryoungia desertarenae sp. nov. The type strain is ADMK78T (= MCC 3400T; KACC 21383T; JCM 33657T). We also proposed the reclassification of Rhizobium daejeonense, R. naphthalenivorans and R. selenitireducens, into the genus Ciceribacter, based on core gene phylogeny and AAI values.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introductions
The family Rhizobiaceae Conn 1938, at present, consists of 16 genera summarized in the List of Prokaryotic Names with Standing in Nomenclature (www.bacterio.net) (Parte 2020). Most Rhizobiaceae genera like, Rhizobium, Agrobacterium, Ensifer (Sinorhizobium), Allorhizobium, Liberibacter, Pararhizobium, Neorhizobium, and Shinella are associated with plants. However, members of Rhizobiaceae have been reported from diverse ecosystems, including from aquatic and marine ecosystems (Peix et al. 2005; Cao et al. 2020). Members of the genera like Georhizobium, Hoeflea, Lentilitoribacter, Martelella, Rhizobium, and Pseudorhizobium have been reported from marine or aquatic environments (Peix et al. 2005; Rivas et al. 2005; Park et al. 2013; Kimes et al. 2015; Cao et al. 2020; Chaudhari et al. 2020). High-throughput amplicon sequence-based study suggested the dominance of the members of Rhizobiaceae in the brackish water Pangong Lake (Chaudhari et al. 2020).
Rhizobium is one of the main genera in the family Rhizobiaceae, and it was first proposed in 1889 (Frank 1889; Kuykendall et al. 2005). Presently, the genus Rhizobium comprises of 91 validly published species (https://lpsn.dsmz.de/genus/rhizobium). The majority of Rhizobium species are known for their symbiotic fixation of nitrogen within the root nodules of leguminous plants (Lindström et al. 2010, 2015). However, non-symbiotic and free-living members of Rhizobium have been found in various niches viz. soils, including the rhizosphere, bioreactor, lake water, arsenic-rich groundwater, and beach sand (Quan et al. 2005; Panday et al. 2011; Ramana et al. 2013; Sheu et al. 2016; Li et al. 2017a, b; Mohapatra et al. 2017). Recently, multiple new genus names such as Allorhizobium, Pararhizobium, Neorhizobium, and Pseudorhizobium have been proposed by dissecting Rhizobium (Mousavi et al. 2014; Mousavi et al. 2015; Kimes et al. 2015; Hördt et al. 2020).
Rann of Kachchh is reputed to be the largest salt desert in the world and is a transitional area between marine and terrestrial ecosystems (Pandit et al. 2015). The region experiences diagonal fluctuations of temperature, in summers it reaches up to 50 °C and drops down below zero during winters. Due to the hot and hypersaline environment, there is a vast possibility of isolating novel halophilic and halotolerant microorganisms with high economic and industrial potential (Ruginescu et al. 2020). During the investigations on bacterial diversity of the saline desert soil collected from the Rann of Kachchh, Gujarat, India, the strain ADMK78T was isolated with 16S rRNA gene sequence similarity less than 98.7%. The present study aimed to demonstrate the taxonomic position of strain ADMK78T through a polyphasic and genomic analysis.
Materials and methods
Isolation of bacterium
A bacterial strain ADMK78T was isolated on Zobell Marine Agar following the serial dilution of the saline desert soil (23.7337° N, 69.8597° E) collected from the Rann of Kachchh, Gujarat, India. The newly isolated strain ADMK78T was maintained on Zobell Marine Agar at 37 °C, and preserved at − 80 °C as a suspension in 20% (v/v) glycerol and by lyophilization with 20% (w/v) skimmed milk.
16S RNA phylogeny
High-quality genomic DNA was extracted from the strain following the manual bacterial genomic DNA isolation protocol using CTAB (Minas et al. 2011). The 16S rRNA gene sequence was amplified using universal primers (27F: 5′-AGAGTTTGATCCTGGCTCAG-3′and 1492R: 5′-TACGGCTACCTTGTTACGACTT-3′) according to the methods described by Gulati et al (2008), and the amplified product was purified and subjected to DNA sequencing using the ABI PRISM Big Dye Terminator v3.1 Cycle Sequencing kit on a 3730xl Genetic Analyzer (Applied BioSystems, Thermo Scientific, USA). The similarity search for the 16S rRNA gene sequence of strain ADMK78T was performed against the type strains of prokaryotic species in the EzBioCloud’s valid species database (Yoon et al. 2017). The 16S rRNA gene sequence of strain ADMK78T was also used as queries to closely related gene sequences using the NCBI BLASTn tool against the non-redundant nucleotide database (Altschul et al. 1990). Multiple alignments of sequences of strain ADMK78T and its nearest neighbours retrieved from EzBioCloud’s server and NCBI GenBank were performed using ClustalW (Larkin et al 2007). Phylogenetic analysis of the 16S rRNA was performed using the neighbour-joining, maximum-parsimony, and maximum-likelihood algorithms in the MEGA software (version 10.2.1) (Kumar et al. 2018). Bootstrap values were determined based on 1000 replications (Efron et al. 1996). The newly generated 16S rRNA gene sequence was deposited with NCBI GenBank under accession MK942856.
Whole genome sequencing and core-genes based phylogeny
The strain was grown on Zobell Marine liquid medium incubated at 28 °C for 32 h and genomic DNA was harvested following the JGI protocol version 3 for bacterial genomic DNA isolation using CTAB (Minas et al. 2011). Genome sequencing was performed using a hybrid approach of two platforms, first on an Illumina MiSeq platform with 2 × 250 bp v2 chemistry, followed by sequencing with Oxford Nanopore Technology (ONT) on a minION platform. The Nanopore reads were assembled using Canu v. 2.0 with default settings (Koren et al. 2017). The overlaps between the ends of circular contigs were identified using NUCmer v. 3.1 (Kurtz et al. 2004) and removed using a custom Perl script. Two rounds of polishing were performed using the paired-end Illumina reads. In each round, the Illumina reads were mapped to the genome assembly using bowtie2 v. 2.3.4.1 with default parameters, followed by polishing using Pilon v. 1.23 with default settings (Langmead and Salzberg 2012; Walker et al. 2014). Whole-genome sequences were annotated using the RAST web server (http://rast.nmpdr.org/rast.cgi) (Aziz et al. 2008). The biosynthetic gene clusters (BGCs) for various secondary metabolites, were identified by using an online genome mining pipeline antiSMASH 5.0 (Blin et al. 2019). The complete genome sequence of strain ADMK78T was deposited with NCBI under the accession number CP058350- CP058352.
Genome sequences from species of the different genera of the family Rhizobiaceae were downloaded in FASTA format from the NCBI database (www.ncbi.nlm.nih.gov). To find genome sequences of closely related strains, gene sequence of rpoB gene of strain ADMK78T was used as a search query against RefSeq select proteins database of Rhizobiaceae group (taxid:356). A total of 197 genomes were used for core-gene phylogeny (Table S1). The core gene-based phylogenetic analysis was carried out using the UBCG pipeline from the concatenated sequences of 92 core genes extracted by UBCG, and a maximum-likelihood phylogenetic tree was inferred using RAxML version 8.2.8 with the GTRGAMMA model and 100 bootstrap replications (Stamatakis et al. 2014; Na et al. 2018). The core-gene phylogenetic tree was displayed using iTOL (Letunic and Bork 2019). The Average Nucleotide Identity (ANI) was determined between strain ADMK78T and 81 strains of the Rhizobiaceae family using FastANI (Jain et al. 2018) with the default settings (kmer = 16, fragment length = 3000, minimum shared fraction = 0.2). Digital DDH values and confidence intervals were calculated using the recommended settings of the GGDC 2.1 (Meier-Kolthoff et al. 2013). The percentage of conserved proteins (POCP) was calculated based on the previously described approach (Qin et al. 2014). The pairwise amino acid identity (AAI) was calculated using the ComparM software package (https://github.com/dparks1134/CompareM). The results of ANI and AAI were displayed as a heatmap using HeatMap Illustrator of TBtools (Chen et al. 2020). The AAI values were plotted for the strain ADMK78T and type species of each genera of family Rhizobiaceae using ggplot function in R.
Physiology and chemotaxonomy
The type strains of Rhizobium ipomoeae LMG 27163T and Ciceribacter lividus DSM 25528T were obtained from the BCCM/LMG Bacteria Collection, Belgium (LMG) and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) respectively. Both type strains were used as reference strains and evaluated together under identical experimental conditions to those for strain ADMK78T.
Morphological, physiological, and biochemical tests for strain ADMK78T were performed on Zobell Marine Agar plates incubated under aerobic conditions. Gram-staining (K001, Himedia, India) was used following the manufacturer's instructions. The hanging drop technique was used to check the motility (Tittsler and Sandholzer 1936). Scanning electron microscopy was performed to observe cell morphology as described in Rahi et al. (2017). Oxidase disc (DD018, Himedia, India) was used for testing oxidase activity, and catalase activity was determined by bubble formation in a 3% (v/v) H2O2 solution. Growth at different temperatures (4, 10, 15, 20, 28, 37, 45 and 55 °C), NaCl concentrations [0–2% (w/v) at 0.5% intervals] and pH values (4.0–11.0 at 1.0 pH unit intervals) was examined after incubation in Zobell Marine broth for 7 days in automated microbial growth analyzer (Bioscreen C, OY Growth Curves, Finland). The initial pH of the inoculation broth was adjusted using 1 M HCl and 1 M NaOH. Biochemical characteristics, enzyme activities, and oxidation/or reduction of carbon sources were performed using the API 20E and API ZYM systems (07584D and 25,200, bioMérieux, France) and Biolog GN III system (OmniLog, Biolog, USA) following manufacturer’s instructions.
For analysis of chemotaxonomic features, the strain was grown on Zobell Marine Agar, while Rhizobium ipomoeae LMG 27163T and Ciceribacter lividus DSM 25528T were grown on TSA, and incubated at 28 °C, and cell biomass was harvested after 24 h. Preparation and analysis of fatty acid methyl esters were performed as described by Sasser (2001) using the Microbial Identification System (MIDI) and the Microbial Identification software package (Sherlock version 6.1; MIDI database, TSBA6). Whole-cell proteins were extracted using ethanol/formic acid after 24 h growth on TSA, to generate the Mean Spectral Profile (MSP). The proteins ranging from 2 to 20 KDa were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF MS) autoflex speed (Bruker Daltonik GmbH, Germany) with Biotyper 3.0 database (Kurli et al. 2018). A total of 27 replicate spectra were used to generate a MALDI-TOF MS mean spectra profile (MSP) of strain ADMK78T, which was compared with MSPs of the reference strains Rhizobium ipomoeae shin9-1T and Ciceribacter lividus MSSRFBL1T generated during this study following the same procedure (Kurli et al. 2018).
Results and discussion
16S RNA phylogeny
Strain ADMK78T showed the highest similarity to Rhizobium wuzhouense W44T (98.7%), followed by Rhizobium ipomoeae LMG 27163T (97.9%) in the search against type strain database of EzBioCloud. The sequence search in the NCBI nucleotide database resulted in more than 99% sequence similarity for two sequences. The first one was from an alpha-proteobacterium (EU770254.1) associated with Microcystis aeruginosa culture, and the second one was from Ciceribacter sp. strain AIY3W (MH463946.2) isolated from low salinity lakes on the Tibetan plateau.
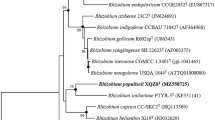
Phylogeny-based on 16S rRNA gene sequences placed the strain ADMK78T branch along with closely related sequences of an alphaproteobacterium and Ciceribacter sp. AIY3W in the cluster of Ciceribacter species (Fig. S1). The 16S rRNA gene phylogeny for only type strains placed strain ADMK78T along with Rhizobium wuzhouense W44T, though the bootstrap value was below 50%, and its phylogenetic position was not consistent across trees obtained with the maximum likelihood, maximum parsimony, and neighbour-joining methods (Fig. 1). The 16S rRNA gene phylogeny also revealed the scattered branching of Rhizobium species. Several species of Rhizobium were distantly placed from the core clade that contained the type species of the genus making it a non-monophyletic group (Hördt et al. 2020).
Phylogenetic tree based on 16S rRNA gene sequences of strain ADMK78T and type strains of members of the family Rhizobiaceae inferred by using the Maximum Likelihood method and Kimura 2-parameter model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Empty circles indicate branches of the tree that were also recovered using the neighbour-joining method, and black filled circles indicate that all three methods recovered the corresponding nodes. Bar, 0.1 substitutions per nucleotide position
Genome features
The genome of ADMK78T had a size of 4,342,374 bp, which is smaller than the genome size of symbiotic members of Rhizobium and in the range of the sizes of the non-symbiotic strains of Rhizobium (Table 1). It consisted of a circular chromosome of 3,590,542 bp and two circular plasmids of 708,533 and 43,299 bp. The overall genome sequencing coverage for the strain ADMK78T was 147.5x, with an N50 value of 3,590,542 bp. The genome sequence quality of strain ADMK78T was as per the genome standards proposed by Chun et al (2018), and the detailed genome features are provided in Table 1. The genome of strain ADMK78T contains 4377 protein-coding sequences (CDS), of which 64 genes were assigned to the stress response functions, including heat and cold shock, hyperosmotic stress, and protection from reactive oxygen. We could not find any nitrogen-fixation and nodulation genes in the genome of strain ADMK78T. Four secondary metabolite regions were identified in the main chromosome, while two in the plasmid pPRADMK78_01, using anti-SMASH pipeline (Table S5). The putative secondary metabolite clusters include bacteriocin, terpene, homoserine lactone cluster, beta-lactone-containing protease inhibitor and TfuA-related RiPPs. Members of the genus Rhizobium have been shown to produce antimicrobial compounds like bacteriocins (Oresnik et al. 1999), and quorum-sensing molecules, including homoserine lactones (He et al. 2003).
The core-gene based phylogenetic analysis placed strain ADMK78T as an independent branch with Rhizobium ipomoeae shin9-1 T as the closest neighbour and followed by Rhizobium wuzhouense W44T, R. rosettiformans W3T, and recently described Rhizobium rhizophilum 7209-2T (Gao et al. 2020) and “R. glycinendophyticum” CL12T (Wang et al. 2020) (Fig. 2). Strain ADMK78T was placed distantly from the species of Ciceribacter, and Rhizobium naphthalenivorans, R. selenitireducens and R. daejeonense. The core-gene phylogeny also exhibited that strain ADMK78T is distant from the members of Allorhizobium, Agrobacterium and Rhizobium (Fig. 2). The clade consisting of the strain ADMK78T along with Rhizobium ipomoeae, “R. glycinendophyticum”, R. wuzhouense, R. rosettiformans, R. rhizophilum and 21 non-type strains represents a new genus. Previous studies, proposed four new genera by dissecting the genus Rhizobium, based on the phylogeny of housekeeping genes (de Lajudie et al. 1998, Kathiravan et al. 2013; Mousavi et al. 2015; Mousavi et al. 2015). However, the Genome Taxonomy Database (GTDB) a resource that provides a comprehensive genome-based taxonomy of all prokaryotes, placed all members of Ciceribacter and the putative new genus clade containing ADMK78T under Allorhizbium (Parks et al 2020). Phylogenomic analysis has been considered as a key tool in defining genera (Espariz et al. 2016; Chun et al. 2018), the core-gene phylogeny exhibited clear delineation of all genera of Rhizobiaceae (Fig. 2). The core-gene phylogeny also grouped Rhizobium daejeonense, R. naphthalenivorans and R. selenitireducens along with members of genus Ciceribacter. The possibility of transferring Rhizobium naphthalenivorans and R. selenitireducens in the genus Ciceribacter has already been discussed (Hördt et al. 2020). However, a formal proposal was not made, citing the low resolution and difficulties to infer any taxonomic conclusions (Hördt et al. 2020).
Although the core-gene phylogeny provided an excellent measure to group together the phylogenetically related members, the taxonomy reorganization also requires a set of criteria to define the diversity within genera. The genus boundary has been proposed at 50% POCP (Qin et al. 2014). The POCP value inter-generic comparisons were more than 50% for the genera belonging to the Rhizobiaceae family (Table S2). Hence, this universal cut-off of 50% POCP was not applied to the Rhizobiaceae family. Several studies have reported the POCP cut-off failure for genus-level circumscription for the members of Bacillaceae, Burkholderiaceae, Neisseriaceae, and Rhodobacteraceae (Aliyu et al. 2016; Li et al. 2017a, b; Lopes-Santos et al. 2017; Wirth and Whitman 2018). These studies confirmed that a single cut-off value of POCP is unlikely to be a universal threshold for delimiting prokaryotic genera.
Several studies have indicated that AAI represents a powerful tool for genome-based taxonomy assignments. The AAI values to delimit genera typically vary between 60–80% and do not exceed 85% (Luo et al. 2014). The AAI values calculated for the Rhizobiaceae family members indicate a cut-off at 75% delimits all its genera (Figs. 3, 4). All the strains grouped under the new genus clade in the core-gene phylogeny (Fig. 2), were clustered together in AAI plots above 75% cut-off value (Fig. 3). Therefore we adopted this cut-off for defining the genus boundaries, which led us to propose a new genus Peteryoungia by including four Rhizobium species, R. ipomoeae, R. rosettiformans, R. wuzhouense and R. rhizophilum, and the strain ADMK78T. In addition to this, the core-gene phylogeny (Fig. 2) and pairwise AAI values (Figs. 3, 4) provided sufficient evidence to reclassify three Rhizobium species, namely R. daejeonense, R. naphthalenivorans and R. selenitireducens, into the genus Ciceribacter.
Average amino acid identity (AAI) plots using the type species of different genera of the family Rhizobiaceae. Peteryoungia desertarenae ADMK78T, Peteryoungia ipomoeae shin9-1T, Ciceribacter lividus MSSRFBL1T, Allorhizobium undicola ORS992T, Agrobacterium tumefaciens CCBAU3354T, Neorhizobium galegae HAMBI 540T, Pseudorhizobium pelagicum R1-200B4T and Rhizobium leguminosarum USDA 2370T
The genomic DNA G + C content of strain ADMK78T was 58.6 mol%, which is slightly lower than its closely related species included in the newly proposed genus Peteryoungia (ranging between 60.0–61.7 mol%) (Table 1). The ANI and dDDH relatedness values of ADMK78T with all species reclassified under the genus Peteryoungia are lower than (Table 1; Fig. S2), the proposed species boundaries of 95–96% for ANI and 70% for dDDH (Chun et al. 2018; Wang et al. 2016). This supported our claim that the strain ADMK78T is a putative novel species of this newly proposed genus.
Physiology and chemotaxonomy
The colonies of strain ADMK78T, were circular and translucent on Zobell Marine Agar (Fig. S4). Microscopic observation revealed that strain ADMK78T is a rod-shaped bacterium with cell size ranging from 0.3–0.5 × 1.5–2 µm, Gram-stain-negative (Fig. S3), and non-motile. It is interesting to note that all the species included in the newly proposed genus Peteryoungia are able to tolerate at least 2% of NaCl (Table 1). Details on the phenotypic characteristics, including results of carbon source utilization and chemical sensitivity in Biolog GENIII plates (Table S5), and enzyme activity in API-Zym strips (Table S6), and the differences with respect to the closely-related phylogenetic neighboring species of Peteryoungia and Ciceribacter, are shown in Table 2.
The primary fatty acids detected in strain ADMK78T were C18:1 ω7c and C18:0 (Table S3). Small proportions of C16:0, C18:1 ω7c 11-methyl, C18:0 3-OH, C20:1 ω7c, and summed feature 2 (C12:0 aldehyde) were also detected for strain ADMK78T. The strain ADMK78T exhibits higher proportions of C18:0, which was present in relatively lower amounts in the other species of Peteryoungia (Table S3). Contrary to this, a similar proportion of C18:0 was detected for Ciceribacter lividus MSSRFBL1T. However, strain ADMK78T and all other members of Peteryoungia do not possess C19:0 cyclo ω8c, which is the key fatty acid for Ciceribacter lividus. The mean spectra profile of strain ADMK78T has 33 unique peaks in comparison to the closely related taxa out of a total of 70 peaks (Table S4), which attributed to the discrimination of ADMK78T from the closely related species.
In conclusion, the core-gene phylogeny and pairwise AAI analyses showed a separate clade for five Rhizobium species with strain ADMK78T, which indicated their membership to a novel genus. Based on these results, we propose a new genus Peteryoungia in the family Rhizobiaceae by reclassifying five Rhizobium species. The genotypic and phenotypic data generated for the strain ADMK78T revealed that the strain represents a novel species in the newly proposed genus Peteryoungia, for which the name Peteryoungia desertarenae sp. nov. is proposed. In addition to this, we also propose the transfer of three rhizobium species into Ciceribacter.
Description of Peteryoungia gen. nov.
(Pe.ter.young’i.a. N.L. fem. n. Peteryoungia, named to honour Prof J. Peter W. Young, who contributed extensively towards the taxonomy and genomics of the family Rhizobiaceae).
Cells are Gram-stain-negative, aerobic, rod- to oval-shaped, and non-motile. Moderately halophilic. Positive for oxidase and catalase. All members can utilize α-d-Lactose. The major fatty acid (> 10% of total fatty acids) is C18:1 ω7c. The DNA G + C content is 58.6–61.7 mol %. A member of the family Rhizobiaceae, class Alphaproteobacteria according to 16S rRNA gene sequence analysis and core-gene phylogeny. The type species for the genus is Peteryoungia ipomoeae.
Description of Peteryoungia ipomoeae comb. nov.
Peteryoungia ipomoeae (i.po.moe′ae. N.L. gen. n. ipomoeae, of the water convolvulus Ipomoea; pertaining to the isolation of the type strain from a water convolvulus field).
Basonym: Rhizobium ipomoeae (Sheu et al. 2016).
The description is the same as for R. ipomoeae (Sheu et al. 2016). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Peteryoungia. The type strain is shin9-1 T (= LMG 27163 T = KCTC 32148 T).
Description of Peteryoungia desertarenae sp. nov.
(de.sert.a.re’nae. L. neut. n. desertum desert; L. fem. n. arena sand; N.L. gen. n. desertarenae of desert sand).
Cells are Gram-negative, straight rods with round ends (0.3–0.5 × 1.5–2 µm), and non-motile. Colonies grown on Zobell Marine Agar are 1–3 mm in diameter, circular, raised with an entire margin, and translucent opacity. The optimal temperature for growth is 28 °C and the optimal pH is 7.0. Growth occurs in the absence of NaCl with up to 2% tolerance in Zobell Marine broth. It is oxidase and catalase positive. The strain showed positive results in Biolog GN III analyses for utilization of d-maltose, d-trehalose, d-cellobiose, d-gentiobiose, sucrose, d-turanose, α- d-lactose, d-melibiose, β-methyl-d-glucoside, d-salicin, N-acetyl- d-glucosamine, N-acetyl-β-d-mannosamine, N-acetyl-d-galactosamine, α-d-glucose, d-mannose, d-fructose, d-galactose, d-fucose, l-fucose, l-rhamnose, inosine, d-sorbitol, d-mannitol, d-arabitol, myo-inositol, glycerol, d-glucose-6-phosphate, d-fructose-6-phosphate, d-aspartic acid, Glycyl-l-proline, glycyl-l-proline, l-alanine, l-arginine, l-aspartic acid, l-glutamic acid, l-histidine, l-pyroglutamic acid, l-serine, pectin, d-galacturonic acid, d-gluconic acid, d-glucuronic acid, glucuronamide, mucic acid, d-saccharic acid, p-hydroxy-phenylacetic acid, d-lactic acid methyl easter, l-lactic acid, citric acid, α-keto-glutaric acid, d,l-malic acid, bromo-succinic acid, Tween 40, γ-amino-butyric acid, α-hydroxy-butyric acid, α-hydroxy-d,l butyric acid, α-keto-butyric acid, acetoacetic acid, propionic acid, acetic acid, formic acid, sodium lactate, tetrazolium violet and blue, nalidixic acid, lithium chloride (Table S3). Positive results in API ZYM strips for leucine arylamidase, trypsin, naphthol-AS-BI-phosphohydrolase, α-glucosidase, N-acetyl-β-glucosaminidase activities (Table S4). C18:0 and C18:1 ω7c are the predominant cellular fatty acids. The DNA G + C content of the type strain is 58.6 mol%.
The type strain ADMK78T (= MCC 3400T; KACC 21383T; JCM 33657T) was isolated from saline desert sand collected from the Kutch District of Gujarat, India. The GenBank sequence accession number of the genome sequence is CP058350-CP058352, and the 16S rRNA gene sequence of strain ADMK78T is MK942856.
Description of Peteryoungia rosettiformans comb. nov.
Peteryoungia rosettiformans (ro.set.ti.for′mans. N.L. fem. n. rosetta (from L. fem. n. rosa rose) rosette; L. pres. part. formans forming; N.L. part. adj. rosettiformans rosette-forming, referring to the ability of the organism to form rosette-shaped structures).
Basonym: Rhizobium rosettiformans (Kaur et al. 2011).
The description is the same as for R. rosettiformans (Kaur et al. 2011). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Peteryoungia. The type strain is W3T (= CCM 7583 T = MTCC 9454 T).
Description of Peteryoungia wuzhouensis comb. nov.
Peteryoungia wuzhouensis (wu.zhou.en'sis. N.L. fem. n. wuzhouensis pertaining to Wuzhou, a city in China, where the type strain was isolated).
Basonym: Rhizobium wuzhouensis
Homotypic synonym: Rhizobium wuzhouense (Yuan et al. 2018).
The description is the same as for R. wuzhouense (Yuan et al. 2018). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Peteryoungia. The type strain is W44T (= CCTCC AB 2017179T = GDMCC 1.1257T = KCTC 62194T).
Description of Peteryoungia rhizophila comb. nov.
Peteryoungia rhizophila (rhi.zo'phi.la. Gr. fem. n. rhiza root; Gr. masc. adj. philos loving; N.L. fem. adj. rhizophila root-loving).
Basonym: Rhizobium rhizophila
Homotypic synonym: Rhizobium rhizophilum (Gao et al. 2020).
The description is the same as for R. rhizophilum (Gao et al. 2020). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Peteryoungia. The type strain is 7209-2T (= CGMCC 1.15691T = DSM 103161T).
Emended description of the genus Ciceribacter Kathiravan et al. 2013
The description is the same as given by Kathiravan et al. (2013) with the following amendments. Colonies vary in colour from bluish-black to semi-translucent. A few species can fix di-nitrogen. Catalase-positive and oxidase-negative. The major fatty acid is C18: 1ω7c, and the DNA G + C content ranges between 60.5–63.5 mol %. The type species is Ciceribacter lividus.
Description of Ciceribacter daejeonensis comb. nov.
Ciceribacter daejeonensis (dae.jeon.en'sis. N.L. masc. adj. daejeonensis pertaining to Daejeon, a city in Korea, where the type strain was isolated).
Basonym: Rhizobium daejeonensis
Homotypic synonym: Rhizobium daejeonense (Quan et al. 2005).
The description is the same as for R. daejeonense (Quan et al. 2005). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Ciceribacter. The type strain is L61T (= KCTC 12121T = IAM 15042T = CCBAU 10050T).
Description of Ciceribacter selenitireducens comb. nov.
Ciceribacter selenitireducens (se.le.ni.ti.re.du’cens N.L. n. selenis -itis, selenite; L. pres. part. reducens, converting to a different state; N.L. part. adj. selenitireducens, selenite reducing, referring to the organism’s ability to reduce the selenium oxyanion selenite to elemental selenium).
Basonym: Rhizobium selenitireducens (Hunter et al. 2007).
The description is the same as for R. selenitireducens (Hunter et al. 2007). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Ciceribacter. The type strain is B1T (= ATCC BAA-1503T = LMG 24075T = NRRL B-41997T).
Description of Ciceribacter naphthalenivorans comb. nov.
Ciceribacter naphthalenivorans (naph.tha.le.ni.vo’rans. N.L. neut. n. napthalenum, naphthalene; L. pres. part. vorans, devouring; N.L. part. adj. naphthalenivorans, naphthalene-devouring).
Basonym: Rhizobium naphthalenivorans (Kaiya et al. 2012).
The description is the same as for R naphthalenivorans (Kaiya et al. 2012). Phylogenetic analysis of the core-genes and pairwise provided strong evidence for the placement of this species in the novel genus Ciceribacter. The type strain is TSY03bT (= KCTC 23252T = NBRC 107585T).
Data availability
The GenBank accession number for the 16S rRNA gene sequence of strain ADMK78T is MK942856. The draft genome sequence has been deposited in GenBank under the accession number CP058350-CP058352.
References
Aliyu H, Lebre P, Blom J, Cowan D, De Maayer P (2016) Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst Appl Microbiol 39:527–533
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75–15
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acid Res 47:W81–W87
Cao J, Wei Y, Lai Q, Wu Y, Deng J, Li J, Liu R, Wang L, Fang J (2020) Georhizobium profundi gen. nov., sp. nov., a piezotolerant bacterium isolated from a deep-sea sediment sample of the New Britain Trench. Int J Syst Evol Microbiol 70:373–379
Chaudhari D, Dhotre D, Jani K, Sharma A, Singh Y, Shouche Y, Rahi P (2020) Bacterial communities associated with the biofilms formed in high-altitude brackish water Pangong Tso located in the Himalayan Plateau. Curr Microbiol 77:4072–4084
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbio 68:461–466
Conn HJ (1938) Agricultural and industrial bacteriology. J Bacteriol 36:320–321
Efron B, Halloran E, Holmes S (1996) Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci USA 93(23):13429
Espariz M, Zuljan FA, Esteban L, Magni C (2016) Taxonomic identity resolution of highly phylogenetically related strains and selection of phylogenetic markers by using genome-scale methods: the Bacillus pumilus group case. PLoS ONE 11:e0163098
Frank B (1889) Über die Pilzsymbiose der Leguminosen. Ber Dtsch Bot Ges 7:332–346 (in German)
Gao JL, Wang LW, Xue J, Tong S, Peng G, Sun YC, Zhang X, Sun J-G (2020) Rhizobium rhizophilum sp. nov., an indole acetic acid-producing bacterium isolated from rape (Brassica napus L.) rhizosphere soil. Int J Syst Evol Microbio 70:5019–5025
Gulati A, Rahi P, Vyas P (2008) Characterization of phosphate-solubilizing fluorescent pseudomonads from the rhizosphere of seabuckthorn growing in the cold deserts of Himalayas. Curr Microbiol 56:73–79
He X, Chang W, Pierce DL, Seib LO, Wagner J, Fuqua C (2003) Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J Bacteriol 185(3):809–822
Hördt A, López MG, Meier-Kolthoff JP, Schleuning M, Weinhold L-M, Tindall BJ, Gronow S, Kyrpides NC, Woyke T, Göker M (2020) Analysis of 1000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front Microbiol 11:468
Hunter WJ, Kuykendall LD, Manter DK (2007) Rhizobium selenireducens sp. nov.: a selenite-reducing alpha-Proteobacteria isolated from a bioreactor. Curr Microbiol 55:455–460
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of Meier-Kolthoff 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114
Kaiya S, Rubaba O, Yoshida N, Yamada T, Hiraishi A (2012) Characterization of Rhizobium naphthalenivorans sp. nov. with special emphasis on aromatic compound degradation and multilocus sequence analysis of housekeeping genes. J Gen Appl Microbiol 58:211–224
Kathiravan R, Jegan S, Ganga V, Prabavathy VR, Tushar L, Sasikala Ch, Ramana ChV (2013) Ciceribacter lividus gen. nov., sp. nov., isolated from rhizosphere soil of chick pea (Cicer arietinum L.). Int J Syst Evol Microbiol 63:4484–4488
Kaur J, Verma M, Lal R (2011) Rhizobium rosettiformans sp. nov., isolated from a hexachlorocyclohexane dump site, and reclassification of Blastobacter aggregatus Hirsch and Muller 1986 as Rhizobium aggregatum comb. nov. Int J Syst Evol Microbiol 61:1218–1225
Kimes NE, Lopez-Perez M, Flores-Felix JD, Ramirez-Bahena MH, Igual JM, Peix A, Rodriguez-Valera F, Velazquez E (2015) Pseudorhizobium pelagicum gen. nov., sp. nov. isolated from a pelagic Mediterranean zone. Syst Appl Microbiol 38:293–299
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH et al (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kurli R, Chaudhari D, Pansare AN, Khairnar M, Shouche YS, Rahi P (2018) Cultivable microbial diversity associated with cellular phones. Front Microbiol 9:1229
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M et al (2004) Versatile and open software for comparing large genomes. Genome Biol 5(2):R12
Kuykendall LD (2005) Rhizobiales ord. nov. In Bergey's Manual of Systematic Bacteriology, 2nd edn, vol. 2 (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria), pp. 324–361. Edited by Brenner DJ, Krieg NR, Staley JT, Garrity GM, Springer, New York.
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Li Y, Lei X, Xu Y, Zhu H, Xu M et al (2017) Rhizobium albus sp. nov., isolated from lake water in Xiamen Fujian Province of China. Curr Microbiol 74:42–48
Li Y, Xue H, Sang SQ, Lin CL, Wang XZ (2017) Phylogenetic analysis of family Neisseriaceae based on genome sequences and description of Populibacter corticis gen nov, sp nov, a member of the family Neisseriaceae, isolated from symptomatic bark of Populus x euramericana canker. PLoS One 12:e0174506
Lindström K, Mousavi SA (2010) Rhizobium and other N-fixing symbioses. In: Encyclopedia of Life Science (ELS). Chichester: John Wiley and Sons.
Lindström K, Amsalu AA, Mousavi SA (2015) Evolution and taxonomy of nitrogen-fixing organisms with emphasis on rhizobia. In: de Bruijn FJ (ed) Biological nitrogen fixation. Wiley, Hoboken, NJ, pp 21–38
Lopes-Santos L, Castro DBA, Ferreira-Tonin M, Correa DBA, Weir BS et al (2017) Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie Van Leeuwenhoek 110:727–736
Luo C, Rodriguez-R LM, Konstantinidis KT (2014) MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42(8):e73
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat 14:60
Minas K, McEwan NR, Newbold CJ, Scott KP (2011) Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett 325(2):162–169. https://doi.org/10.1111/j.1574-6968.2011.02424.x
Mohapatra B, Sarkar A, Joshi S, Chatterjee A, Kazy SK et al (2017) An arsenate-reducing and alkane-metabolizing novel bacterium, Rhizobium arsenicireducens sp. nov., isolated from arsenic-rich groundwater. Arch Microbiol 199:191–201
Mousavi SA, Osterman J, Wahlberg N, Nesme X, Lavire C, Vial L, Paulin L, de Lajudie P, Lindstrom K (2014) Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215
Mousavi SA, Willems A, Nesme X, de Lajudie P, Lindstrom K (2015) Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90
Na SI, Kim YO, Yoon S-H, Ha S, Baek I et al (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Oresnik IJ, Twelker S, Hynes MF (1999) Cloning and characterization of a Rhizobium leguminosarum gene encoding a bacteriocin with similarities to RTX toxins. Appl Environ Microbiol 65(7):2833–2840
Panday D, Schumann P, Das SK (2011) Rhizobium pusense sp. nov., isolated from the rhizosphere of chickpea (Cicer arietinum L.). Int J Syst Evol Microbiol 61:2632–2639
Pandit AS, Joshi MN, Bhargava P, Shaikh I, Ayachit GN et al (2015) A snapshot of microbial communities from the Kutch: one of the largest salt deserts in the World. Extremophiles 19:973–987
Park S, Lee JS, Lee KC, Yoon JH (2013) Lentilitoribacter donghaensis gen. nov., sp. nov., a slowly-growing alphaproteobacterium isolated from coastal seawater. Antonie Van Leeuwenhoek 103:457–464
Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P (2020) A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol 38:1079–1086
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612
Peix A, Rivas R, Trujillo ME, Vancanneyt M, Velazquez E, Willems A (2005) Reclassification of Agrobacterium ferrugineum LMG 128 as Hoeflea marina gen. nov., sp. nov. Int J Syst Evol Microbiol 55:1163–1166
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196(12):2210–2215
Quan ZX, Bae HS, Baek JH, Chen WF, Im WT et al (2005) Rhizobium daejeonense sp. nov. isolated from a cyanide treatment bioreactor. Int J Syst Evol Microbiol 55:2543–2549
Rahi P, Kurli R, Khairnar M, Jagtap S, Pansare AN et al (2017) Description of Lysinibacillus telephonicus sp. nov., isolated from the screen of a cellular phone. Int J Syst Evol Microbiol 67:2289–2295
Ramana CV, Parag B, Girija KR, Raghu Ram B, Venkata Ramana V et al (2013) Rhizobium subbaraonis sp. nov., an endolithic bacterium isolated from beach sand. Int J Syst Evol Microbiol 63:581–585
Rivas R, Sanchez-Marquez S, Mateos PF, Martinez-Molina E, Velazquez E (2005) Martelella mediterranea gen. nov., sp. nov., a novel alpha-proteobacterium isolated from a subterranean saline lake. Int J Syst Evol Microbiol 55:955–959
Ruginescu R, Gomoiu I, Popescu O, Cojoc R, Neagu S, Lucaci I, Batrinescu-Moteau C, Enache M (2020) Bioprospecting for novel halophilic and halotolerant sources of hydrolytic enzymes in brackish, saline and hypersaline lakes of Romania. Microorganisms. 8(12):1903.
Sasser M (2001) Technical note # 101 identification of bacteria by gas chromatography of cellular fatty acids. Stat:1–6.
Sheu SY, Chen ZH, Young CC, Chen WM (2016) Rhizobium ipomoeae sp. nov., isolated from a water convolvulus field. Int J Syst Evol Microbiol 66:1633–1640
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tittsler RP, Sandholzer LA (1936) The use of semi-solid agar for the detection of bacterial motility. J Bacteriol 31:575–580
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. https://doi.org/10.1371/journal.pone.0112963
Wang Q, Zhu W, Wang ET, Zhang LS, Li X et al (2016) Genomic identification of rhizobia-related strains and threshold of ANI and core-genome for family, genus and species. Int J Environ Agri Res 2:76–86
Wang C, Li A, Yuan T, Bao G, Feng G, Zhu H (2020) Rhizobium glycinendophyticum sp. nov., isolated from roots of Glycine max (Linn. Merr.). Antonie Van Leeuwenhoek 113:147–154
Wirth JS, Whitman WB (2018) Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int J Syst Evol Microbiol 68:2393–2411
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y et al (2017) Introducing EzBioCloud: a taxonomically United database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Yuan T, Liu L, Huang S, Taher AH, Tan Z et al (2018) Rhizobium wuzhouense sp. nov., isolated from roots of Oryza officinalis. Int J Syst Evol Microbiol 68:2918–2923
Acknowledgements
The authors thank Prof. Aharon Oren, Department of Plant and Environmental Sciences, The Hebrew University of Jerusalem, Israel, and Prof. Bernhard Schink, Department of Biology, University of Konstanz, Germany, for their expert suggestions concerning the correct species name, species epithet and Latin etymology. Authors also thank Prof J Peter W Young, University of York, to grant us permission to propose new genus Peteryoungia. The authors acknowledge the financial support of the Department of Biotechnology (BT/Coord. II/01/03/2016).
Funding
Financial support by the Department of Biotechnology (BT/Coord. II/01/03/2016) and (BT/PR8218/BCE/8/1044/2013).
Author information
Authors and Affiliations
Contributions
MK: methodology, formal analysis, validation; AH and AP: methodology, formal analysis; AN and KJ methodology, isolation; DM and YS funding acquisition; PR conceptualization, methodology, data curation, formal analysis, supervision, and writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
The experiments reported in this manuscript did not involve human participants and/or animals.
Consent for publication
The manuscript is submitted with the consent of all authors.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rahi, P., Khairnar, M., Hagir, A. et al. Peteryoungia gen. nov. with four new species combinations and description of Peteryoungia desertarenae sp. nov., and taxonomic revision of the genus Ciceribacter based on phylogenomics of Rhizobiaceae. Arch Microbiol 203, 3591–3604 (2021). https://doi.org/10.1007/s00203-021-02349-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02349-9