Abstract
Strain YIM B00363T, a Gram-positive, aerobic, non-motile, rod-shaped, spore-forming bacterium, was isolated from saline soil samples collected from a salt lake in Xinjiang province, north-west China, and was characterized using a polyphasic approach. The optimum growth temperature was 37 °C and the optimum pH was 7.5–8.0. The major menaquinone was MK-7; anteiso-C15:0 (53.52%), iso-C15:0 (15.04%) and C16:0 (12.76%) were the predominant cellular fatty acids. The diagnostic diamino acid of the cell wall peptidoglycan was meso-diaminopimelic acid. The phospholipids were phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, unidentified phospholipids, unidentified glycolipids and unknown lipids. The DNA G + C content of the type strain was 50.4 mol%. Phylogenetic analysis based on 16S rRNA gene sequences indicated that the strain YIM B00363T belonged to a cluster comprising species of the genus Paenibacillus. The nearest relatives were P. residui MC-246T and P. senegalensis JC66T, with 93.2% and 92.8% gene sequence similarities, respectively. On the basis of its phenotypic characteristics and phylogenetic distinctivenes, strain YIM B00363T represents a novel species of the genus Paenibacillus, for which the name Paenibacillus turpanensis sp. nov. is proposed. The type strain is YIM B00363T (= CGMCC 1.17507T = KCTC 43184T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
On the basis of the analysis of the 16S rRNA gene sequences of group 3 bacilli, the genus Paenibacillus was proposed by Ash et al. (1993) and then emended by Shida et al. (1997). At the time of writing, the names of more than 255 species of the genus Paenibacillus have been validly published (https://www.bacterio.net/paenibacillus.html). Some species had been reported to have the capacity of fixing nitrogen, such as P. polymyxa, P. taohuashanense, P. azotofixans, P. sabinae, P. sonchi, P. forsythiae and P. sophorae (Grau and Wilson 1962; Xie et al. 2012; Seldin et al. 1984; Ma et al. 2007; Hong et al. 2009; Ma and Chen 2008; Jin et al. 2011). Some Paenibacillus strains play a significant role in agriculture and industry (Seldin 2011). Previous studies have reported that microorganisms found in extreme environments can produce a variety of natural compounds and their specific mechanisms for adapting to extreme environments (Tang et al. 2002). In order to explore more multifunctional, special mechanisms and valuable microbes, we targeted halophilic microorganisms in hypersaline ecosystems. In the investigation of microbial resources of salt environment in Xinjiang province, one novel strain YIM B00363T was isolated from the hypersaline sediment of Wuzunbulake salt lake. In this study, we report the novel strain YIM B00363T belonging to the genus Paenibacillus by means of a polyphasic taxonomic study.
Methods and materials
Bacterial isolation and cultivation
Strain YIM B00363T was isolated from saline soil samples collected from Wuzunbulake salt lake, Turpan city, Xinjiang province, by the spread plate method on Nitrogen Fixing agar (NFA) medium (0.2 g KH2PO4, 0.2 g MgSO4, 0.2 g NaCl, 5.1 g CaCO3, 10 g mannitol, 15 g agar, 1L H2O and pH 7.0). The plates were then incubated at 37 °C for 7 days. Strain YIM B00363T was one of the isolates that appeared on the NFA plates under aerobic condition. Single colonies were purified by transferring them onto tryptic soy agar (TSA) medium plates. It was maintained on TSA at 4 °C and as a glycerol suspension (20%, v/v) at − 80 °C.
Phylogenetic and genotypic analysis
Genomic DNA for PCR amplification was prepared using TIANamp Bacteria DNA Kit (TIANGEN BIOTECH (BEIJING) CO., LTD), according to the manufacturer’s instructions. The 16S rRNA gene sequence of YIM B00363T was amplified using the universal bacterial primers PA (5′-CAGAGTTTGATCCTGGCT-3′) and PB (5′-AGGAGGTGATCCAGCCGCA-3′), synthesized by Sangon Biotech Co. Ltd (Shanghai, PR China). The amplification programme was used as described previously (Ueda et al. 2013). The PCR products were cloned into pEASY®-T5 Zero Cloning Vector (Kit, TransGen Biolech). The sequence was determined by the Tsingke Company (Beijing, PR China). Sequence similarities to the related type strains were calculated by using the EzBiocloud (https://www.ezbiocloud.net/) (Yoon et al. 2017). Multiple alignments with sequences of the most closely relatives were carried out using the CLUSTAL X 1.8 program (Thompson et al. 1997). Phylogenetic trees were reconstructed by the neighbour-joining (Saitou and Nei 1987), maximum parsimony (Fitch 1971) and maximum likelihood (Felsenstein 1981) tree-making algorithms using the software packages MEGA version 7.0 (Kumar et al. 2016). The stability of relationships was assessed by performing bootstrap analyses with 1000 resamplings (Felsenstein 1985).
The genome of YIM B00363T was sequenced using a PacBio + Illumina Hiseq at Shanghai Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China). The sequenced reads were assembled using SOAPdenovo software version 2.04 (https://soap.genomics.org.cn/soapdenovo.html). The DNA G + C mol% value was obtained from the genomic sequences.
Phenotypic, physiological and biochemical characteristics
Cell morphology was examined by light microscopy (BX41; Olympus) and transmission electron microscopy (JEM-2100, JEOL) using cultivated on TSA at 37 °C for 2 days. The presence of endospores was investigated by using the phase contrast microscope (BX51M; Olympus) and transmission electron microscopy (JEM-2100, JEOL) when cells were cultivated on TSA plates at 37 °C for 10 days. Gram staining was performed by the Burke method (Nam et al. 2008) and the result was confirmed by the KOH test (Baron and Finegold 1990). Motility was analysed by the wet-mount method (Nam et al. 2008). Anaerobic growth was determined using the GasPak anaerobic system (BBL) according to the manufacturer’s instructions. Growth at different temperatures (4, 10, 15, 20, 25, 30, 35, 37, 40, 45, 50 and 55 °C) and NaCl concentrations (0–10%, w/v, at 1% intervals) were examined by using TSA medium at 37 °C for 14 days. The ability of the strain to grow at different pH values (4.0–10.0, at 0.5 intervals by using the buffer system described by Tang et al. 2010). Catalase activity was determined by production of bubbles after adding 3% H2O2 to the tested bacteria (Tarrand and Gröschel 1982). Tests for hydrolysis of gelatin, starch, Tween 20, 40 and 80, urease activity, nitrate reduction, and hydrogen disulphide production were determined by using traditional methods (Dong and Cai 2001). Carbon sources utilization and chemical sensitivity were determined by using BIOLOG GEN III MicroPlate. Using API ZYM systems tested enzyme activities. Acid production was detected by 50CH systems. Additional biochemical tests were performed using two Gallery systems (bioMérieux): API 20 NE and API 20 E according to the manufacturer’s instructions. Antibiotic susceptibility tests were performed using discs impregnated with various antimicrobial compounds.
Chemotaxonomic characteristics
Biomass for chemical and molecular studies was obtained by cultivation in TSA without agar (pH 7.0) at 37 °C and collected when the bacteria reached their mid-exponential phase. Analyses of diaminopimelic acid in the cell wall and sugars of whole-cell hydrolysates were performed according to the procedures described by Lechevalier and Lechevalier (1970) and Tang et al. (2009). Cellular fatty acids were extracted, methylated and analysed by using the Sherlock Microbial Identification System (MIDI) according to the manufacturer’s instructions. Fatty acid methyl esters were analysed by using the Microbial Identification software package (Sherlock Version 6.1; MIDI databaseTSBA6) (Sasser 1990). The respiratory quinones of YIM B00363T were extracted from lyophilized cells (Collins et al. 1977), purified by TLC and then analysed by HPLC according to the methods of Xie and Yokota (2003). Polar lipids were extracted, examined by two-dimensional TLC and identified using the procedures described by Collins and Jones (1980) and Minnikin et al. (1979).
Results and discussion
Molecular phylogenetic analysis
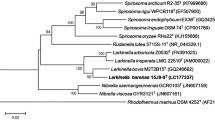
An almost complete 16S rRNA gene sequence (1536 bp) of strain YIM B00363T was generated. YIM B00363T was most moderately related to P. hodogayensis SGT (Takeda et al. 2005) and P. ginsengarvi Gsoil 139T (Yoon et al. 2007) with 93.8% sequence similarity, which is far below 97%. The available data indicate that organisms having less than 97.0% 16S rRNA gene sequence similarity will not exhibit more than 60% reassociation, irrespective of the hybridization method applied (Stackebrandt and Goebel 1994; Keswani and Whitman 2001). Therefore, neither wet lab nor in-silico DNA-DNA hybridization were carried out. Meanwhile, these strains clustered in the genus Paenibacillus stably after phylogenetic analyses of the 16S rRNA gene sequences (Fig. 1, S1, S2). The NJ tree clustered YIM B00363T with P. residui MC-246T (93.2%), P. senegalensis JC66T (92.8%) and P. thermoaerophilus TC22-2bT (92.8%) (Fig. 1). The ML and MP trees showed similar results (Figs S1 and S2). According to 16S rRNA gene sequence similarities and topology of the phylogenetic trees, the literature data of above five strains, P. hodogayensis, P. ginsengarvi, P. residui, P. senegalensis and P. thermoaerophilus, were selected for comparison in this study. The DNA G + C content of strain YIM B00363T was 50.4 mol%. These data supported the finding that strain YIM B00363T represents a different genomic species of the genus Paenibacillus.
Neighbour-joining tree based on 16S rRNA gene sequences showing the relationships between strain YIM B00363T and some members of the genus Paenibacillus. Bootstrap values (> 50%) based on 1000 replicates are shown at the branch nodes. Asterisks indicate that the corresponding branches were also recovered in trees generated with the maximum parsimony and maximum likelihood methods. Bacillus subtilis DSM 10T was used as an outgroup. Bar, 1% sequence divergence
Phenotypic, physiological and biochemical characteristics
YIM B00363T was Gram positive, aerobic, rod shaped without peritrichous flagella (Fig S3a), swollen by ellipsoidal spores and approximately 2–3 µm long and 0.5–1.5 µm wide (Fig S3b). It could grow at 20–45 °C (optimum at 37 °C), at pH 6.0–9.5 (optimum at 7.5–8.0) and in the presence of 0–5% (w/v) NaCl (optimum without NaCl). Other physiological characteristics are given in Table 1, Table S1 and in the species description.
Chemotaxonomic characteristics
Amino acids contained meso-diaminopimelic acid as the cell wall diamino acid, with glucose (11.2%), galactose (10.8%), xylose (12.7%), arabinose (20%) and fucose (18.1%) as the major whole-cell sugars (> 10%). The predominant menaquinone was MK-7. The polar lipids were phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), unidentified phospholipids (PL), unidentified glycolipids (GL) and unknown lipids (UL). (Fig S4). The cellular fatty acid profile of strain YIM B00363T contained anteiso-C15:0 (53.52%), iso-C15:0 (15.04%) and C16:0 (12.76%) as major fatty acids (> 10%), and C14:0 (5.62%), iso-C16:0 (3.66%), anteiso-C17:0 (2.07%), iso-C14:0 (2.00%), iso-C17:0 (1.95%) and C12:0 (1.52%) as minor fatty acids. Compared with the data of other two reference strains, iso-C16:0 was minor fatty acid of YIM B00363T, but major fatty acid (> 10%) of others (Table 2).
All chemotaxonomic data conform to the characteristics of the genus Paenibacillus, which was described by Logan et al. (2009) and Tindall et al. (2010). However, there are some differences between strain YIM B00363T and four reference strains, for example, the content of iso-C16:0. Based on the phenotypic, genotypic, phylogenetic and chemotaxonomic data presented here, strain YIM B00363T should belong to the genus Paenibacillus. Thus, it represents a novel species, for which the name Paenibacillus turpanensis sp. nov. is proposed.
Description of Paenibacillus turpanensis sp. nov.
Paenibacillus turpanensis (tur.pan.en’sis. N.L. masc. adj. turpanensis relating to Turpan, the name of a city in Xinjiang, China, the geographical origin of isolation of the type strain).
Paenibacillus turpanensis is Gram stain-positive, non-motile, aerobic, rod-shaped, spore-forming bacterium. Cells are rods with a width of 0.5–1.5 μm and length of 2–3 μm. It can grow at 20–45 °C (optimum 37 °C), at pH 6–9.5 (optimum pH 7.5–8) and in the presence of 0–5% (w/v) NaCl (optimum without NaCl). It is positive for catalase activity, nitrate reduction, aesculin ferric citrate, hydrolysis of starch, and Tween 20, 40 and 80 and negative for production of H2S, production of indole, Voges–Proskauer test, hydrolysis of gelatin, l-arginine and urea. d-glucose, l-arabinose and maltose are assimilated. For enzyme activities (API ZYM system), it is positive for alkaline phosphatase, esterase C4, esterase lipase C8, lipase C14, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase. For the utilization of carbon sources (API Biolog GEN III systems), the following substrates are utilized for growth: dextrin, d-maltose, d-trehalose, d-cellobiose, gentiobiose, sucrose, d-turanose, Stachyose, d-raffinose, α-d-lactose, d-melibiose, d-salicin, N-acetyl-d-glucosamine, α-d-glucose, d-fructose, d-galactose, l-rhamnose, pectin, d-galacturonic acid, l-galactonic acid lactone, d-gluconic acid, d-glucuronic acid, glucuronamide and acetic acid as substrates. With API 50CH, acid is produced from glycerol, l-arabinose, d-ribose, d-xylose, methyl-βd-xylopyranoside, d-galactose, d-glucose, fructose, d-mannose, l-rhamnose, methyl-αd-glucopyranoside, aesculin, salicin, d-cellobiose, d-maltose, d-lactose, d-melibiose, d-saccharose, d-trehalose, d-melezitose, d-raffinose, amidon (starch), glycogen, d-gentiobiose, d-turanose, d-tagatose and potassium 5-ketogluconate and weakly produced from l-sorbose, amygdalin, d-lyxose and l-fucose. It is sensitive to kanamycin (30 μg), gentamicin (10 μg), neomycin (30 μg), amoxicillin/clavulanic acid (10 μg), erythromycin (5 μg), carbenicillin (100 μg), chloramphenicol (30 μg), tetracycline (30 μg), novobiocin (5 μg) and streptomycin (300 μg). The whole-cell hydrolysates contain meso-diaminopimelic acid as the cell wall diamino acid. Glucose, galactose, xylose, arabinose and fucose are the major whole-cell sugars. The polar lipids are phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, unidentified phospholipids, unidentified glycolipids and unknown lipids. The predominant menaquinone is MK-7. Anteiso-C15:0 (53.52%), iso-C15:0 (15.04%) and C16:0 (12.76%) are the major fatty acids. The DNA G + C content of the type strain is 50.4 mol%.
The type strain is YIM B00363T (= CGMCC 1.17507T = KCTC 43184T), isolated from the saline soil of Turpan city in Xinjiang province, north-west China. The GenBank accession numbers of the 16S rRNA gene and the genome sequence of YIM B00363T are MT032315 and WTLI00000000, respectively.
Abbreviations
- NFA:
-
Nitrogen Fixing agar
- TSA:
-
Tryptic soy agar
- DPG:
-
Diphosphatidylglycerol
- PG:
-
Phosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- PL:
-
Unknown phospholipid
- GL:
-
Unidentified glycolipids
- UL:
-
Unknown lipids
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260
Baron EJ, Finegold SM (1990) Bailey and Scott’s diagnostic microbiology, 8th edn. Mosby, St Louis
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Dai XL, Shi KX, Wang X, Fan J, Wang R, Zheng SX, Wang GJ (2019) Paenibacillus flagellatus sp. nov., isolated from selenium mineral soil. Int J Syst Evol Microbiol 69:183–188
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Grau FH, Wilson PW (1962) Physiology of nitrogen fixationby Bacillus polymyxa. J Bacteriol 83:490–496
Hong YY, Ma YC, Zhou YG, Gao F, Liu HC, Chen SF (2009) Paenibacillus sonchi sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Sonchus oleraceus. Int J Syst Evol Microbiol 59:2656–2661
Jin HJ, Lv J, Chen SF (2011) Paenibacillus sophorae sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Sophora japonica. Int J Syst Evol Microbiol 61:767–771
Kampfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Keswani J, Whitman WB (2001) Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int J Syst Evol Microbiol 51:667–678
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lechevalier MP, Lechevalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Logan NA, Berge O, Bishop AH, Busse HJ, de Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Ma YC, Chen SF (2008) Paenibacillus forsythiae sp. nov., a nitrogen-fixing species isolated from rhizosphere soil of Forsythia mira. Int J Syst Evol Microbiol 58:319–323
Ma Y, Xia Z, Liu X, Chen S (2007) Paenibacillus sabinae sp., nov., a nitrogen-fixing species isolated from the rhizosphere soils of shrubs. Int J Syst Evol Microbiol 57:6–11
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Mishra AK, Lagier JC, Rivet R, Raoult D, Fournier PE (2012) Non-contiguous finished genome sequence and description of Paenibacillus senegalensis sp. nov. Stand Genom Sci 7:70–81
Nam JH, Bae W, Lee DH (2008) Oceanobacillus caeni sp. nov., isolated from a Bacillus-dominated wastewater treatment system in Korea. Int J Syst Evol Microbiol 58:1109–1113
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note, vol 10 1. MIDI Inc, Newark
Seldin L (2011) Paenibacillus, nitrogen fixation and soil fertility. In: Logan NA, De Vos P (eds) Endospore-forming soil bacteria, soil biology, vol 27. Springer, Berlin, Heidelberg, pp 287–307
Seldin L, Van Elsas JD, Penido EGC (1984) Bacillus azotofixans sp. nov., a nitrogen-fixing species from Brazilian soils and grass roots. Int J Syst Bacteriol 34:451–456
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Takeda M, Suzuki I, Koizumi J (2005) Paenibacillus hodogayensis sp. nov., capable of degrading the polysaccharide produced by Sphaerotilus natans. Int J Syst Evol Microbiol 55:737–741
Tang SL, Nuttall S, Ngui K, Fisher C, Lopez P, Dyall-Smith M (2002) HF2: a double-stranded DNA tailed haloarchaeal virus with a mosaic genome. Mol Microbiol 44:283–296
Tang SK, Wang Y, Chen Y, Lou K, Cao LL, Xu LH, Li WJ (2009) Zhihengliuella alba sp. nov., and emended description of the genus Zhihengliuella. Int J Syst Evol Microbiol 59:2025–2032
Tang SK, WangY ZH, Lee JC, Lou K, Kim CJ, Li WJ (2010) Haloechinothrix alba gen. nov., sp. nov., a halophilic, filamentous actinomycete of the suborder Pseudonocardineae. Int J Syst Evol Microbiol 60:2154–2158
Tarrand JJ, Gröschel DH (1982) Rapid, modified oxidase test for oxidase-variable bacterial isolates. J Clin Microbiol 16:772–774
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P (2010) Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:249–266
Ueda J, Yamamoto SC, Kurosawa N (2013) Paenibacillus thermoaerophilus sp. nov., a moderately thermophilic bacterium isolated from compost. Int J Syst Evol Microbiol 63:3330–3335
Vaz-Moreira I, Figueira V, Lopes AR, Pukall R, Sproer C, Schumann P, Nunes OC, Manaia CM (2010) Paenibacillus residui sp. nov. isolated from urban waste compost. Int J Syst Evol Microbiol 60:2415–2419
Xie CH, Yokota A (2003) Phylogenetic analyses of Lampropedia hyalina based on the 16S rRNA gene sequence. J Gen Appl Microbiol 49:345–349
Xie JB, Zhang LH, Zhou YG, Liu HC, Chen SF (2012) Paenibacillus taohuashanense sp., nov. a nitrogen-fixing species isolated from rhizosphere soil of the root of Caragana kansuensis Pojark. Antonie Van Leeuwenhoek 102:735–741
Yoon MH, Ten LN, Im WT (2007) Paenibacillus ginsengarvi sp. nov., isolated from soil from ginseng cultivation. Int J Syst Evol Microbiol 57:1810–1814
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
This study was performed with the support of the National Natural Science Foundation of China (31760003, 31500011 and 31270055), the Natural Science Foundation of Yunnan Province (2017FB039) and the South and Southeast Asia Cooperation Base on Microbiological Resource Prevention and Utilization (2018IA100)
Author information
Authors and Affiliations
Contributions
LY and H-WH: carried out the data analysis, part of chemical classification and wrote the manuscript. YW, Y-RK and MY: performed the polyphasic taxonomy except chemical classification. YL, X-QW and G-FZ: prepared the experiments and isolated the novel strain. W-YZ and S-KT: designed the separation medium and directed the classification and takes full responsibility for the final submission. All the authors reviewed and approved the final version of the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The NCBI GenBank accession number for the 16S rRNA gene sequence of strain YIM B00363T is MT032315. The draft whole-genome sequence for YIM B00363T has been deposited at DDBJ/ENA/GenBank under accession number WTLI00000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, L., Huang, HW., Wang, Y. et al. Paenibacillus turpanensis sp. nov., isolated from a salt lake of Turpan city in Xinjiang province, north-west China. Arch Microbiol 203, 77–83 (2021). https://doi.org/10.1007/s00203-020-02003-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02003-w